Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability (original) (raw)
Abstract
Proteasome activator PA200 enhances proteasome-mediated cleavage after acidic residues in vitro; however, its role within cells is not known. Here, we show that, in response to ionizing radiation, PA200 forms hybrid proteasomes with 19S caps and 20S core proteasomes that accumulate on chromatin, leading to an increase in proteolytic activity. Unlike many other proteins that respond to DNA damage, the response of PA200 appears to be independent of Ataxia Telangiectasia Mutated and p53, but dependent on DNA-dependent protein kinase activity. Nonetheless, PA200 is critical because PA200-knockdown cells show genomic instability and reduced survival after exposure to ionizing radiation. This phenotype is reproduced by specific inhibition of postglutamyl activity of proteasomes, but combined treatment with PA200 siRNA and postglutamyl inhibitor does not show additive effects on survival. Together, these data suggest a unique role for PA200 in genomic stability that is likely mediated through its ability to enhance postglutamyl cleavage by proteasomes.
Keywords: ATM, chromatin, DNA-dependent protein kinase, ionizing radiation, DNA damage
Proteasomes are responsible for the degradation of a large number of protein targets throughout eukaryotic cells (1). The function of proteasomes is regulated by activators that bind to the ends of the 20S core catalytic particle in a single or double fashion and open the ends of the core proteasome to allow entry of protein targets (2). Four proteasome activators (PAs) have been identified in mammalian cells: PA700 (also known as 19S), PA28α/β, PA28γ, and PA200 (1, 3). To date, the cellular role for PA200 has not been defined, although it has been suggested to play a role in response to ionizing radiation (IR)-induced damage (4).
The cellular response to IR exposure is a complex process that requires the coordinated effort of many proteins to complete successfully three fundamental steps: sensing DNA breaks, regulating cell cycle progression, and repairing the DNA lesion (5–7). The initial response to IR is governed by the recruitment and activation of PI3-related kinases such as Ataxia Telangiectasia Mutated (ATM), ATM and Rad3-related (ATR), and DNA-dependent protein kinase (DNA-PK) (8). ATM is critical for the phosphorylation of many proteins required for cell cycle control and DNA repair, including NBS1, RAD51, checkpoint kinases Chk1 and Chk2, and the tumor suppressor p53 (9–12). Many of these proteins (i.e., p53, H2AX, and NBS1) can also be phosphorylated by DNA-PK, suggesting some redundancy in this pathway (13–15). If damage is too extensive, cells may initiate cell death potentially through p53-dependent mechanisms (16). PA200 relocalizes to nuclear foci upon exposure to IR (4); however, its role in the above described pathway is unclear.
PA200 is an evolutionarily conserved, nuclear localized, multiple HEAT-repeat protein that associates with the ends of mature 20S proteasomes and enhances peptide hydrolysis, most notably after acidic residues (postglutamyl cleavage) in vitro (4, 17, 18). Both mammalian PA200 and its ortholog in Saccharomyces cerevisiae (known as Blm10) are reported to form hybrid proteasomes consisting of one 19S cap on one end and one Blm10/PA200 cap on the other end of a core proteasome (4, 19). PA200 forms nuclear foci and accumulates on chromatin after IR exposure, indicating that PA200 responds to damage induced by IR and may function in DNA repair (4, 20). However, two observations challenge the idea that PA200 plays a role in DNA repair. First, yeast that are null for Blm10 do not show marked sensitivity to DNA damage induced by IR exposure or bleomycin (19). Second, mice lacking PA200 show normal lymphocyte development, a process that requires DNA repair proteins to rearrange antigen receptor gene segments (21). Based on these observations, the importance of PA200 in response to DNA damage and in DNA repair have been questioned. However, it is conceivable that PA200 plays an important role in response to IR-induced damage but that this role occurs after DNA repair is complete, for example. Consistent with the notion that PA200 can impact the cellular response to DNA damage, Schmidt et al. (19) observed bleomycin sensitivity of yeast expressing a deletion mutant lacking the C terminus of Blm10.
In the current work, we characterized how PA200 responds to IR and determined the importance of PA200 in survival from IR-induced damage. We provide evidence that PA200-19S hybrid proteasomes are enhanced after IR exposure and accumulate on chromatin. Furthermore, PA200 and postglutamyl proteolytic activity are required for the cellular response to IR and maintenance of genomic integrity.
Results
Elevated Levels of PA200-19S Hybrid Proteasomes After IR Exposure.
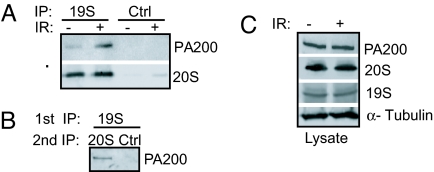
To demonstrate that PA200 and 19S caps could interact with either end of the same core proteasome to form hybrid PA200-20S-19S proteasomes (henceforth referred to as PA200-19S hybrid proteasomes) we immunoprecipitated 19S cap subcomplexes and show associated PA200 and core proteasomes (Fig. 1A). Given that PA200 associates with the end of core proteasomes and within cells, virtually all detectable PA200 is associated with core proteasomes (18–20, 22) the observed association of PA200 with 19S caps likely reflects a hybrid proteasome comprising a core proteasome with a 19S cap on one end and a PA200 cap on the other end. To confirm the existence of PA200-19S hybrid proteasomes, we immunoprecipitated the complex by using an antibody specific for a 19S cap subunit (S10B) and assessed the associated proteins for PA200–core proteasome complexes by immunoprecipitation of core proteasomes followed by Western blotting for PA200 (Fig. 1B). Upon exposure to IR, the levels of PA200-19S hybrid proteasomes are enhanced (Fig. 1A), suggesting that they play a role in the cellular response to DNA damage. Total levels of the 19S subunit (S10B), PA200 and 20S core proteasomes are not enhanced by IR exposure (Fig. 1C), arguing that the increase in PA200-19S hybrid proteasomes is likely caused by enhanced formation or stabilization of this hybrid complex rather than increased synthesis of the individual subunits. Together, these data demonstrate that IR-induced DNA damage enhances the cellular levels of PA200-19S hybrid proteasomes.
Fig. 1.
PA200-19S hybrid proteasomes are enhanced after IR exposure. Twenty-four hours after mock treatment (−) or 50-Gy irradiation (+) is shown. (A) Equivalent numbers of live HeLa cells were subjected to immunoprecipitation (IP) for a subunit (S5a) of the 19S cap (19S) or by using a control antibody (Ctrl). Bound proteins were immunoblotted with PA200-specific antiserum or MCP20 antibody (20S). (B) HeLa cells were immunoprecipitated (19S, 1st IP) as in A. Beads were eluted in 0.4 M NaCl and re-IP (2nd IP) for core proteasomes or by using a control antibody (Ctrl). Bound proteins were immunoblotted with PA200-specific antiserum. (C) Western blot analysis of lysates.
Enhanced PA200-20S-19S Protein Levels and Proteasome Activity on Chromatin After IR Exposure.
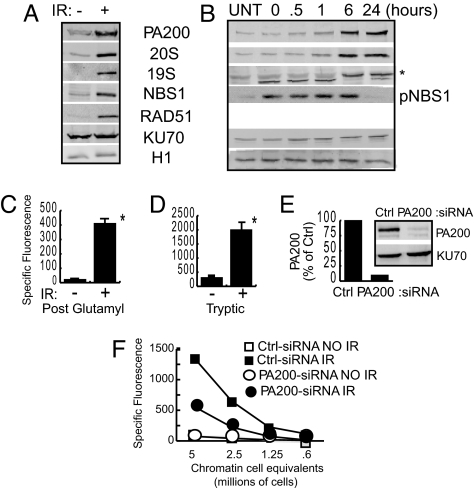
To elucidate possible mechanisms of how PA200 and proteasomes respond to IR, their presence on chromatin was compared with known DNA repair proteins. Isolation of chromatin-associated proteins shows that Rad51 and NBS1 protein levels are enhanced 24 h after IR exposure, whereas histone H1 levels and KU70 remain unchanged (Fig. 2A). Similar to these DNA damage-responsive proteins, PA200, 20S core proteasomes, and 19S caps all accumulate on chromatin 24 h after IR exposure (Fig. 2A). Next, we determined the kinetics of PA200 and proteasome accumulation relative to a known rapid responder to DNA damage. Consistent with existing literature, we detected accumulation of phosphorylated NBS1 (pNBS1) as early as we could harvest and lyse cells after IR exposure [time 0; ≈5 min after exposure, Fig. 2B (23, 24)]. This pNBS1 is maintained until at least 6 h after IR exposure but disappears by 24 h (Fig. 2B). In contrast, PA200 and 20S proteasomes showed delayed accumulation (i.e., not before 6 h after irradiation) on chromatin, and their presence was maintained 24 h after IR exposure (Fig. 2B).
Fig. 2.
Increased hybrid proteasomes and proteasome activity on chromatin after IR exposure. (A) HeLa cells were mock treated (−) or exposed to 50-Gy IR (+), and chromatin-bound proteins were immunoblotted 24 h after IR exposure. (B) Normal BJ fibroblasts were irradiated with 5 Gy and assessed at various time points as in A. Each figure shown is representative of at least three independent experiments. (C and D) An aliquot of the chromatin-bound proteins from A were assessed for proteasome activity 24 h after IR exposure (*, P < 0.001). Specific fluorescence was calculated by subtraction of the fluorescence released in the presence of inhibitor from the absence of inhibitor. Error bars are SD of triplicates. (E and F) HeLa cells were transfected with control (Ctrl) or PA200-specific (PA200) siRNA. (E) Depletion of PA200 was confirmed by immunoblotting of cell lysates. (F) Cells were exposed to 50-Gy IR, and chromatin-bound proteins were assessed for postglutamyl proteolytic activity as described above.
If PA200 and proteasomes accumulate on chromatin after IR exposure, we reasoned that proteasome activity should also be elevated. Chromatin-associated tryptic activity is increased by 6-fold after IR exposure, which is consistent with the 5- to 8-fold increase in 20S core proteasomes on the chromatin (Fig. 2D). Interestingly, postglutamyl cleavage, which is the specificity that is enhanced by PA200 most strikingly in vitro (4), is enhanced 19-fold on chromatin (Fig. 2C). These data suggest that the increase in postglutamyl cleavage activity is caused by more efficient peptide hydrolysis, possibly mediated by the presence of PA200. Consistent with this idea, in cells in which PA200 protein is knocked down by 90% (Fig. 2E), IR-induced accumulation of chromatin-associated postglutamyl activity is diminished to 7-fold (Fig. 2F). This 7-fold IR-induced accumulation of postglutamyl activity in PA200-knockdown cells is similar in magnitude to that observed for core proteasome protein levels and for tryptic activity, suggesting that the remaining postglutamyl activity in PA200-knockdown cells is caused solely by core proteasome accumulation. Together, these data indicate that PA200 and proteasomes respond relatively late to IR exposure by accumulating on chromatin to enhance postglutamyl proteasome activity.
PA200 Accumulation on Chromatin Is Independent of ATM and p53 but Dependent on DNA-PK Activity.
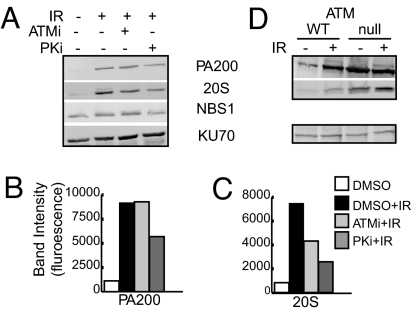
ATM is a PI3-related kinase that initiates signaling after IR exposure to arrest the cell cycle and to recruit and maintain DNA repair proteins (8, 25). Because PA200 and proteasomes respond to DNA damage, we assessed whether their response depended on ATM activity. Treatment with the ATM-specific small molecule inhibitor, KU-55933 (26) (labeled ATMi in Fig. 3) reduces core proteasome accumulation on chromatin by 35–40% [Fig. 3 A and C and supporting information (SI) Fig. S1 A, C, D, and F] and decreases both phospho-NBS1 and Rad51 accumulation on chromatin after IR exposure (Fig. S1_D_). Quantification of the fluorescence emitted from each band and normalized to KU70 fluorescence demonstrates that PA200 accumulation on chromatin after IR exposure is unchanged in the presence of ATM inhibitor (Fig. 3A, compare second and third lanes, quantified in Fig. 3B, and Fig. S1 A and B). PA200 exists on chromatin of A-T cells (deficient in ATM), even in the absence of exposure to IR, further supporting the idea that PA200 responds to DNA damage independent of ATM (Fig. 3D, compare first and third lanes). It is notable that baseline levels (i.e., cells not exposed to IR) of PA200 on chromatin of A-T cells is greater than that observed in their wild-type counterparts (Fig. 3D, compare first and third lanes). IR exposure induces accumulation of PA200 on chromatin of wild-type cells, but the high level of PA200 on A-T chromatin is not increased further by IR exposure (Fig. 3D, compare first and second lanes, and third and fourth lanes). IR-induced PA200 accumulation on chromatin also occurs in p53-deficient HCT-116 cells and p53-deficient mouse embryonic fibroblasts (Fig. S2). Thus, PA200 accumulation on chromatin appears to occur independently of ATM and p53.
Fig. 3.
PA200 accumulation on chromatin is independent of ATM but dependent on DNA-PK activity. Chromatin-bound proteins were immunoblotted for the indicated proteins. (A) HeLa cells were mock (−) or 50-Gy-irradiated (+) in the presence or absence of the ATM-specific inhibitor (ATMi) KU-55933 (10 μM) or the DNA-PK-specific inhibitor (PKi) NU7026 (10 μM). (B and C) The band intensities in A were quantified as fluorescence normalized to KU70 protein levels. (D) Normal BJ fibroblasts (WT) or ATM-null GM-5823 cells (null) were mock (−) or 5-Gy-irradiated (+) and analyzed after 24 h.
DNA-PK is another PI3-related kinase that responds to IR-induced damage by phosphorylation of key DNA repair proteins (27, 28). Inhibition of DNA-PK with the small molecule NU7026 (labeled PKi in Fig. 3A) diminished PA200 accumulation on chromatin by ≈34–40% (Fig. 3 A and B and Fig. S1 A, B, D, and E). Core proteasome accumulation on chromatin is also reduced (by 30–60%) by the DNA-PK inhibitor (Fig. 3 A and C and Fig. S1 A, C, D, and F). Thus, although proteasome accumulation on chromatin can be prevented by inhibition of either ATM or DNA-PK, accumulation of PA200 on chromatin appears to depend mainly on DNA-PK activity.
PA200 and Proteasome Activity are Required for Survival After IR Exposure.
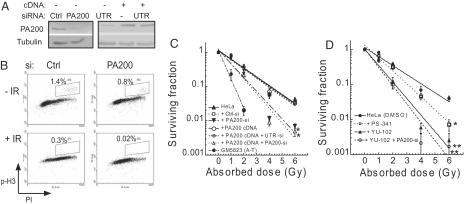
Given that PA200 appears to respond to IR exposure, we tested the importance of PA200 to the cellular response to DNA damage. Therefore, we depleted 90% of PA200 protein in HeLa cells by using PA200-specific siRNA (Fig. 4A). After IR exposure, HeLa cells arrest exclusively at G2/M because of their p53 status (Fig. S3). Although PA200-knockdown HeLa cells did not show any gross defects in G2/M cell cycle arrest 24 h after IR exposure (Fig. S3), more detailed analysis using the mitotic marker phosphohistone H3 (_P_-H3) reveals a reduction in the fraction of cells in mitosis compared with control knockdown cells (Fig. 4B, compare +IR plots). Even in the absence of IR exposure, the fraction of PA200-knockdown cells in mitosis is decreased compared with control siRNA-transfected cells (Fig. 4B, compare −IR plots). Consistent with this defect in the G2/M checkpoint, long-term survival after IR exposure is impaired in PA200 siRNA-treated cells (Fig. 4C, inverted filled triangles, P < 0.05) compared with control siRNA cells or untransfected cells (Fig. 4C, open squares and filled triangles). To verify that PA200 siRNA effects were mediated by PA200 knockdown, we transfected PA200 cDNA that is insensitive to siRNA targeting the 3′-UTR (UTR-si), but sensitive to siRNA targeting the PA200-coding region (PA200-si). The survival defect was restored by transfection of PA200 cDNA in UTR siRNA-treated cells (Fig. 4C, open circles), but the defect was not restored in PA200 siRNA-treated cells that target the PA200-coding region (Fig. 4C, open triangles). Because PA200 enhances proteasome-mediated cleavage after glutamic acid residues (4), we hypothesized that inhibition of this activity would also lead to decreased survival after IR exposure. The proteasome inhibitor YU-102 is a small molecule inhibitor known to inhibit specifically postglutamyl activity of proteasomes, but not chymotryptic or tryptic activities (29). Cells treated with YU-102 show a highly significant (Fig. 4D, filled triangles, P < 0.01) decrease in cell survival after IR exposure (Fig. 4D). The effect of YU-102 on cell survival is slightly greater than that observed for PA200 knockdown, which may be caused by more effective inhibition of postglutamyl activity because PA200-knockdown cells still contain some residual PA200 protein (≈10%). The proteasome inhibitor PS-341, which inhibits chymotryptic and tryptic activities of the proteasome and the bulk of protein degradation in the cell (29), does not affect cell survival after IR exposure as dramatically as YU-102 (Fig. 4D, open squares, P = 0.05). Cells treated with a combination of both PA200 siRNA and YU-102 show survival defects comparable with YU-102 treatment alone (Fig. 4D), suggesting that YU-102 and PA200 siRNA function in the same pathway. These data argue that PA200, through enhancement of postglutamyl specificity of proteasomes, is essential for optimal survival after IR exposure.
Fig. 4.
PA200 and postglutamyl peptide hydrolysis are essential for survival from IR-induced damage. (A–D) HeLa cells were transfected with control (Ctrl) siRNA, PA200-specific (PA200) siRNA, siRNA specific for the 3′-UTR of PA200 (UTR) with or without a PA200 cDNA expression plasmid. (A) Depletion of PA200 and restoration by PA200 cDNA was confirmed by immunoblotting of cell lysates. (B) Staining for phosphorylated histone H3 (_P_-H3) and DNA content (PI) of control siRNA and PA200 siRNA cells exposed to 50-Gy IR (+IR) or not (−IR). Percentage of gated cells within the indicated region (R3) is indicated for each siRNA and treatment. (C) Indicated cells were exposed to varying doses of IR and monitored for cell survival. Error bars indicate SD of three independent experiments. (*, P < 0.05, by χ2 analysis). (D) Indicated cells were treated with DMSO (Ctrl) or proteasome inhibitors (1 nM PS-341 or 1 μM YU-102) for 24 h and assessed as in C. Cells treated with YU-102 exhibit significantly reduced cell survival after IR exposure compared with control cells (**, P < 0.01, by χ2 analysis; *, P < 0.05). Error bars indicate SD from three independent experiments.
PA200 and Postglutamyl Proteasome Activity Contribute to Genomic Stability.
Because sensitivity to IR, as measured by decreased cell survival after IR exposure, is often correlated with genomic instability (30), we speculated that PA200 deficiency and diminished postglutamyl proteasome activity may result in genomic instability. In support of this idea, the number of chromosome aberrations (end-to-end associations, breaks and gaps, bridges) in PA200-depleted cells were significantly greater compared with control treated cells even in the absence of IR exposure (Table 1). Correspondingly, cells treated with YU-102 showed dramatically (P < 0.01) increased genomic instability as measured by micronuclei formation (Table 2). Because both PA200 knockdown and inhibition of postglutamyl proteasome activity led to genetic instability and decreased survival after IR, these findings further support the notion that PA200 functions together with core proteasomes during the cellular response to IR-induced DNA damage.
Table 1.
Chromosome abnormalities on PA200 siRNA
| Cell type | Chromosome end associations (per 200 metaphases) | Bridges (per 300 anaphases) | Chromosome gaps + breaks (per 200 metaphases) | Chromatid gaps + breaks (per 200 metaphases) |
|---|---|---|---|---|
| HeLa | 19 | 10 | 3 | 5 |
| HeLa (Ctrl siRNA) | 21 | 14 | 3 | 6 |
| HeLa (PA200 siRNA) | 40* | 31* | 11* | 15* |
| GM5823 (A-T) | 64* | 51* | 16* | 12* |
Table 2.
Proteasome inhibitors induce genetic instability
| Cell type | Micronuclei (per 100 cells) | Mitotic index, % |
|---|---|---|
| HeLa | 3 | 100 |
| HeLa (control) | 4 | 97 |
| HeLa (YU-102) | 15** | 69** |
| HeLa (PS-341) | 9** | 81* |
Discussion
Overall, we have presented several lines of evidence indicating that PA200 responds to DNA damage in a manner that is essential for cell survival and that PA200 is critical to maintain genomic stability. Our analysis shows that PA200 responds to IR by increasing levels of PA200-20S-19S hybrid proteasomes and that these complexes accumulate and enhance proteolytic activity on chromatin part because of DNA-PK activity.
PA200 is required for optimal cell survival after IR exposure in HeLa cells. Although the initial characterization of PA200 demonstrated that it responds to radiation, ES cells derived from PA200-null mice do not show an apparent increase in sensitivity to IR (21). Similarly, Blm10-deficient yeast do not show defects in survival when exposed to DNA damaging agents (19). It is currently unclear whether differences between yeast and mammalian responses to DNA damage or cell type differences between mammalian cells may explain these discrepancies. Indeed, the defect we observe is subtle in that it is in the IR dose required to reduce survival by 90–99%. Nevertheless, the range of the defect in PA200-knockdown cells is similar to that observed in cells knocked down for bona fide DNA damage-responsive proteins such as Rad51 or 14-3-3σ (31, 32), arguing that the impact of PA200 is both statistically and biologically significant.
Together, these data support the idea that PA200 is essential for optimal survival from IR exposure, yet a direct role in repair of DNA damage per se is unlikely for several reasons. First, lymphocyte development, which depends on DNA repair proteins of the NHEJ pathway such as KU70 for appropriate recombination of V, D, and J genomic segments, occurs normally (21). Second, no defects in DNA repair or recombination were detectable in PA200-null cell lines (21). Third, we observe that the kinetics of PA200 accumulation on chromatin after IR exposure occurs well after most DNA repair is thought to be completed (23, 33).
The pathway that regulates PA200 accumulation may involve DNA-PK because inhibition of DNA-PK diminished accumulation of PA200 on chromatin after IR exposure. This regulation by DNA-PK but not by ATM is striking because many of the proteins targeted during DNA repair, such as H2AX, 53BP1, and KAP1, can be phosphorylated by either ATM or DNA-PK (13, 15, 34). Although DNA-PK inhibition diminishes PA200 on chromatin by 34–40%, there may also be other regulators (such as ATR) of PA200 responsiveness to IR-induced damage.
The regulation of core proteasome accumulation on chromatin after IR exposure appears to be more complex than that observed for PA200. Although the DNA-PK inhibitor blocks both core proteasome and PA200 accumulation on chromatin, ATM inhibition diminishes core proteasome, but not PA200, accumulation. These data suggest that although PA200 is associated with core proteasomes, core proteasomes that are not associated with PA200 also accumulate on chromatin, and their regulation may be different from PA200-containing proteasomes. Indeed, IR induces core proteasome accumulation on chromatin of PA200-knockdown cells (data not shown), suggesting that PA200 is not required for proteasome localization to chromatin. Rather, the presence of PA200 appears to enhance the efficiency of postglutamyl proteolytic cleavage of core proteasomes. This is suggested by the observation that PA200-knockdown cells show an IR-induced accumulation of postglutamyl cleavage of 7-fold compared with 19-fold in PA200-sufficient cells. The remaining 7-fold accumulation of postglutamyl cleavage after IR exposure of PA200-knockdown cells is similar in magnitude to that observed for tryptic activity in PA200-sufficient cells and to core proteasome protein levels in PA200-sufficient cells. Therefore, although the precise requirements for accumulation of PA200 and core proteasomes after IR exposure still remain to be defined, it appears that PA200 enhances the efficiency of postglutamyl cleavage on chromatin after IR exposure.
The accumulation of PA200, core proteasomes, and 19S caps on chromatin after IR exposure suggests a requirement for proteolytic machinery in the response to IR exposure and DNA damage. This along with existing literature that yeast 20S and 19S proteasome subunits are recruited to sites of DNA repair supports the idea that degradation machinery may be a crucial step of the response (35). Furthermore, inhibition of postglutamyl proteasome activity with YU-102 reproduces the phenotype of PA200-knockdown cells in impaired survival after IR exposure and increased genomic instability (Fig. 4D). YU-102 treatment showed more dramatic results than PS-341, which inhibits chymotryptic and tryptic activities of proteasomes. This observation is striking given that inhibition of postglutamyl proteasome activity by YU-102 does not alter degradation of a GFP reporter protein, arguing that YU-102 does not affect global protein degradation. The same GFP reporter protein is substantially protected by PS-341 (29). Finally, treatment of cells with both YU-102 and PA200 siRNA does not have a greater effect on survival than each treatment alone, suggesting that the impairment of cell survival by PA200 siRNA is caused by inhibition of postglutamyl activity. Collectively, these observations argue for an important role for PA200, likely through its ability to enhance postglutamyl activity of proteasomes, in the response to IR exposure, and these responses are important for genomic stability. Future studies focusing on defining targets for PA200-19S hybrid proteasomes will likely shed light on the precise mechanism of how PA200 impacts survival after IR exposure and how it affects genomic stability.
Materials and Methods
Cell Culture, Reagents, and Antibodies.
All cell lines were maintained in DMEM supplemented with 10% serum (20, 36). The ATM inhibitor KU-55933 and the DNA-PK inhibitor NU7026 have been described (26, 37). The following antibodies have been described: anti-PA200 antisera, MCP20 (anti-core proteasome subunit, α6), anti-histone H1-H4, KU70, and RC23V (anti-flu peptide) (20). The antibodies specific for the following proteins were purchased: 19S proteasome (subunit S10B) from Calbiochem; Rad51 (clone 3C10) and phosphorylated (Ser-10) histone H3 from Millipore; NBS1 and _P_-NBS1 from Novus Biologicals; human p53 (DO-1) from Sigma; murine p53 (pAb240) from Abcam; PUMA (Ab-1) from Calbiochem; and α-tubulin (DM 1A) from Sigma.
Immunoprecipitations and Western Blot Analysis.
Cell lysis, immunoprecipitations, and fluorescence-based Western blot analysis were performed as described in ref. 20. Quantification of band intensities was performed by using ImageQuant software version 5.2 (Molecular Dynamics) by line analysis and integration of area under the curve.
IR Treatment, DNA Content Analysis, and Isolation of Chromatin-Bound Proteins.
Cells were plated 24 h before irradiation. Adherent cells were exposed to Cs irradiation at a dose rate of 65.93 rad/min; after the indicated time, cells were harvested, and chromatin-bound proteins were isolated as described in ref. 20.
Proteasome Activity Assay.
Cells were lysed in 1% Triton X-100 containing 0.5 mM EDTA, and chromatin-bound proteins were isolated (20). The proteins eluted from the chromatin were then incubated with 100 μM methyl coumarin amide (MCA)-linked peptide substrates (Leu-Leu-Glu-MCA for postglutamyl activity or Leu-Arg-Arg-MCA for tryptic activity; Peptides International) in the presence or absence of the proteasome inhibitor MG132 (2.5 μM). Cleavage of MCA was detected by fluorescence emission at 460 nm after excitation at 360 nm using a Molecular Dynamics fluorometer.
Transfection with siRNA.
The PA200-specific and control siRNA (Lo GC duplex, 1.4 μM final) were obtained directly from Invitrogen. For chromatin accumulation studies, cell cycle analysis and _P_-H3 staining, cells were transfected by using Amaxa nucleofection technology twice at 48-h intervals and irradiated 96 h after initial transfection.
Survival Assay.
Cell survival was performed by the procedure described (31, 38). Seventy-two hours after siRNA transfection (with Lipofectamine) or 24 h after proteasome inhibitor treatment, cells were plated as single cells, incubated for 6 h, and subsequently exposed to IR. After 12 or more days, cell colonies were fixed, stained with crystal violet, and counted.
Phosphorylated Histone H3 Staining.
Ninety-six hours after initial transfection, PA200 siRNA and control siRNA HeLa cells were mock-treated or irradiated with 50 Gy as described above. Twenty-four hours after irradiation cells were analyzed for _P_-H3 as described in ref. 39. Cells were analyzed on a Coulter FC500 flow cytometer. Data were analyzed by using WinMDI software (Joe Trotter, Scripps Research Institute). The percentage of gated cells within the region (R3) is shown for each siRNA and treatment.
Metaphase Chromosome Damage Analysis.
Knockdown cells and AT cells (GM5823) were monitored for the chromosome abnormalities indicated in the absence of IR exposure. Metaphase chromosome spreads were prepared by procedures described in ref. 31.
Micronuclei Analysis.
HeLa cells were treated with DMSO (control) or proteasome inhibitor PS-341 (1 nM) or YU-102 (1 μM) for 24 h, and micronuclei formation was determined by procedures described in ref. 38. Each experiment was performed three times. Mitotic index was determined by the procedure described in ref. 31.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Amy Stewart for radiation treatments. p53 wild-type and null cells were generously provided by Keshav Singh, the PUMA antibody was a gift from Myron Czuczman, and the murine p53 antibody was donated by Thomas Tomasi (all from Roswell Park Cancer Institute). This work was supported by investigator start-up funds (to N.B.), National Institutes of Health Grants CA10445 and CA123232 (to T.K.P.), National Institutes of Health T32 Training Grant CA085183 (to J.B.), and National Cancer Institute/National Institutes of Health Cancer Center Support Grant CA016056 (to Roswell Park Cancer Institute).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: Cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khanna KK, Jackson SP. DNA double-strand breaks: Signaling, repair, and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 6.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 8.Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: Variations on a theme? Curr Opin Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 9.Bartek J, Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490:117–122. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- 10.Canman CE, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 11.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11–Rad50–Nbs1 complex. Nat Struct Mol Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 12.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Someya M, et al. Association of ionizing radiation-induced foci of NBS1 with chromosomal instability and breast cancer susceptibility. Radiat Res. 2006;166:575–582. doi: 10.1667/RR0638.1. [DOI] [PubMed] [Google Scholar]
- 15.Stiff T, et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 16.Xie S, et al. PIk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J Biol Chem. 2001;276:43305–43312. doi: 10.1074/jbc.M106050200. [DOI] [PubMed] [Google Scholar]
- 17.Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J Struct Biol. 2004;146:425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Ortega J, et al. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J Mol Biol. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt M, et al. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 20.Blickwedehl J, et al. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat Res. 2007;167:663–674. doi: 10.1667/RR0690.1. [DOI] [PubMed] [Google Scholar]
- 21.Khor B, et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwanczyk J, et al. Structure of the Blm10–20S proteasome complex by cryoelectron microscopy: Insights into the mechanism of activation of mature yeast proteasomes. J Mol Biol. 2006;363:648–659. doi: 10.1016/j.jmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatei M, et al. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 25.Shiloh Y. ATM and ATR: Networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 26.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 27.Block WD, Yu Y, Lees-Miller SP. Phosphatidylinositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32-kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004;32:997–1005. doi: 10.1093/nar/gkh265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Q, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myung J, Kim KB, Lindsten K, Dantuma NP, Crews CM. Lack of proteasome active site allostery as revealed by subunit-specific inhibitors. Mol Cell. 2001;7:411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 30.Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 31.Dhar S, Squire JA, Hande MP, Wellinger RJ, Pandita TK. Inactivation of 14-3-3σ influences telomere behavior and ionizing radiation-induced chromosomal instability. Mol Cell Biol. 2000;20:7764–7772. doi: 10.1128/mcb.20.20.7764-7772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takata M, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White DE, et al. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 2006;66:11594–11599. doi: 10.1158/0008-5472.CAN-06-4138. [DOI] [PubMed] [Google Scholar]
- 35.Krogan NJ, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 36.Wood LD, et al. Characterization of ataxia telangiectasia fibroblasts with extended life-span through telomerase expression. Oncogene. 2001;20:278–288. doi: 10.1038/sj.onc.1204072. [DOI] [PubMed] [Google Scholar]
- 37.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 38.Hunt CR, et al. Genomic instability and enhanced radiosensitivity in Hsp701- and Hsp703-deficient mice. Mol Cell Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X, Darzynkiewicz Z. Cytometric assessment of histone H2AX phosphorylation: A reporter of DNA damage. Methods Mol Biol. 2006;314:73–80. doi: 10.1385/1-59259-973-7:073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information