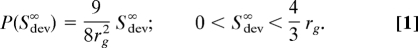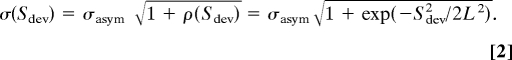Emergence of symmetry in homooligomeric biological assemblies (original) (raw)
Abstract
Naturally occurring homooligomeric protein complexes exhibit striking internal symmetry. The evolutionary origins of this symmetry have been the subject of considerable speculation; proposals for the advantages associated with symmetry include greater folding efficiency, reduced aggregation, amenability to allosteric regulation, and greater adaptability. An alternative possibility stems from the idea that to contribute to fitness, and hence be subject to evolutionary optimization, a complex must be significantly populated, which implies that the interaction energy between monomers in the ancestors of modern-day complexes must have been sufficient to at least partially overcome the entropic cost of association. Here, we investigate the effects of this bias toward very-low-energy complexes on the distribution of symmetry in primordial homooligomers modeled as randomly interacting pairs of monomers. We demonstrate quantitatively that a bias toward very-low-energy complexes can result in the emergence of symmetry from random ensembles in which the overall frequency of symmetric complexes is vanishingly small. This result is corroborated by using explicit protein–protein docking calculations to generate ensembles of randomly docked complexes: the fraction of these that are symmetric increases from 0.02% in the overall population to >50% in very low energy subpopulations.
Keywords: evolution, protein structure, Monte Carlo Simulation, modeling
The origin of the striking symmetry of homooligomeric protein assemblies has been the subject of much speculation. The potential benefits of symmetric assemblies to an organism include increased coding economy (1), greater stability, reduced aggregation, robustness to errors in synthesis (2, 3), and amenability to allosteric regulation (3). It has also been proposed that structural symmetry arises because it makes proteins more designable and foldable, or is a result of domain swapping (4–7). Monod and colleagues argued that symmetrical arrangements enhance the sensitivity of selection because “effects of single mutations occurring in a symmetrical oligomer … should be greatly amplified as compared to the effects of similar mutations in a monomer or non-symmetrical oligomer,” since each mutation at the interface effectively counts twice (3). In analogy to atomic clusters it has been suggested that symmetric oligomeric arrangements may, in general, be low in energy (8). Shakhnovich and coworkers argued that homodimers are favored over heterodimers because the mean energies of homodimeric interfaces are likely to be lower than those of heterodimeric interfaces (9, 10). Many of these arguments are discussed in greater detail in an excellent review by Goodsell and Olsen (2).
As with many evolutionary questions, it is difficult to assess which of the factors described above contributed to the symmetry of modern-day assemblies and to what extent. However, with the advent of molecular modeling, it is now possible to assess the sufficiency of quantitative hypotheses in this area. In this article we develop a quantitative model for the emergence of symmetry in biological assemblies that is based on explicit docking calculations on realistic protein models.
The key premise underlying our approach is that selection is only likely to operate on primordial complexes with sufficient initial interaction energy to at least partially overcome the entropic costs of association of the monomers; evolution can only optimize a complex that is populated sufficiently to confer a benefit to the organism. For example, contrast the effects of a mutation that stabilizes an interface by 1 kcal/mol on the population of binding modes with interaction energies of 0 and −8 kcal/mol, respectively. Assuming that the free-energy cost arising from the entropy loss in association is 10 kcal/mol, the mutation increases the population of the first binding mode from 5 ×10−8 to 3 ×10−7, and the second, from 0.035 to 0.18. Because the population of the first binding mode is vanishingly small even after mutation, it is unlikely to contribute to fitness, and hence, the second, much lower energy binding mode is a more likely starting point for evolutionary optimization by mutation and selection.
We investigate the extent to which such a bias toward very-low-energy primordial complexes could have contributed to the emergence of the highly symmetric homooligomeric complexes ubiquitous in nature today. We show that there are two competing factors that determine the fraction of complexes with a given degree of symmetry in a subpopulation: for complexes of increasing symmetry, the number of possible configurations decreases while the variance in their interaction energy grows. We show by using both simple analytic models and explicit protein–protein docking simulations that the combination of these two counteracting factors leads to a predominance of symmetric structures in very low energy binding modes. In contrast with most previous suggestions, in our model, symmetrical complexes are not favored because they provide specific functional advantages, but because the initial pool of protein complexes available for competitive selection is heavily biased toward symmetrical binding modes.
Results and Discussion
To determine whether a bias toward low energy alone can give rise to highly symmetric complexes, we first quantify the extent of symmetry in randomly docked complexes, and show that a majority of naturally occurring complexes are more symmetric than 99.98% of random complexes. Next, we show that the variance in the interaction energy distribution for randomly docked complexes increases with increasing symmetry. We then combine these results and find that increasingly stringent bias toward low-energy structures yields increasingly symmetric populations. At each step, the results from simple model calculations are corroborated by using explicit protein–protein docking simulations; in particular, we find that the fraction of symmetric complexes generated in protein docking simulations (11) increases from 0.02% in the overall population to over 50% in very-low-energy subpopulations.
Extent of Symmetry in Randomly Generated Complexes.
We begin by comparing the symmetry of naturally occurring homodimeric proteins to that of randomly generated homodimeric complexes. To quantify the notion of nearly symmetric requires a metric for deviations from perfect symmetry in homodimer complexes. Let A and B be any two atoms in one protein monomer, and A′ and B′ be the corresponding atoms in the other monomer. If the dimer is perfectly symmetric, then
We define the symmetry measure _S_dev to be the average of |dist(A, B′) − dist(A′, B)| over all pairs of α-carbon atoms. This atomic-distance-based measure is useful, among other reasons, because the system energy is largely a function of interatomic distances. Perfect symmetry corresponds to _S_dev = 0.
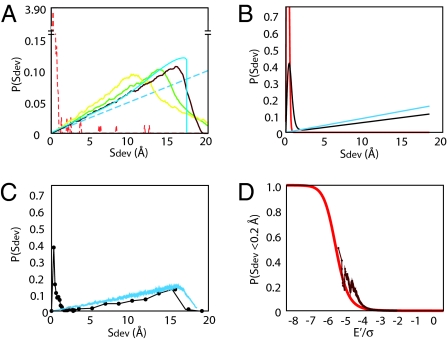
Estimates of the _S_dev probability distribution, P(_S_dev), for random homodimers were obtained by randomly orienting pairs of monomeric protein structures (Table 1) followed by a translation to bring them into atomic contact with no energy minimization. The _S_dev probability distributions for different proteins (Fig. 1A, yellow, green, and brown lines) are very similar, with an approximately linear increase in probability density with increasing _S_dev starting from zero at perfect symmetry. To investigate the generality of this result, we determine P(_S_dev) for a simple model system composed of a pair of identical spheres of radius rg with atoms uniformly distributed on their surfaces. For finite distance between the two spheres, this calculation (supporting information (SI) Appendix) involves an integral that must be evaluated numerically. In the limit of infinite distance between the spheres, the integral can be evaluated analytically, and the result is simple:
As shown in Fig. 1A, the interacting sphere model (blue lines) qualitatively recapitulates the results for the random protein complexes (yellow, green, and brown lines). The infinite-distance solution (dashed blue line) is a reasonable approximation to the exact solution even at small intermonomer distances (solid blue line), in particular, when the deviation, _S_dev, from perfect symmetry is small.
Table 1.
Increase in symmetry produced by refinement and selection
| Protein monomer | PDB ID code | Length | P(_S_dev < 0.2 Å) in % | ||
|---|---|---|---|---|---|
| Random ensemble | Refined ensemble (low/high resolution) | Low-energy ensemble (lowest 2.5%) | |||
| Protein L | 1hz6 | 61 | 0.02 | 0.35/0.31 | 2.9 |
| Ubiquitin | 1ogw | 76 | 0.03 | 0.18/0.19 | 1.9 |
| Ribosome binding factor | 1tif | 59 | 0.03 | 0.29/0.35 | 2.9 |
| cheY | 2chy | 128 | 0.02 | 0.10/0.19 | 3.2 |
| Fatty acid binding protein | 1lfo | 127 | 0.01 | 0.10/0.15 | 2.6 |
Fig. 1.
Emergence of symmetry from ensembles of randomly docked homodimeric complexes. (A) Comparison of _S_dev distributions for interacting spheres, complexes formed by randomly docking monomeric proteins, and native protein homodimers. Solid blue, numerical solution of supplementary Eq. 1 for contacting spheres; dashed blue, analytic solution (Eq. 1) for spheres at infinite distance; Yellow (1hz6), green (1hz6) and brown (2chy), numerical results for random protein homodimer complexes generated by explicit protein docking. Red, _S_dev distribution of 796 naturally occurring protein homodimer structures. The y axis is broken to accommodate the sharp peak near _S_dev = 0 for the naturally occurring complexes. (B) Increase in symmetry in random homodimeric complexes with increasingly stringent energy-based selection obtained by numerically integrating Eq. 3. Blue, no energy threshold; black, energy E < −4.6 σ; red, E < − 10.0σ (A more negative threshold implies a tighter binding is required for function.) (C) Numerical results from Rosetta all-atom protein docking calculations with 2chy. Blue, _S_dev distribution for randomly generated docked complexes (no energy threshold); black, _S_dev distribution for very-low-energy docked complexes with E < −4.6σ below the mean. (D) Increase in the fraction of symmetric structures (_S_dev < 0.2 Å) in population after energy-based selection; brown, result from 2chy docking simulations; red, simple model result (SI Appendix, Eq. S8). As discussed in the SI Appendix, we estimate σ to be typically ≈1 kcal/mol (Table S1).
The empirical _S_dev distribution for a set of 796 native protein homodimers contrasts strikingly with that of randomly generated dimeric complexes as illustrated in Fig. 1A (red line). The majority of native structures are more symmetric than >99.98% of randomly docked complexes; the probability of observing a random complex with _S_dev < 0.2, as is typical for native homodimers, is ≈1/5,000.
The contrast between the high symmetry of native complexes and low symmetry of random homodimers frames the central objective of this article: to establish a quantitative explanation for the high symmetry of oligomeric assemblies observed in nature.
Interaction Energy Distribution for Randomly Generated Complexes.
Consider a set of randomly generated homodimeric interfaces, each of which contains N independent interactions across the interface. Supposing each of these interactions is drawn independently from a distribution with standard deviation E, the distribution for the total interaction energy has a standard deviationσasym=NE. For the special case of symmetric homodimer interfaces, there are N/2 distinct interactions, each of which occurs twice, and the total interaction energy has standard deviationσasym=N/2(2E)=2σasym. For higher-order symmetric oligomers, the same argument yields _j_-fold greater variance in the interaction energy distribution for _j_-fold symmetric oligomers over their asymmetric counterparts. This is a generalization of the result obtained by Shakhnovich and coworkers for flat disks containing a single type of interaction center in their study of the evolutionary advantages of homodimers over heterodimers (9, 10).
We tested the prediction of a broader energy distribution for symmetrical protein arrangements by using docking simulations. As shown in Fig. S1, the distribution of interaction energies in randomly generated C2 symmetric complexes is indeed broader than that of the randomly generated asymmetric complexes described above; the variance is ≈1.8 times greater in the symmetric case, reasonably close to the theoretical estimate of 2. The distributions are Gaussian, as might be anticipated from the Central Limit Theorem applied to a sum of random interactions (see SI Appendix for discussion).
The analysis is readily extended to complexes that deviate from perfect symmetry. The interaction energy is the sum of pairs of (A, B′) and (A′, B) interactions; in the _S_dev = 0 Å limit these are perfectly correlated and for large values of _S_dev they are completely uncorrelated. In the intervening partially symmetric regime, the interaction energies are partially correlated. The difference in the AB′ distance and the A′B distance is on average _S_dev, and hence the correlation ρ(_S_dev) between the AB′ and A′B interaction energies decays as _S_dev approaches the characteristic length of the interaction potential. Assuming for concreteness a Gaussian decay in the interaction energy correlation over length L (≈1 Å), we obtain (see SI Appendix and Fig. S2)
The Gaussian energy distribution for complexes with symmetry _S_dev is then
Emergence of Symmetry.
Combining Eqs. 1 and 3, we find that the frequency of complexes with interaction energy E and symmetry _S_dev is
Evolutionary selection can operate only on complexes sufficiently populated to contribute to function; thus, we are interested in the distribution of _S_dev for complexes with binding energy strong enough to at least partially overcome the 10–15 kcal/mol entropic cost of association (12). We obtain the _S_dev distribution as a function of the binding energy threshold E′ required for a contribution to fitness by numerically integrating P(_S_dev, E) from −∞ to E′. As shown in Fig. 1B, with increasingly stringent thresholds (i.e., more negative E′), the probability distribution shifts from predominantly asymmetric (i.e., high-_S_dev) complexes in the random population (blue) to increasingly symmetric (black); when the energy threshold drops below −10σ (red), the distribution is similar to that of the highly symmetric natural homodimers (Fig. 1A, red). In the intermediate case (black), the distribution is bimodal with dominantly asymmetric and dominantly symmetric subpopulations; we quantify the abruptness of this transition between the red and blue modes further below.
We investigated whether this striking emergence of symmetry is manifested in explicit protein docking calculations. The randomly docked complexes described above were refined by using the Rosetta Monte Carlo protein docking protocol (11) to produce well packed protein-like interfaces. For each of the five protein test cases, this energy-based refinement alone considerably increased the extent of symmetry in the population (Table 1; see the SI Appendix and Fig. S3 for discussion). We then selected the subset of refined structures with energies below different cutoff thresholds and determined the _S_dev distribution in each low-energy subpopulation. As shown in Fig. 1C for a typical protein, this energy-based enrichment (black) shifts the original random distribution (blue) dramatically toward low values of _S_dev, and closely parallels the analytic model results (Fig. 1B).
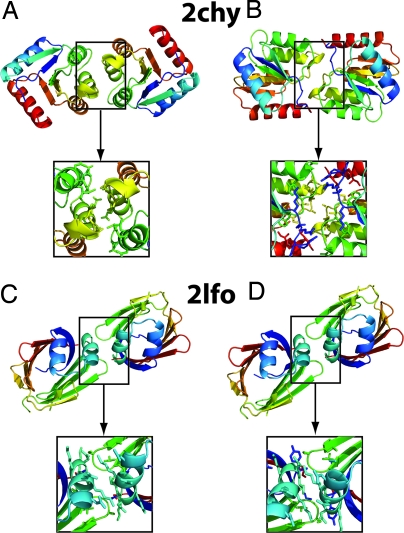
The increase in the fraction of nearly symmetric structures (_S_dev < 0.2 Å) with more stringent energy-based selection threshold is illustrated in Fig. 1D. The simple model predicts a relatively abrupt phase-transition behavior as the energy cutoff for selection drops below −5σ (Fig. 1D, red). This increase in symmetry after energy-based selection is recapitulated in the populations of refined docked protein complexes: increasingly stringent bias toward low-energy structures produces populations with increasing fractions of nearly symmetric (_S_dev < 0.2 Å) structures (Fig. 1D, brown line, and Table 1). Examples of very-low-energy docked complexes with high symmetry are shown on the left side of Fig. 2.
Fig. 2.
Very-low-energy complexes generated by random docking of monomeric proteins are predominantly highly symmetrical. (Left) The most symmetrical binding mode of the three lowest-energy structures after high-resolution docking of the normally monomeric proteins 2chy (A) and 2lfo (C). (Right) Low-energy symmetric binding modes after sequence evolution (see SI Appendix): 2chy (B) and 2lfo (D). Sequence optimization lowers the interaction energies and can change the relative energy of different binding modes, but does not fundamentally change their symmetry.
The preceding analysis suggests that the primordial complexes with populations sufficient to contribute to fitness and hence be subject to natural selection were very likely to be symmetrical. In our model, these states will form the starting point for subsequent evolutionary optimization by mutation and selection. As discussed earlier, Monod et al. (3) suggested that symmetrical interfaces have a further advantage at this evolutionary optimization stage because the effect of a mutation is effectively amplified by symmetry. Indeed, the argument presented in the paragraphs preceding Eq. 2 carries over immediately to the mutation case: the standard deviation of the interaction energy associated with single mutations at interfaces is2-fold greater for symmetric complexes than for asymmetric complexes because each mutation effectively occurs twice. However, this effect is counteracted to some extent by the lower frequency of mutations at symmetric interfaces because asymmetric interfaces can contain up to twofold more independently mutable residues. We investigated the trade-off between these two effects by carrying out explicit simulations of sequence evolution on the randomly generated protein complexes described above. As described in the SI Appendix and Fig. S4, in some but not all cases, cycles of sequence evolution and selection increase the extent of symmetry in the population. Examples of low-energy symmetric structures resulting from cycles of mutation and selection are shown on the right side of Fig. 2.
The analysis in this article has focused primarily on oligomeric forms in which there is a single type of interaction patch between monomers. Routes to more complex oligomers have been discussed by Monod et al. (3), Hansen (13), Teichmann et al. (14, 15), and Bennett et al. (4, 5). Teichmann and coworkers have shown that higher-order oligomers are often built up of smaller symmetric dimers and trimers that associate by using additional interaction patches (15). Our findings of increased likelihood of symmetry among lowest-energy complexes are relevant to both the early stage and late stage in acquisition of oligomer structure in their model; for example, in a tetramer, the initial forming of dimers from monomers, and subsequently the formation of tetramer from interacting dimers.
As noted in the introduction, Shakhnovich and coworkers have studied a question related to the problem investigated here; the energetic advantage of homodimeric versus heterodimeric complexes (9, 10). In the two-dimensional interacting disk model used in those studies, homodimeric complexes are necessarily symmetric, so the problem of the origin of symmetry of homodimeric complexes did not arise. For this model the authors found a twofold greater variance in the homodimer energy distribution than in the heterodimer energy distribution, which parallels our finding for symmetric versus asymmetric homodimeric complexes in this article. Although provocative, in the absence of a model for the decrease in the number of states with increasing symmetry, the earlier work does not indicate whether selection for low energy alone is sufficient to result in the emergence of symmetric structures. (See SI Appendix for further discussion.)
The importance of estimating the prior probability of symmetric complexes is illustrated by the apparent inconsistency between the prevalence in nature of homodimers (and dimers of dimers) (15) and the _j_-fold increase in the variance in energy in cyclic oligomers with J subunits as noted in the above analysis of the interaction energy distribution for random complexes, which should favor the higher-order symmetric structures in the low-energy subpopulation. As emphasized earlier, the fraction of symmetric structures in the low-energy population depends not only on the energy variance, but also on the prior probability of forming a symmetric complex. The probability of randomly forming a symmetric complex is much smaller for higher-order cyclic oligomers††, and the balance between the two factors favors symmetric dimers over higher-order symmetric oligomers.
The key result of this article is that symmetric assemblies can arise solely from a bias toward complexes with sufficiently favorable interaction energy to be significantly populated despite the entropic cost of association. Particularly notable is the almost complete shift toward the _S_dev distribution of native complexes (Fig. 1A, red) produced by selecting the lowest-energy complexes (Fig. 1C, black) from large populations of randomly docked complexes (Fig. 1C, blue) generated in Rosetta all-atom docking simulations. This enrichment occurs because the increase in the variance of the interaction energy with increasing symmetry more than compensates for the decrease in the density of states with increasing symmetry. As shown in the SI Appendix, if the variance in the interaction energy becomes large, as perhaps is the case of partially folded monomers that can reconfigure to optimize interactions with other monomers, symmetry can arise at equilibrium in the absence of selection, which may have relevance to amyloid formation. Similar considerations may also have relevance to the nucleation of symmetry from intrinsically nonsymmetric monomers in nonbiological assemblies such as crystals and clusters. Finally, although we have shown that the pool of randomly arising candidates for eventual competitive selection are a priori extremely biased toward symmetry, it must be emphasized that, as in any argument about history, it is nearly impossible to evaluate the relative contributions of different potential causes; along with the initial bias described here, the factors noted by Monod et al. (3, 16) and others summarized in the introduction likely contributed to the high symmetry of modern homooligomers.
Methods
The protein-modeling software Rosetta was used to perform the protein–protein docking and protein design calculations (11). Rosetta uses real-space Monte Carlo minimization to find the lowest-energy conformation of binding partners. The first step in the docking protocol consists of a low-resolution search where only the backbone atoms and a single “centroid” atom representing the side chains are modeled. Random starting points for docking are generated by randomizing the rotational degrees of freedom of the binding partners, and then translating them into contact. Fig. 1A and column 4 of Table 1 was generated at this stage. In the subsequent high-resolution docking stage, a detailed physically realistic all-atom energy function (17) is optimized by using Monte Carlo minimization protocol. Each step in this protocol consists of (i) a small random perturbation to the rigid body degrees of freedom, (ii) discrete optimization of the side-chain rotamer conformations, and (iii) quasi-Newton optimization of the side chain and rigid body degrees of freedom (11).
The SI Appendix contains a more detailed description of the methodology, notably a more detailed description of the docking and design calculations, derivations of the analytical results, detailed descriptions of the numerical simulations used to generate Fig. 1, and simulations investigating the effect of protein shape on the distribution of _S_dev (Fig. S5).
Supplementary Material
Supporting Information
Acknowledgments.
We thank Ora Schueler-Furman, John Moult, Rhiju Das, and David Eisenberg for stimulating conversations about the origin of symmetry. This work was supported by a Knut and Alice Wallenberg Foundation postdoctoral fellowship (to I.A.), by Defense Threat Reduction Agency Contract MIPR7KO8970172 (to C.E.M.S.), in part by Spanish MEC grant SAB2006-0089 and project FPA2006-05423 (to D.B.K.), by the regional Comunidad de Madrid HEPHACOS project (D.B.K.), and by Department of Energy Grant DE-FG02-00ER41132 (to D.B.K.), and by the National Institutes of Health and Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
††
The number of degrees of freedom required to specify an oligomer structure is 6 [(number of subunits) − 1] for asymmetric complexes and 4 for symmetric complexes, so the fraction of randomly formed complexes that are symmetric is exponentially decreasing with exponent [6 (number of subunits) − 10]. Dimers of dimers have 6 degrees of freedom versus the 4 degrees of freedom for cyclic tetramers, which may also contribute to their greater prevalence. The loss of entropy on binding also favors dimers over higher-order oligomers because the entropic cost of forming a higher-order oligomer increases linearly with the number of subunits.
References
- 1.Crick FH, Watson JD. Structure of small viruses. Nature. 1956;177(4506):473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- 2.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 3.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 4.Bennett MJ, Choe S, Eisenberg D. Domain swapping: Entangling alliances between proteins. Proc Natl Acad Sci USA. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4(12):2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy Y, Cho SS, Shen T, Onuchic JN, Wolynes PG. Symmetry and frustration in protein energy landscapes: a near degeneracy resolves the Rop dimer-folding mystery. Proc Natl Acad Sci USA. 2005;102:2373–2378. doi: 10.1073/pnas.0409572102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Helling R, Tang C, Wingreen N. Emergence of preferred structures in a simple model of protein folding. Science. 1996;273(5275):666–669. doi: 10.1126/science.273.5275.666. [DOI] [PubMed] [Google Scholar]
- 8.Wolynes PG. Symmetry and the energy landscapes of biomolecules. Proc Natl Acad Sci USA. 1996;93:14249–14255. doi: 10.1073/pnas.93.25.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukatsky DB, Shakhnovich BE, Mintseris J, Shakhnovich EI. Structural similarity enhances interaction propensity of proteins. J Mol Biol. 2007;365(5):1596–1606. doi: 10.1016/j.jmb.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukatsky DB, Zeldovich KB, Shakhnovich EI. Statistically enhanced self-attraction of random patterns. Phys Rev Lett. 2006;97(17):178101. doi: 10.1103/PhysRevLett.97.178101. [DOI] [PubMed] [Google Scholar]
- 11.Gray JJ, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331(1):281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 12.Brady GP, Sharp KA. Entropy in protein folding and in protein-protein interactions. Curr Opin Struct Biol. 1997;7(2):215–221. doi: 10.1016/s0959-440x(97)80028-0. [DOI] [PubMed] [Google Scholar]
- 13.Hanson KR. Symmetry of protein oligomers formed by isologous association. J Mol Biol. 1966;22(2):405–409. doi: 10.1016/0022-2836(66)90149-5. [DOI] [PubMed] [Google Scholar]
- 14.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453(7199):1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA. 3D complex: A structural classification of protein complexes. PLoS Comput Biol. 2006;2(11):e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monod J. In: Symmetry and Function of Biological Systems at the Macromolecular Level. Engstrom A, Strandberg B, editors. New York: Wiley; 1969. pp. 15–29. [Google Scholar]
- 17.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information