Structural definition of a conserved neutralization epitope on HIV-1 gp120 (original) (raw)
Abstract
The remarkable diversity, glycosylation and conformational flexibility of the human immunodeficiency virus type 1 (HIV-1) envelope (Env), including substantial rearrangement of the gp120 glycoprotein upon binding the CD4 receptor, allow it to evade antibody-mediated neutralization. Despite this complexity, the HIV-1 Env must retain conserved determinants that mediate CD4 binding. To evaluate how these determinants might provide opportunities for antibody recognition, we created variants of gp120 stabilized in the CD4-bound state, assessed binding of CD4 and of receptor-binding-site antibodies, and determined the structure at 2.3 Å resolution of the broadly neutralizing antibody b12 in complex with gp120. b12 binds to a conformationally invariant surface that overlaps a distinct subset of the CD4-binding site. This surface is involved in the metastable attachment of CD4, before the gp120 rearrangement required for stable engagement. A site of vulnerability, related to a functional requirement for efficient association with CD4, can therefore be targeted by antibody to neutralize HIV-1.
Supplementary information
The online version of this article (doi:10.1038/nature05580) contains supplementary material, which is available to authorized users.
HIV's hidden weakness
Human immunodeficiency virus type 1 (HIV-1) evades host defences thanks to its protective layers of immune camouflage. But b12 antibody pierces these barriers, so its interactions with HIV's gp120 envelope glycoprotein are of interest to vaccine and drug designers. The conformational flexibility of gp120 complicates analysis, but this obstacle has been overcome by using gp120 molecules stabilized in the CD4-bound state. Biophysical analysis with these 'frozen' gp120s shows receptor binding to occur in two steps: an initial 'handshake' followed by a shape-shifting 'bear hug'. The antibody by-passes gp120's protective shape shifting by exploiting an unexpected vulnerability of the handshake: the need for a substantial 'onrate' to efficiently engage CD4. The cover X-ray crystallographic image catches b12 antibody (green) as it grabs onto this newly identified site of vulnerability (yellow).
Supplementary information
The online version of this article (doi:10.1038/nature05580) contains supplementary material, which is available to authorized users.
Main
The human immunodeficiency virus type 1 (HIV-1) crossed from chimpanzees to humans early in the twentieth century and has since infected ∼1% of the world’s adult population1,2. This spread and the absence of an effective vaccine are to a large degree a consequence of the ability of HIV-1 to evade antibody-mediated neutralization3,4,5. On HIV-1, the only viral target available for neutralizing antibodies is the envelope spike, which is composed of three copies of the gp120 exterior envelope glycoprotein and three gp41 transmembrane glycoprotein molecules6,7. Genetic, immunological and structural studies of the HIV-1 envelope glycoproteins have revealed extraordinary diversity, manifest in a variety of immunodominant loops, as well as multiple overlapping mechanisms of humoral evasion, including self-masquerading glycan and conformational masking8,9,10,11. These evolutionarily honed barriers of diversity and evasion have confounded traditional vaccine development.
Two strategies have been proposed to surmount these barriers: examination of known broadly neutralizing antibodies (2F5, 2G12, 4E10 and b12) to identify susceptible targets of neutralization12, and analysis of functional constraints to identify potential sites of vulnerability13. To facilitate viral entry, the gp120 glycoprotein must bind to cell-surface CD4 (ref. 14), alter its conformation to reveal a site for co-receptor attachment15, and trigger conformational rearrangements in the gp41 glycoprotein to mediate fusion of viral and host cell membranes16,17. Constraints on envelope (Env) variation and exposure, associated with required functions of viral entry, provide potential footholds for broad, antibody-mediated neutralization.
Here we combine analysis of function with clues from antibody. We constructed stabilized gp120 molecules, constrained to stay in the CD4-bound conformation even in the absence of CD4, and tested the effect of this stabilization on the binding kinetics of CD4 and on antibodies reactive with sites of receptor binding. We then determined the crystal structure of the broadly neutralizing antibody b12 in complex with one of the stabilized gp120 molecules. Analysis of this structure, combined with detailed antigenic analyses of gp120 molecules stabilized to various extents in the CD4-bound conformation, not only reveals the functionally conserved surface that allows for initial CD4 attachment, but also provides an atomic-level description of the b12 epitope, which serves as a key target for humoral neutralization of HIV-1.
Recognition of the CD4-binding site
Conformational flexibility of HIV-1 gp120 complicated analysis of antibody recognition. To circumvent this complication, we used an iterative structure-based scheme to stabilize gp120 in its CD4-bound state (Table 1, Supplementary Fig. 1 and Supplementary Tables 1 and 2). The CD4-bound state of gp120 comprises an inner domain, outer domain and four-stranded bridging sheet mini-domain18. Five disulphides and four cavity-altering substitutions were created to restrict interdomain movements and to stabilize bridging sheet formation. Crystallographic analysis of gp120 variants with these substitutions in complex with CD4 and antibody 17b at 2.0–2.2 Å resolution showed that four of five disulphides formed and that disulphide and cavity-altering substitutions induced minimal structural perturbation. We also analysed a two-disulphide variant at 2.5 Å resolution as well as a three-disulphide variant at 2.8 Å resolution (Table 1). In both structures, all potential disulphides formed.
Table 1.
Characterization of stabilized gp120 cores.
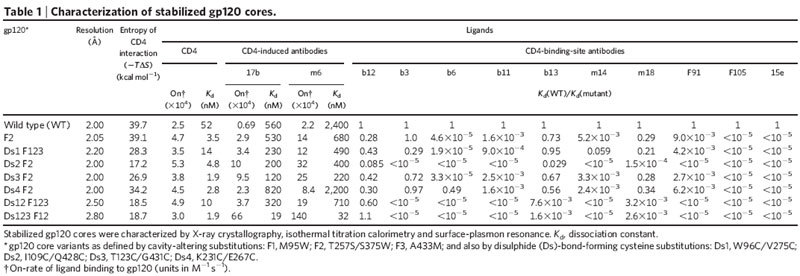
To measure the degree of conformational fixation, we used isothermal titration calorimetry to assess the entropy of interaction between the different stabilized gp120 cores and CD4 (Table 1 and Supplementary Table 3). Disulphides had a more substantial effect than cavity-altering substitutions. Tethering the centre two strands of the bridging sheet (by linking residues 123–431) or the termini-proximal ends of the domains (96–275 or 231–267) reduced the entropy of interaction by 15–30%. A more substantial effect (a 60% reduction) was gained by tethering the bridging sheet to the inner domain (109–428). Thermodynamic analysis thus quantified the degree of variant gp120 stabilization and demonstrated substantial remaining flexibility, despite the presence of up to three stabilizing disulphides.
Because the epitope for CD4-induced antibodies is only formed in the CD4-bound state10,19,20, we could use the increase in antibody on-rate to assess the degree to which stabilization ‘preformed’ the variant gp120 molecules in the CD4-bound state. Reasonable correlations were observed between the on-rate for CD4-induced antibodies and the entropy of CD4 binding (r = 0.74, P < 0.036 for antibody 17b (ref. 19); r = 0.76, P < 0.029 for antibody m6 (ref. 21)), with the most extreme change for the three-disulphide variant (a 95-fold increase in on-rate for antibody 17b and a 65-fold increase for antibody m6) (Table 1 and Supplementary Fig. 1c).
Notably, almost no change in CD4 on-rate was observed with the stabilized gp120 molecules (Table 1, Supplementary Table 4 and Supplementary Fig. 1c), suggesting that the initial contact surface recognized by CD4 is present in unliganded gp120 before the CD4-induced conformational change. Similarly invariant on-rates have recently been reported for CD4 binding to gp120 molecules restrained from achieving a CD4-bound conformation by removing a flexible loop22. Thus, in contrast to the CD4-induced antibodies, a conformational change is not required to expose an initial site of contact for CD4. Rather, it serves to lock CD4 into place once contact has been made.
To determine the degree to which fixation of gp120 in its CD4-bound conformation restricts antigenic recognition, we tested binding of a panel of CD4-binding-site antibodies to the stabilized gp120 variants (Table 1). The average overall difference in free energy of binding for CD4-binding-site antibodies to wild type and stabilized variants correlated well with the change in entropy of variant gp120 binding to CD4 (r = 0.89, P < 0.0035) (Supplementary Fig. 1c). Conformational constraint thus effectively hides the CD4-bound conformation of gp120 from recognition by antibodies that target the site of CD4 binding. Indeed, of all the CD4-binding-site antibodies tested, only the unique, broadly neutralizing antibody b12 was able to bind well to stabilized gp120.
Structure of a b12–gp120 complex
Although gp120 is the primary target of neutralizing antibodies elicited during natural infection23, most gp120-reactive antibodies are ineffective at neutralizing primary HIV-1 isolates (reviewed in refs 4, 24). Only two gp120-reactive antibodies (b12, 2G12) with effective neutralization activity against diverse primary HIV-1 isolates have thus far been identified25,26,27. In vivo, b12 can protect macaques against vaginal challenge from pathogenic simian–human immunodeficiency virus (SHIV)28. Although the structure of the entire, unbound b12 immunoglobulin (IgG1) has been determined29, a molecular description of its interaction with gp120 remained elusive.
To facilitate a structural understanding of b12 neutralization of HIV-1, we screened crystallizations of b12 in complex with various forms of gp120. The lack of b12 conformational fixation of gp120 (ref. 10), coupled to inherent gp120 flexibility, complicated crystallization of a b12–gp120 complex; to overcome this complication, we used the mutationally stabilized gp120 molecules. Complexes with wild-type core or with single-disulphide variants (96–275 or 123–431) failed to produce crystals suitable for structural analysis. However, diffraction to 2.3 Å was obtained from hexagonal crystals of the antigen-binding fragment (Fab) of b12 in complex with a two-disulphide-stabilized gp120 core. We solved the complex structure by molecular replacement and refined it to an _R_-value of 19.3% (_R_free = 25.5%) (Supplementary Table 5).
Notably, only the heavy chain of b12 interacted with gp120, with each of the three heavy-chain complementarity-determining regions (CDRs) making extensive contact (Fig. 1, Supplementary Table 6 and Supplementary Figs 2 and 3). Despite this unusual heavy-chain-only usage, the surface areas of interaction were in the range typical for antibody–protein interfaces (reviewed in ref. 30), with a total of 1,480 Å2 buried31 in the interaction (737 Å2 on gp120 and 743 Å2 on b12).
Figure 1. Structure of b12 in complex with an HIV-1 gp120 core.
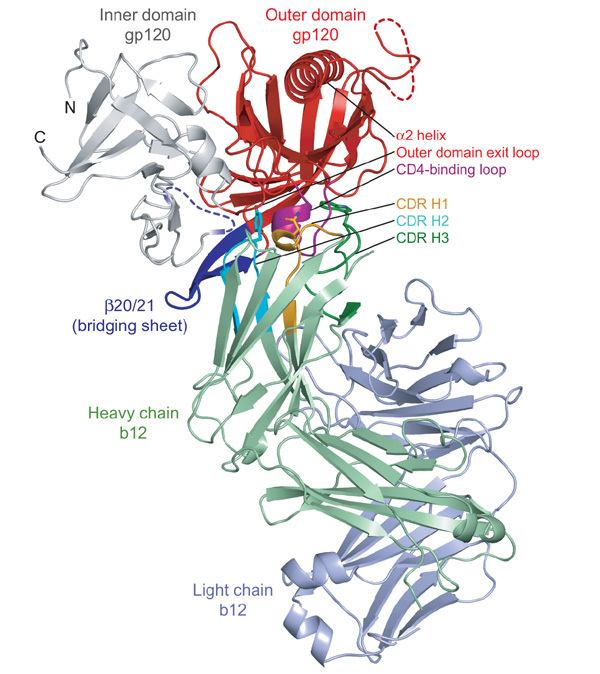
Polypeptide chains are depicted in ribbon representation, with disordered regions as dashed lines. The gp120 inner domain is grey, and the outer domain is red, except for the CD4-binding loop, which is purple. The strands and associated loops, which in the CD4-bound conformation correspond to the bridging sheet, are blue. The b12 light chain is blue–grey and the b12 heavy chain is green, with associated CDRs highlighted in orange (H1), cyan (H2) and dark green (H3). The heavy-chain dominance of the binding interaction is apparent, with the nearest light-chain approach separated by ∼10 Å from gp120. Heavy-chain-only interactions are rare, although heavy-chain interactions predominate in a number of viral Env–antibody complexes, including those from SARS coronavirus49 and influenza virus haemagglutinin50. Three b12 residues (Asn 31, Tyr 53 and Trp 100) from each of the heavy-chain CDRs are depicted in stick representation. Together, these three residues combine to form ∼40% of the b12 contact surface. They can be seen gripping the CD4-binding loop, the central focus of the b12 interaction with gp120.
The gp120 surface bound by b12 was confined largely to the gp120 outer domain, consistent with predictions from alanine substitutions (Supplementary Table 7 and ref. 32). We examined this surface for features that might make it amenable to antibody recognition. The outer domain is composed of two barrels, stacked end to end (Fig. 2). The termini-distal barrel comprises seven anti-parallel β-strands; the other contains six β-strands embracing the α2 helix. b12 binds at the barrel–barrel juncture to the gp120 face opposite the α2 helix. Although most of the gp120 backbone is involved in secondary structure interactions, the barrel juncture contains a number of structural elements with unpaired backbone, often used by antibodies in recognition. Principal among these are two parallel loops—the CD4-binding loop and the outer domain exit loop—that extend from the only two parallel β-strands in the entire gp120 core. b12 takes advantage of this backbone reactivity, forming six direct and four water-mediated hydrogen bonds with the main-chain atoms of these two loops (Supplementary Fig. 2). Overall, the outer domain comprises 82% of the gp120 contact surface with b12, with the CD4-binding loop forming a little over one-half of this surface.
Figure 2. Conformational states of gp120.
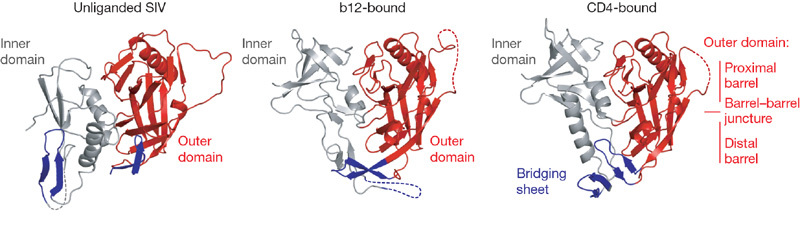
The unliganded, b12- and CD4-bound conformations of gp120 are depicted, with polypeptide in ribbon representation and disordered regions as dashed lines. Inner domains are grey, outer domains are red and regions that in the CD4-bound state correspond to the bridging sheet are blue. Both b12- and CD4-bound conformations are of the Ds12 F123 variant of HIV-1, whereas the unliganded structure is of simian immunodeficiency virus (SIV). Comparison of these three gp120 conformations highlights not only the structural plasticity of the inner domain and bridging sheet, but also the conformational stability of the outer domain.
Comparison of b12- and CD4-binding
Both CD4 and b12 bind primarily to the outer domain of gp120, which is remarkably well preserved between unliganded, b12- and CD4-bound states (Figs 2 and 3). Their entropies of interaction, however, are very different, with CD4 inducing a 40–50 kcal mol-1 change, and b12 inducing only a 6 kcal mol-1 change10. This difference in conformational fixation of gp120 is seen in the divergent atomic-mobility values of the domains (Fig. 3b).
Figure 3. b12 and CD4 recognition of gp120.
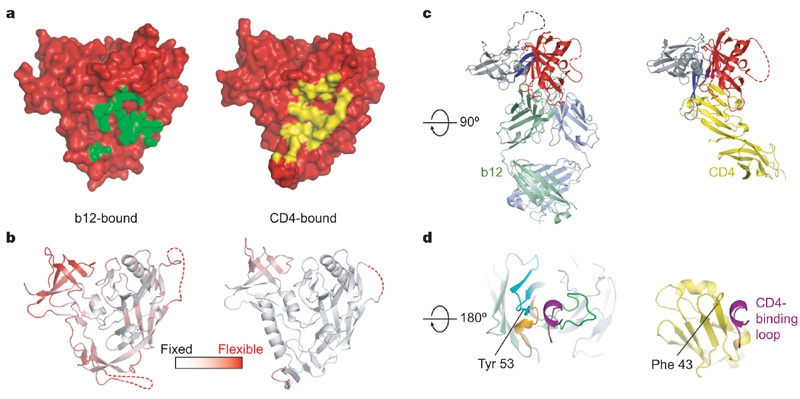
The orientation of gp120 in a and b is the same as in Fig. 2, with c and d rotated about a horizontal axis by 90° and 180°, respectively. a, Molecular surface of gp120 in red, with the b12-contact surface in green (left) and the CD4-contact surface in yellow (right). b, Ribbon diagram of the b12- and CD4-bound gp120 coloured according to the atomic mobility of the polypeptide, with white for fixed and red for flexible. In the b12-induced conformation, only the outer domain is fixed by interaction with antibody, with the average atomic mobility of the outer domain about one-half that of the inner domain. By contrast, CD4 fixes the entire core, resulting in outer and inner domains of similar overall atomic mobility. c, Comparison of b12 and CD4 angles of attachment. The polypeptide chains are depicted in ribbon representation, and coloured according to Fig. 1, with CD4 in yellow. The similarity in angle of attachment as well as the binding focus of b12 on the outer domain (red) are evident in this orientation. d, The CD4-binding loop of gp120 (purple) is shown with b12 (left) and CD4 (right). There are parallels between several key CD4 contact residues and those of b12. For example, Phe 43 of CD4 (shown in stick model) inserts into a critical juncture at the nexus of the inner, outer and bridging sheet regions of gp120, whereas Tyr 53 of b12 (also shown in stick model) inserts at a similar position, although displaced by ∼3 Å.
The angles of approach of CD4 and b12 to gp120 are similar, although not precisely the same (Fig. 3c). If the gp120 outer domains of b12- and CD4-bound structures were superimposed to orient equivalently b12 and CD4, about one-half of CD4 domain 1, which makes all of the contacts with gp120, is not encompassed by the b12 Fab (despite the volume of the Fab variable domains being twice as large as that of domain 1 of CD4). The projection of CD4 outside of the angle of b12 approach suggests that the parameters that sterically restrict b12 binding to the functional spike are not overly stringent.
The contact surfaces of CD4 and b12 on gp120 have considerable overlap (Fig. 3a and Supplementary Fig. 4). Most of this overlap is on the outer domain, where the CD4-binding loop is a central focus of binding for both CD4 and b12. CD4 and b12, however, interact with the CD4-binding loop quite differently (Fig. 3d). b12 uses all three of its CDR heavy-chain loops to grasp virtually all surface-exposed portions of the loop. In contrast, CD4 only binds to one side of the loop, making anti-parallel hydrogen bonds between CD4- and gp120-main-chain atoms.
The primary difference between b12 and CD4 interactions with gp120 involves the conformationally mobile β20/21. For b12, these interactions are peripheral to the binding surface, with alanine substitution of the primary b12 contact with β20/21 (at b12 residue Asn 56) having little impact on overall b12 binding33. In contrast, CD4 interactions with β20/21 form an integral part of the binding surface, burying 160 Å2 of surface area and forming a topologically contiguous contact surface with the CD4-binding loop.
To delineate further the differences in binding between b12 and CD4, we characterized binding interactions with an HIV-1 gp120 fragment, termed OD1 (ref. 34). This fragment comprises residues 252–482 and encompasses the entire outer domain including V3 as well as the β20/21 excursion. Binding of b12 to OD1 showed nearly identical rates of association compared to binding of b12 to HXBc2 core gp120, although the dissociation rate was about 15-fold more rapid (Supplementary Fig. 5). We were unable to detect binding of CD4 to OD1 (Supplementary Fig. 5). Because CD4 demonstrated virtually no change in on-rate when tested on conformationally stabilized gp120 molecules, we turned to a dodecameric variant of CD4 (D1D2-Igαtp (ref. 35)), as avidity from multivalent binding provides an effective means by which to reduce off-rate. We observed that D1D2-Igαtp binds with virtually identical rates of association to both OD1 and core gp120 (Supplementary Fig. 5).
The results suggest the following series of molecular interactions for b12 and CD4 binding to gp120 (Supplementary Fig. 6). Initial contact by CD4 occurs with the structurally invariant outer domain, to a surface constitutively exposed on the envelope spike. In primary isolates, which are generally resistant to neutralization by soluble CD4, this interaction is not stable and CD4 readily ‘falls off’. However, at the cell surface (or with dodecameric CD4), multiple CD4 molecules can bind simultaneously to the viral spike and use avidity to enhance stability. The avidity-enhanced outer domain–CD4 complex provides a receptive contact surface for the bridging sheet. A highly coordinated rearrangement of the inner domain allows for formation of the bridging sheet, which welds CD4 into place.
Contact by b12 occurs at the same constitutively exposed surface initially recognized by CD4. However, b12 is able to latch onto this outer domain surface with high affinity, without additional gp120 conformational change. This absence of conformational constraint allows b12 to bind and neutralize primary isolates that otherwise would be protected by conformational masking. In this manner, b12 uses the functionally conserved initial contact site for CD4 on gp120 to neutralize effectively HIV-1 (Fig. 4).
Figure 4. Structural definition of a conformationally invariant, antibody-accessible portion of the CD4-binding site.
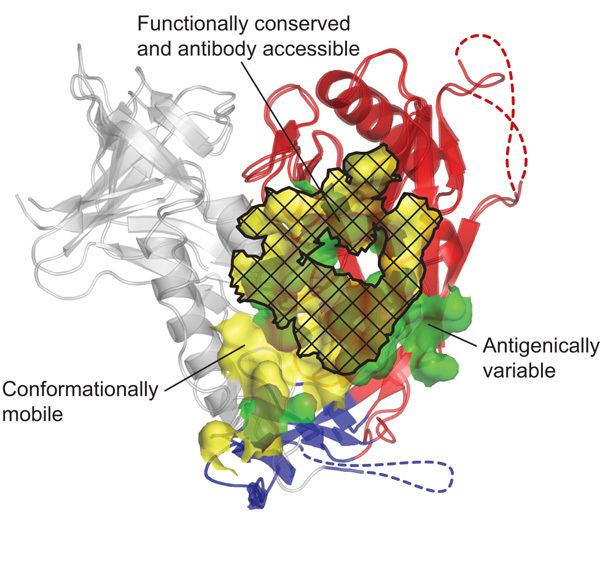
The b12- and CD4-bound conformations of gp120 are shown in ribbon representation, after superposition of outer domains (red). A semitransparent molecular surface shows the contact surfaces of b12 (green) and CD4 (yellow). Subsets of these surfaces, corresponding to regions of conformational flexibility (for example, of the inner domain (grey) or bridging sheet (blue)), are delineated, as are regions of b12 contact outside of the conserved CD4-binding site. As can be seen, functional analysis serves to transcend the particulars of b12 binding, whereas antibody defines accessibility. Although we have formally shown only the b12 contact surface to be accessible in the context of a functional viral spike, the highly effective neutralization of D1D2-Igαtp and the kinetics of its association with both core and OD1 variants of gp120 suggest that the CD4-binding surface on the outer domain is accessible.
A site of HIV-1 vulnerability
Wedged between the glycan-shielded silent face and the conformationally flexible inner domain/bridging sheet, with its projecting variable loops, the identified site is recessed, although it must be accessible to a molecule as large as CD4 to serve its function. Otherwise, its size and reactive loops make it suitable for recognition by antibody. Recent serological analysis of long-term non-progressors attributes the broad and potent neutralizing properties of some of these sera to antibodies directed against this site36. The potential of receptor-binding-site antibodies to effect broad neutralization has been recognized since the first viral structures revealed conserved sites of receptor binding37,38. Here we show how an unanticipated requirement for the maintenance of a substantial receptor on-rate allows b12 to access and disable this most difficult of targets, the highly protected HIV-1 Env.
Methods
Structure-based stabilization of gp120 in its CD4-bound conformation
Cavity-altering substitutions were identified by examination of highly conserved residues that bordered interdomain cavities, whereas all Cβ-Cβ distances between 3–7 Å were examined to identify potential stabilizing disulphides. A high-throughput expression system, coupling transient expression with swainsonine, endoglycosidase H and concanavalin A, was devised from related procedures39 to produce gp120 suitable for crystallization. Briefly, codon-optimized variants were constructed in the context of core gp120 (HXBc2) and expressed transiently in 293 cells under control of the CMV/R promoter40. Swainsonine (20 mg l-1; Biomol) was added to the cell-culture medium 2 h before DNA transfection, and supernatants were collected after 48 and 96 h. Secreted gp120 molecules were purified by 17b-affinity chromatography and deglycosylated with endoglycosidase H (Endo Hf; New England Biolab) at pH 5.9, 37 °C, with gp120 containing residual uncleaved glycan moieties removed by passage over concanavalin A Sepharose (Sigma-Aldrich). Ternary complexes of deglycosylated core gp120, d1d2 of CD4 and 17b Fab were prepared as described previously41, with _P_2221 crystals grown (Supplementary Table 1) around conditions identified for the _Drosophila_-produced wild-type core18. Because crystals were small (rods, typical diameter 40 μm) and subject to high radiation dosages during data collection, difference Fourier maps comparing the initial and final swatches of data were inspected to identify radiation-induced disulphide breakage42, and the refined models (Supplementary Table 1) adjusted to reflect the initial, radiation-damage-free structure.
Surface-plasmon resonance
A Biacore 3000 surface-plasmon resonance spectrometer was used to measure kinetic constants (see Supplementary Table 4 and Supplementary Fig. 5 for details).
Isothermal titration calorimetry (ITC)
Wild-type core and variant gp120 molecules were dialysed in PBS buffer and titrated with d1d2 of CD4 on a MicroCal VP-ITC at 37 °C. Resultant data were examined using Origin software (MicroCal).
Structural determination of a b12–gp120 complex
The antigen-binding fragment (Fab) of b12 was produced by papain digestion, and purified with Superdex S200 chromatography (0.35 M NaCl, 2.5 mM Tris pH 7.1, 0.02% NaN3). The Fab peak was pooled and mixed with deglycosylated gp120, which was produced by transient transfection as described above, and the resultant complexes purified by S200 chromatography. Crystals of Fab b12 and a two-disulphide variant (Ds12 F123) were grown by mixing 0.5 μl of complex (4 mg ml-1) in S200 buffer with 0.5 μl of droplet mix (10.5% PEG 8,000, 0.2 M glycine, 105 mM Mg-acetate, 52.5 mM Na-cacodylate pH 6.5) and equilibrating in hanging droplets over reservoirs (droplet mix without glycine) at 20 °C. Hexagonal bi-pyramids (200 μm in length by 90 μm in diameter) were crosslinked43, transferred to 15% PEG 8,000, 150 mM Mg-acetate, 100 mM Na-cacodylate pH 6.5, 30% ethylene glycol, 2.5% 2_R_,3_R_-butandiol, 2.5% trehalose, and flash-frozen in a nitrogen-cryostat stream. Data were collected at 100 K and processed with HKL2000 (ref. 44). Molecular replacement (AMoRe45) identified a 5.2σ peak (15–3 Å data) for the Fab portion (chains H and L) of the b12 IgG (Protein Data Bank 1HZH)29, and phases from the rigid-body refined molecule allowed unambiguous placement of the outer domain in _F_o - _F_c density. Iterative model building (XtalView46), combined with refinement (CNS47, Refmac48), were used to define the remaining ordered parts of gp120.
Supplementary information
Acknowledgements
We thank B. Graham, D. Hamer, S. Harrison, R. Pantophlet, W. Schief, L. Shapiro, I. Wilson and members of the Structural Biology Section, VRC, for discussions and comments on the manuscript; M. Gao for assistance with PDB deposit; H. Katinger for antibody 2G12; G. Lin for suggesting the use of swainsonine; S. Majeed for preparation of Fab 17b; J. Nelson for assistance with b12 ELISAs; M. Posner for antibody F105; J. Robinson for antibodies 17b, 1.5e and F91; J. Stuckey for assistance with figures; M. Venturi for assistance with gp120 production methodology; and the NIH AIDS Research and Reference Reagent Program for CD4. Support for this work was provided by the Intramural Research Program of the NIH, by the International AIDS Vaccine Initiative, by a grant from the Bill and Melinda Gates Foundation Grand Challenges in Global Heath Initiative, and by grants from the NIH. Use of SER-CAT at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science.
Author Contributions T.Z. and P.D.K. carried out structure-based stabilization, SPR analyses and structural determinations; L.X. and G.J.N. constructed gp120 substitutions and developed and implemented a high-throughput gp120-production system suitable for crystallization; B.D. and R.W. carried out ITC characterizations; A.J.H., M.B.Z. and D.R.B. provided b12, b3, b6, b11 and b13, and mutant b12 binding; D.V.R. and J.A. provided D1D2-Igαtp and associated SPR analyses; S.-H.X., X.Y. and J.S. provided OD1 and preliminary design and antigenic analyses; and M.-Y.Z. and D.S.D. provided m6, m14 and m18. All authors contributed to the manuscript preparation.
Coordinates and structure factors have been deposited in the Protein Data Bank and may be obtained from the authors (accession codes 2nxy–2ny6 for the nine variant gp120 molecules with CD4 and 17b; accession code 2ny7 for the b12–gp120 complex)
Competing interests
Coordinates and structure factors have been deposited in the Protein Data Bank and may be obtained from the authors (accession codes 2nxy–2ny6 for the nine variant gp120 molecules with CD4 and 17b; accession code 2ny7 for the b12–gp120 complex). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Korber B, et al. Timing the ancestor of the HIV-1 pandemic strain. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 2.Joint United National Programme on HIV/AIDS. 2006 Report on the global AIDS epidemic. 〈http://www.aids.org/en/HIVdata/2006GlobalReport/〉 (2006)
- 3.Weiss RA, et al. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature. 1985;316:69–72. doi: 10.1038/316069a0. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 5.Parren PW, Moore JP, Burton DR, Sattentua QJ. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13:S137–S162. [PubMed] [Google Scholar]
- 6.Kowalski ML, et al. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 7.Lu M, Blackow S, Kim P. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nature Struct. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 8.Starcich BR, et al. Identification and characterization of conserved and variable regions of the envelope gene HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 9.Wyatt R, et al. The antigenic structure of the human immunodeficiency virus gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 10.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 11.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 12.Burton DR. Antibodies, viruses and vaccines. Nature Rev. Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 13.Luftig MA, et al. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nature Struct. Mol. Biol. 2006;13:740–747. doi: 10.1038/nsmb1127. [DOI] [PubMed] [Google Scholar]
- 14.Dalgleish AG, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 15.Feng F, Broder CC, Kennedy PE, Berger EA. HIV-1 entry co-factor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 17.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 18.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thali M, et al. Characterization of conserved human immunodeficiency virus type 1 (HIV-1) gp120 neutralization epitopes exposed upon gp120–CD4 binding. J. Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 21.Zhang MY, et al. Improved breadth and potency of an HIV-1-neutralizing single-chain antibody by random mutagenesis and sequential antigen panning. J. Mol. Biol. 2004;335:209–219. doi: 10.1016/j.jmb.2003.09.055. [DOI] [PubMed] [Google Scholar]
- 22.Rits-Volloch S, Frey G, Harrison SC, Chen B. Restraining the conformation of HIV-1 gp120 by removing a flexible loop. EMBO J. 2006;25:5026–5035. doi: 10.1038/sj.emboj.7601358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Profy AT, et al. Epitopes recognized by the neutralizing antibodies of an HIV-1-infected individual. J. Immunol. 1990;144:4641–4647. [PubMed] [Google Scholar]
- 24.Burton DR, Montefiori DC. The antibody response in HIV-1 infection. AIDS. 1997;11:587–598. doi: 10.1097/00002030-199705000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 26.Trkola A, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4 IgG. J. Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parren PW, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saphire EO, et al. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 30.Sundberg EJ, Mariuzza RA. Molecular recognition in antibody-antigen complexes. Adv. Protein Chem. 2002;61:119–160. doi: 10.1016/S0065-3233(02)61004-6. [DOI] [PubMed] [Google Scholar]
- 31.Connolly ML. The molecular surface package. J. Mol. Graph. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 32.Pantophlet R, et al. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwick MB, et al. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 2003;77:5863–5876. doi: 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, et al. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 2004;78:12975–12986. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arthos J, et al. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein. Implications for therapeutic and vaccine strategies. J. Biol. Chem. 2002;277:11456–11464. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, et al. Neutralizing specificity mapping in complex polyclonal sera. 2006. [Google Scholar]
- 37.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornoviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 38.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 39.Butters TD, et al. Effects of N-butyldeoxynojirimycin and the Lec3.2.8.1 mutant phenotype on N-glycan processing in Chinese hamster ovary cells: application to glycoprotein crystallization. Protein Sci. 1999;8:1696–1701. doi: 10.1110/ps.8.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barouch DH, et al. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 2005;79:8828–8834. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwong PD, et al. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1) J. Biol. Chem. 1999;274:4115–4123. doi: 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- 42.Weik M, et al. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl Acad. Sci. USA. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lusty CJ. A gentle vapor diffusion technique for cross-linking of protein crystals for cryocrystallography. J. Appl. Crystallogr. 1999;32:106–112. doi: 10.1107/S002188989801053X. [DOI] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. doi: 10.1107/S0108767393007597. [DOI] [Google Scholar]
- 46.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 47.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994) [DOI] [PubMed]
- 49.Prabakaran P, et al. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fleury D, Daniels RS, Skehel JJ, Knossow M, Bizebard T. Structural evidence for recognition of a single epitope by two distinct antibodies. Proteins. 2000;40:572–578. doi: 10.1002/1097-0134(20000901)40:4<572::AID-PROT30>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.