Insights on Escherichia coli Biofilm Formation and Inhibition from Whole-Transcriptome Profiling (original) (raw)
. Author manuscript; available in PMC: 2010 Jan 1.
SUMMARY
Biofilms transform independent cells into specialized cell communities. Here are presented some insights into biofilm formation ascertained with the best-characterized strain, Escherichia coli. Investigations of biofilm formation and inhibition with this strain using whole-transcriptome profiling coupled to phenotypic assays, in vivo DNA binding studies, and isogenic mutants have led to discoveries related to the role of stress, to the role of intra- and interspecies cell signaling, to the impact of the environment on cell signaling, to biofilm inhibition by manipulating cell signaling, to the role of toxin/anti-toxin genes in biofilm formation, and to the role of small RNAs on biofilm formation and dispersal. Hence, E. coli is an excellent resource for determining paradigms in biofilm formation and biofilm inhibition.
Biofilm formation
Biofilms are a community of microorganisms attached to a surface by polysaccharides, proteins, and nucleic acids (Sauer et al., 2007). E. coli biofilm development is a complex process that leads to beautiful structures (Fig. 1) that are important for disease and for engineering applications (note the first engineered biofilm was created to secrete peptide antimicrobials to reduce corrosion (Jayaraman et al., 1999)). These matrices are formed through at least five developmental stages that include (i) initial reversible attachment of planktonic cells to a solid surface, (ii) transition from reversible to irreversible attachment, (iii) early development of biofilm architecture, (iv) development of microcolonies into a mature biofilm, and (v) dispersion of cells from the biofilm to return to the planktonic state (van Houdt and Michiels, 2005). Early steps in biofilm formation require the synthesis of different bacterial surface appendages including flagella that allow reversible attachment (Prüß et al., 2006) and cell motility which is a determinant of biofilm architecture (Wood et al., 2006). For irreversible attachment, flagella synthesis is repressed and adhesive organelles like curli fimbriae, encoded by the csg operon, and type I fimbriae, encoded by fim genes, are important for biofilm formation (Prüß et al., 2006). The mannose-sensitive, type I fimbriae also mediate adherence (Connell et al., 1996) and antibiotic-resistant pod formation (Anderson et al., 2003) that is important for invasion of host cells in some urinary tract infections, and bundle-forming pili and the EspA filament are important for biofilm formation by enteropathogenic E. coli (Moreira et al., 2006). Note that conjugation plasmids increase biofilm formation (Ghigo, 2001) in a manner independent of flagella, type I fimbriae, outer membrane autotransporter Ag43 (promotes autoaggregation), and curli (Reisner et al., 2003) due to an envelope stress response (Yang et al., 2008). This review focuses on E. coli biofilm formation and inhibition based on recent developments in the field (primarily whole transcriptome profiling) with both pathogenic and non-pathogenic strains. More comprehensive reviews of E. coli biofilm formation are available such as the that of Ghigo and colleagues (Beloin et al., 2008).
Fig. 1.
E. coli BW25113 biofilm as viewed using the green-fluorescent-protein-expressing plasmid pCM18, confocal microscopy, and IMARIS software (conditions: Luria broth after 48 hr at 37°C, flow rate of 10 mL/hr). Scale bar (upper right) indicates 10 μm.
Whole-transcriptome profiling and stress response
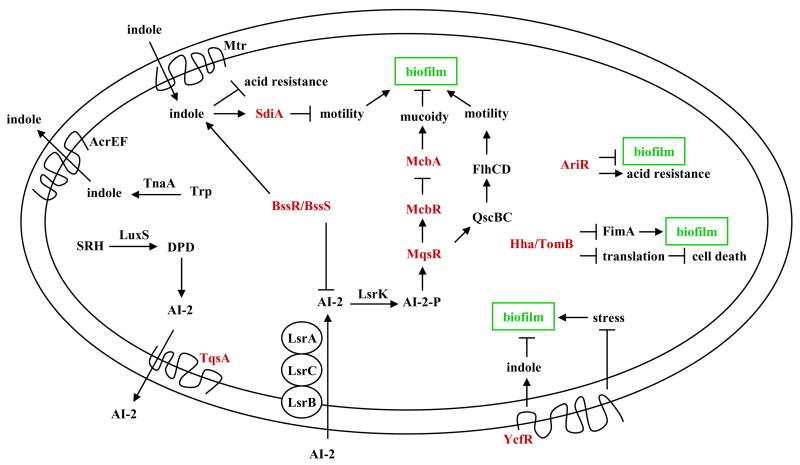
Although DNA microarray technology may miss some aspects of biofilm development related to global averaging of heterogeneous cells (An and Parsek, 2007; Barken et al., 2008), whole-transcriptome profiling has provided robust insights into the biofilm mode of life (a schematic of newly-characterized proteins related to biofilm formation is shown in Fig. 2). For E. coli, five single time point DNA microarrays have been used to explore the genetic basis of its biofilm formation (Schembri et al., 2003; Beloin et al., 2004; Ren et al., 2004b; Junker et al., 2006; Hancock and Klemm, 2007) and one temporal study has been completed (Domka et al., 2007). In the temporal study, six E. coli proteins related to the bacterial signaling molecule cyclic diguanylic acid (c-di-GMP, Fig. 3) were altered in a temporal manner (yaiC, yliF, yciR, yddV, yeaJ, and yjiU); c-di-GMP has been linked to biofilm formation in several strains and its overproduction increases E. coli biofilm formation (Mendez-Ortiz et al., 2006). Of these, YciR has been linked to H-NS and curli formation via c-di-GMP control of the stationary-phase, stress-response, master controller RpoS, and YaiC has been linked to curli and cellulose via c-di-GMP (Weber et al., 2006).
Fig. 2.
Schematic of E. coli proteins related to biofilm formation. Proteins that were identified through whole-transcriptome studies and later characterized as described in this review are shown in red.
Fig. 3.
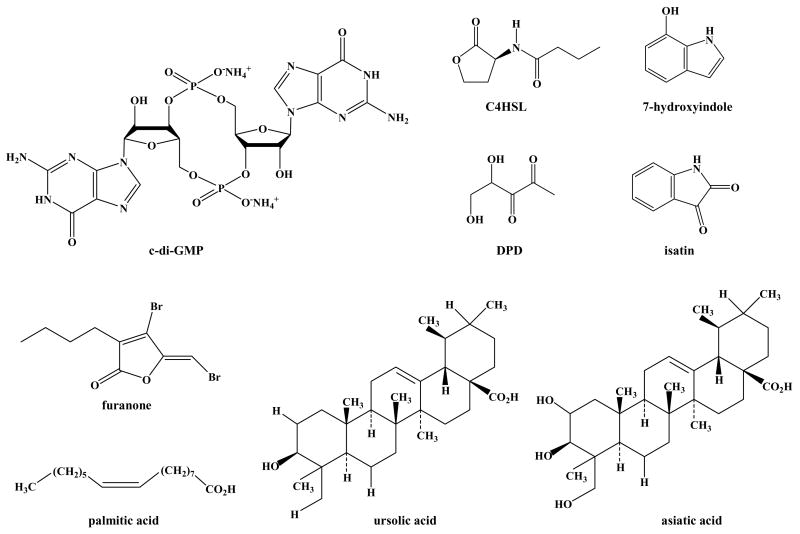
Structure of biofilm-related compounds: cyclic diguanylic acid (c-di-GMP), _N_-butyryl-_L_-homoserine lactone (C4HSL), 4,5-dihydroxy-2,3-pentanedione (DPD), 7-hydroxyindole, isatin, (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone (furanone), palmitic acid, ursolic acid, and asiatic acid.
One common trend in these biofilm transcriptome studies is that stress genes are induced. For example, 42 stress-related genes were identified in the temporal study (Domka et al., 2007), and five induced stress response genes (hslST, hha, soxS, and ycfR) were indentified in 7 h E. coli biofilm cells (Ren et al., 2004b). Follow up studies on the putative outer membrane protein YcfR (renamed BhsA for influencing biofilm formation through hydrophobicity and stress response) indicate this protein mediates the stress response in E. coli by a mechanism that includes inducing indole synthesis (cells that lack BhsA are more sensitive to acid, heat, hydrogen peroxide, and cadmium and have reduced indole concentrations) and that stress itself increases E. coli biofilm formation (Zhang et al., 2007).
In addition, the envelope stress response genes, such as pspABCDE, cpxAR, rpoE, and rseA, were induced in E. coli 8-day-old biofilms cells compared to exponentially-growing planktonic cells regardless of the presence of a conjugative plasmid (Beloin et al., 2004). Furthermore, rpoS plays a key role during biofilm formation because it encodes the sigma S factor which regulates a number of stress-related genes, and yeaGH were also identified as putative stress response genes (Schembri et al., 2003) since they are regulated by RpoS in Salmonella enterica. In addition, cold-shock protein regulators cspABFGI and the heat-shock protein regulator htgA were induced in a temporal fashion during biofilm formation (Domka et al., 2007).
In human urine, stress genes were also induced in asymptomatic bacteriuria E. coli during biofilm formation including both cold shock and heat shock (e.g., cspAGH, ibpAB, pphA, soxS, and yfiD) (Hancock and Klemm, 2007); asymptomatic bacteriuria is a bacterial infection of urine that does not elicit the usual symptoms of an urinary tract infection. Note that stress tolerance is central to the ability of many bacterial pathogens to successfully colonize hostile host environments; for example, Hfq, a protein involved in the stabilization of small, non-coding RNAs (sRNAs), is critical to the ability of uropathogenic E. coli to form biofilms, to colonize effectively, and to persist in the urinary tract (Kulesus et al., 2008). Therefore, the newly-identified relationship between stress tolerance and biofilm formation via whole transcriptome profiling is important.
E. coli genomic tools
To validate the whole transcriptome studies, the isogenic E. coli K-12 library containing all non-lethal deletion mutations (3985 genes) created by the Genome Analysis Project in Japan (Keio collection) (Baba et al., 2006) is invaluable. Each Keio deletion mutant is designed with the ability to eliminate the kanamycin-resistance selection marker by expressing the FLP recombinase protein from pCP20 (Cherepanov and Wackernagel, 1995) since each kanamycin resistance gene is flanked by a FLP recognition target that is excised by FLP recombinase; hence, multiple mutations may be quickly constructed, too, using P1 transduction (Maeda et al., 2008) in a process termed Rapid Gene Knockout. Also available are pCA24N overexpression plasmids which contain His-tagged proteins which facilitate complementation studies as well as easy column chromatography-based methods for protein purification (Kitagawa et al., 2005). Tools for high-throughput genetic screening of two simultaneous gene knockouts are also available for these libraries (Typas et al., 2008). Screening the Keio collection to discern proteins related to biofilm formation identified the importance of 110 genes primarily associated with cell surface structures and cell membrane including genes from flagella, fimbriae, motility, curli, and lipopolysaccharide operons (Niba et al., 2007).
Structure of biofilm regulator AriR
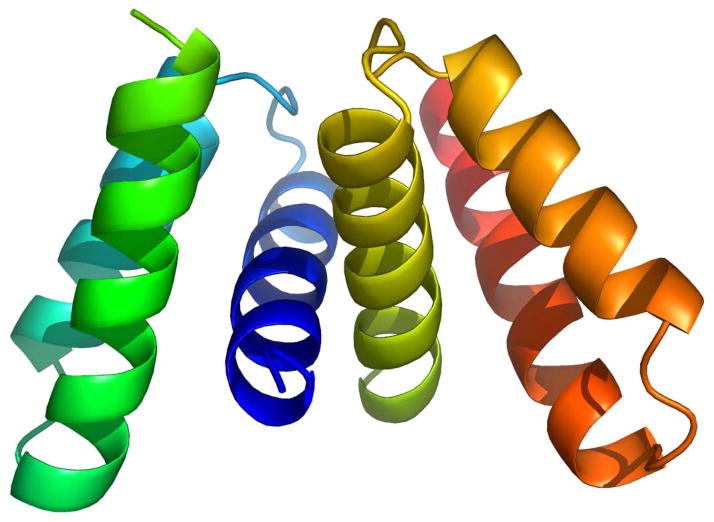
Another development in E. coli biofilms related to stress tolerance is the elucidation of the structure of one of the first biofilm proteins, YmgB, and the discovery that it is part of a fourth acid resistance system in E. coli (YmgB was renamed AriR for regulator of acid resistance influenced by indole) (Lee et al., 2007c) (Fig. 4). Acid resistance is important for E. coli to pass through the low-pH environment of the stomach (pH 2 and below) to colonize the intestinal tract. The E. coli gene cluster ymgABC was identified through whole transcriptome profiling studies related to biofilm development and cell signaling because its expression in many DNA microarrays matched that of well-known acid resistance genes such as gadABC and hdeABD which were differentially regulated like the ymg operon (Lee et al., 2007c). Specifically, the furanosyl borate diester or derivative known as the quorum signal autoinducer 2 (AI-2) repressed ymgAB 3-fold (Ren et al., 2004a); in contrast, the biofilm inhibitor furanone from the algae Delisea pulchra, which masks AI-2 signaling, induced ymgA 2-fold (Ren et al., 2004a). Furthermore, deleting the AI-2 transporter gene tqsA repressed ymgBC 4-fold (Herzberg et al., 2006), ymgABC were induced 14-fold at 15 h relative to 7 h biofilms (Domka et al., 2007), and the stationary-phase biofilm signal indole repressed ymgABC 2- to 5-fold (Lee et al., 2007a). In addition, deleting ymgB represses the acid resistance loci gadABCE and hdeB (Lee et al., 2007c). Therefore, these results suggest strongly that the ymgABC gene cluster, and thus likely the AriR protein itself, plays an important role in E. coli biofilm formation and acid resistance as a result of AI-2 or indole signaling. Corroborating this hypothesis, phenotypic studies showed AriR represses biofilm formation in rich medium containing glucose, decreases cellular motility, and protects the cell from acid confirming that AriR plays a major role in acid-resistance in E. coli (Lee et al., 2007c). The data shows that these phenotypes are potentially mediated through interactions with the important cell signal indole, and gel shift assays suggest that AriR is a non-specific DNA-binding protein. In vivo DNA microarrays also show that AriR binds, either directly or indirectly via a second protein, genes important for biofilm formation. Surprisingly, the structure of the protein (1.8 Å resolution) shows that AriR is a biological dimer that is homologous to the E. coli global regulatory protein Hha (also a non-specific DNA binding protein), despite its low protein sequence identity of only 9%. Note that Hha does not control acid resistance, and whole-transcriptome studies show Hha and AriR control different genes. Hence, AriR influences both acid resistance and biofilm formation and may have other functions, too.
Fig. 4.
Structure of AriR (YmgB) dimer (Lee et al., 2007c). AriR forms a dimer in solution, as determined using size exclusion chromatography and was crystallized as a head-to-head dimer. The final model includes two protein molecules (each containing residues 25–86) and 31 water molecules; the N-terminal 24 amino acids of AriR are spontaneously cleaved. Each subunit of the AriR dimer consists of three α-helices, spanning residues 27–44 (α1), 50–62 (α2) and 67–84 (α3). Helices α2 and α3 are oriented in a near perfect anti-parallel fashion with respect to one another with helix α1 crossing in front of them at nearly a 90° angle. The tertiary structure of the monomer is maintained by an extensive network of hydrophobic interactions consisting almost exclusively of leucine, isoleucine, and valine residues. The peripheral residues of the protein are primarily polar and charged. The dimerization contact is mediated predominantly by residues in helix α1, including Ser31, Leu34, Gly35, Val38, Thr39, Val48, and Met42, and results in the burial of 1326 Å2 of solvent accessible surface. Like AriR, Hha is an all α-helical protein although it has four helices while AriR has three.
Interspecies cell signaling: AHLs
Cell signaling plays a role in the formation of some biofilms (Davies et al., 1998; Stanley and Lazazzera, 2004). In E. coli, acylhomoserine lactones (AHLs) from other bacteria are sensed through SdiA so E. coli can detect signals that it does not synthesize (Michael et al., 2001; van Houdt et al., 2006). For example, SdiA of the close E. coli relative S. enterica is activated by AHLs present in the gastrointestinal tract of turtles (Smith et al., 2008). These exogenous AHLs such as _N_-butyryl-_L_-homoserine lactone (Fig. 3) from P. aeruginosa reduce E. coli biofilm formation (Lee et al., 2007a). In addition, _N_-hexanoyl-_L_-homoserine lactone from strains such as P. syringae increase acid resistance of E. coli by 44% by inducing gadA by 33%; this increase in survival in a harsh (acidic) environment upon detecting other bacteria may give E. coli a competitive advantage (van Houdt et al., 2006).
Intraspecies cell signaling: AI-2
In contrast to AHLs, addition of purified AI-2 increases E. coli biofilm formation (González Barrios et al., 2006). This use of synthesized AI-2 with E. coli was the first direct proof that AI-2 controls biofilm formation as prior studies relied on conditioned medium (which contain substances other than AI-2) and luxS mutations (which affect both signaling and metabolism) to link AI-2 to biofilm formation (Hardie and Heurlier, 2008).
AI-2 is a bacterial species-nonspecific signal used by both Gram-negative and Gram-positive bacteria and synthesized by _S_-ribosylhomocysteine lyase (LuxS) (Schauder et al., 2001). LuxS converts _S_-ribosyl-homocysteine into homocysteine and (S)-4,5-dihydroxy-2,3-pentanedione (DPD, Fig. 3), which forms spontaneously into a family of AI-2 molecules (Hardie and Heurlier, 2008). As a bacterial communication signal, AI-2 appears to be exported by the transporter of quorum sensing signal TqsA (Herzberg et al., 2006) (formerly YdgG, this protein was identified initially by biofilm transcriptome profiling (Ren et al., 2004b)). AI-2 is internalized by a lsr operon-encoded system (Taga et al., 2003), and then controls a variety of genes (DeLisa et al., 2001; Sperandio et al., 2001; Ren et al., 2004a). The lsr operon of seven genes lsrACDBFGE is induced by phospho-AI-2 and regulated by LsrR, LsrK, and GlpDK (Taga et al., 2001; Taga et al., 2003; Xavier and Bassler, 2005a). The regulator LsrR represses the AI-2 uptake operon lsr, which is derepressed by the binding of phospho-AI-2 to LsrR (Taga et al., 2003). Another regulator, LsrK, a cytoplasmic kinase, phosphorylates internal AI-2 into an activated molecule (Xavier and Bassler, 2005a). Note that both LsrK and LsrR control also regulate other genes including sRNAs (Li et al., 2007). The glycerol uptake and metabolism system encoded by glpDFK genes also influences AI-2 signaling by regulating lsr transcription through LsrR (Xavier and Bassler, 2005a).
Some insights have been gained as to how AI-2 controls biofilm formation. In E. coli, AI-2 stimulates biofilm formation and changes its architecture by stimulating flagellar motility via the motility quorum-sensing regulator MqsR (formerly B3022 that was identified through biofilm transcriptome profiling (Ren et al., 2004b)) which acts through the two-component motility regulatory system QseBC (González Barrios et al., 2006) to transcriptionally regulates FlhDC, the master regulator of flagella and motility genes fliLMNOPQR, fliAZ, flhBA, and flgABCDMN (Liu and Matsumura, 1994; Claret and Hughes, 2002; Clarke and Sperandio, 2005b). MqsR also induces expression of the transcription factor YncC (González Barrios et al., 2006); YncC inhibits the expression of periplasmic YbiM, which prevents overproduction of colanic acid (causing mucoidy) and prevents YbiM from inhibiting biofilm formation (Zhang et al., 2008). Colanic acid synthesis is induced in mature biofilms (Domka et al., 2007) and is important for the three dimensional architecture of a biofilm but not for biofilm formation (Danese et al., 2000; Prigent-Combaret et al., 2000). YncC was renamed McbR for MqsR-controlled colanic acid and biofilm regulator, and YbiM was renamed McbA since it is the first gene regulated by McbR (Zhang et al., 2008).
These results are consistent with the recent finding that in the oral bacterium Aggregatibacter actinomycetemcomitans, AI-2 regulates its biofilm formation most likely through its QseBC system (Shao et al., 2007). Also, in the human gastric pathogen Helicobacter pylori, AI-2 controls motility by controlling genes upstream of the motility and flagellar regulator FlhA (Rader et al., 2007). Further proof that AI-2 controls motility in different genera is that AI-2 regulates transcription of the flagellin gene flaA in the human pathogen Campylobacter jejuni (Jeon et al., 2003). In addition, BssR (formerly YliH)/BssS (formerly YceP) (both identified through whole transcriptome profiling (Ren et al., 2004b)) regulate E. coli biofilms by influencing AI-2 and indole concentrations in a divergent manner (Domka et al., 2006).
AI-2 has also been shown to influence enterohemorrhagic E. coli (EHEC) (Bansal et al., 2008) which is not surprising since the gastrointestinal tract is colonized by hundreds of bacterial species (Collier-Hyams and Neish, 2005) that produce a diverse range of signals including AI-2 (Clarke and Sperandio, 2005a). Understanding EHEC infections is important given that there are over 73,000 EHEC infections annually in the U.S. which lead to 2,000 hospitalizations and 60 deaths, the economic cost of which is $405 million (Frenzen et al., 2005). EHEC forms a biofilm on various surfaces (Ryu and Beuchat, 2005; Uhlich et al., 2006), and sloughing of the biofilm may cause contamination (Ryu and Beuchat, 2005); however, an effective means of preventing its biofilm formation has not been elucidated, and there is no effective treatment for EHEC infections since antibiotic treatment increases the risk of hemolytic uremic syndrome and renal failure (Wong et al., 2000; Tarr et al., 2005). It was found that AI-2 attracts EHEC in agarose plug chemotaxis assays, increases swimming motility, and increases EHEC attachment to HeLa cells (Bansal et al., 2008). Whole transcriptome profiling shows exposure to AI-2 alters the expression of 23 locus of enterocyte effacement (LEE) genes directly involved in the production of virulence determinants, as well as other genes associated with virulence (e.g., 46 flagellar/fimbrial genes, 24 iron-related genes), in a temporally-defined manner (Bansal et al., 2008). Another recent study (Kendall et al., 2007) using higher glucose concentrations that may have masked the effects of AI-2 (Wang et al., 2005) observed espA (LEE4) and eae (LEE5) are altered upon exposing EHEC to AI-2, and a proteome analysis showed AI-2 increases EHEC virulence using both epithelial cells and nematodes (Kim et al., 2007). These results suggest that AI-2 is an important signal in EHEC infections of the human GI tract.
Interspecies cell signaling: AI-2
Note that AI-2 signaling also occurs between bacterial species. For example, E. coli senses AI-2 that is produced by Vibrio harveyi to assess changes in its cell population (Xavier and Bassler, 2005b). In addition, P. aeruginosa responds to AI-2 and modulates its gene expression pattern including pathogenicity, although it does not itself produce AI-2 (Duan et al., 2003). AI-2 also regulates at extraordinarily low concentrations the dual-species biofilm formation of two Gram-positive human oral commensal bacterial strains, Actinomyces naeslundii T14V and Streptococcus oralis 34 (Rickard et al., 2006).
Intraspecies cell signaling: indole
E. coli produces indole by tryptophanase (TnaA) that can reversibly convert tryptophan into indole, pyruvate, and ammonia (Newton and Snell, 1965). Indole is a signal (Wang et al., 2001b; Di Martino et al., 2003) that inhibits E. coli biofilms (Lee et al., 2007a) and works in a quorum sensing fashion (Lee et al., 2007b) since it satisfies the four criteria for cell signals (Winzer et al., 2002): (i) the putative signal must be produced during a specific stage (indole is produced primarily in the stationary-phase (Wang et al., 2001b)), (ii) the putative signal must accumulate extracellularly and be recognized by a specific receptor (indole is a known extracellular signal (Wang et al., 2001a; Hirakawa et al., 2005) that is exported by AcrEF (Kawamura-Sato et al., 1999) and is imported by Mtr (Yanofsky et al., 1991)), (iii) the putative signal must accumulate and generate a concerted response (indole has been shown to delay cell division (Chant and Summers, 2007)), and (iv) the putative signal must elicit a response that extends beyond the physiological changes required to metabolize or detoxify the signal (indole has been shown to control biofilms (Lee et al., 2007a) and cell division (Chant and Summers, 2007) which are not related to indole metabolism).
It was first reported that indole stimulates biofilm formation for E. coli S17-1 (Di Martino et al., 2003). However, indole was subsequently shown to decrease significantly the biofilm formation of nine non-pathogenic E. coli strains (Domka et al., 2006; Lee et al., 2007a; Zhang et al., 2007) as well as decrease the biofilm formation of pathogenic E. coli O157:H7 (Lee et al., 2007b). The AHL-binding protein SdiA is necessary for this biofilm response with indole (Lee et al., 2007a; Lee et al., 2008b). Indole decreases E. coli biofilms by reducing motility (Domka et al., 2006; Bansal et al., 2007; Lee et al., 2007a), repressing acid resistance genes (Lee et al., 2007a), reducing chemotaxis (Bansal et al., 2007), and reducing attachment to epithelial cells (Bansal et al., 2007) (note this is opposite that of AI-2). Indole also controls plasmid stability (Chant and Summers, 2007). Whereas AHL increases acid resistance 1.4-fold (van Houdt et al., 2006), indole reduces acid resistance 500-fold (Lee et al., 2007a). Also, indole induces the expression of multidrug exporter genes and increases drug resistance (Hirakawa et al., 2005), and tryptophanase activity has been linked to the killing of nematodes by pathogenic E. coli (Anyanful et al., 2005).
Promiscuous cell signaling: indole
Indole is also a promiscuous signal since it alters the phenotypes of non-E. coli strains. For example, indole increases biofilm formation of P. aeruginosa and P. fluorescens even though these pseudomonads do not produce indole (Lee et al., 2007a). Furthermore, in P. aeruginosa, indole alters extensively gene expression in a manner opposite that of AHLs by repressing genes that encode for the mexGHI_-opmD multidrug efflux pump and by repressing genes involved in the synthesis of QS-regulated virulence factors including pyocyanin (phz operon), 2-heptyl-3-hydroxy-4(1_H)-quinolone (PQS) signal (pqs operon), pyochelin (pch operon), and pyoverdine (pvd operon) (Lee et al., 2008a). Corroborating these whole transcriptome results, indole decreases production of pyocyanin, rhamnolipid, PQS, and pyoverdine and enhances antibiotic resistance (Lee et al., 2008a). Further evidence that indole is a signal that affects bacteria that do not synthesize it is shown in co-cultures of E. coli with P. fluorescens cells engineered to remove indole by oxidizing it; removal of indole results in a 12-fold increase in the number of E. coli cells (Lee et al., 2007a); this engineered dual species biofilm represents the first synthetic gene circuit successfully used to control biofilm formation. Therefore, it appears that the mechanism by which procaryotes manipulate the biofilm signal indole is through the relaxed substrate range of many dioxygenases and monooxygenases found in bacteria that bring about indole hydroxylation (Rui et al., 2005); i.e., some of the oxygenases bacteria use for catabolism (Fishman et al., 2005) have also evolved to regulate concentrations of the cell signal indole by removing it via precipitation: competitors that wish to remove indole simply oxidize it in one step to indigo which is insoluble and hence leaves the system. Furthermore, E. coli may use indole to reduce the virulence of strains such as P. aeruginosa.
Temperature-specific signals
Recently it was found temperature affects indole and AI-2 signaling in E. coli which suggests E. coli may use primarily indole signaling outside the human host and AI-2 signaling inside the host (Lee et al., 2008b). It was found that indole addition results in more extensive differential gene expression at 30°C than at 37°C and that indole reduces biofilm formation (without affecting growth) more significantly at 25°C and 30°C than at 37°C. In contrast to indole, the addition of the AI-2 precursor DPD leads to more extensive differential gene expression at 37°C than at 30°C (Lee et al., 2008b). Furthermore, compared to 37°C, at 30°C indole more significantly decreased flagella-related promoter activity, enhanced antibiotic resistance, and inhibited cell division. Also, the addition of AI-2 induces the transcription of virulence genes in enterohemorrhagic E. coli O157:H7 at 37°C but not at 30°C (Lee et al., 2008b).
Uridine monophosphate and indole and AI-2
By using a whole transcriptome approach, it was also found that indole decreases uridine monophosphate (UMP) biosynthesis (carAB, pyrLBI, pyrC, pyrD, pyrF, and upp) and uracil transport (uraA) at 30°C in E. coli (but not at 37°C) whereas AI-2 induces UMP biosynthesis at 37°C (but not at 30°C) (Lee et al., 2008b). Also, like indole, SdiA represses these same set of genes at 30°C. Additional experiments with P. aeruginosa have demonstrated that uracil addition increases quorum-sensing phenotypes and increases virulence in this strain via UMP biosynthesis (Ueda et al., 2008). These results suggest that in E. coli, a building block of RNA, uracil or a derivative of uracil, may report the status of the bacterial signals, AHLs, AI-2, and indole.
Interkingdom signals
Cell signaling is also promiscuous across kingdoms. Examples were given above for interactions with various signals between different species of bacteria that affect biofilm formation; however, interactions also occur between pathogenic E. coli and its host that affect biofilm formation. As shown by a novel two fluorophore chemotactic assay, EHEC is attracted to the human hormones epinephrine and norepinephrine (as with AI-2) (Bansal et al., 2007). In addition, epinephrine and norepinephrine increase EHEC biofilm formation as well as increase motility and attachment to epithelial cells (Bansal et al., 2007). Norepinephrine also increases adhesion of EHEC to cecal mucosa (Chen et al., 2003), colonic mucosa (Green et al., 2004), and ileum (Vlisidou et al., 2004). Corroborating these results, whole transcriptome profiling of EHEC in biofilms indicates that epinephrine and norepinephrine induce the expression of genes involved in surface colonization and virulence while indole decreases their expression (Bansal et al., 2007). Epinephrine and norepinephrine have also been shown to directly induce virulence genes (e.g., LEE genes) in EHEC (Sperandio et al., 2003) through receptor QseC. Taken together, these results suggest that epinephrine and norepinephrine increase EHEC infection while indole attenuates the process.
Biofilm inhibitors related to cell signaling
Advances in deciphering E. coli cell signaling have led to discoveries for inhibiting biofilms. For example, the recognition that indole inhibits biofilm formation (Lee et al., 2007a) as a quorum-sensing cell signal (Lee et al., 2007b) led to an investigation of the impact of hydroxy indoles on biofilm formation (Lee et al., 2007b). Given that indole controls biofilms (Lee et al., 2007a) and is present up to 700 μM (Domka et al., 2006), it was hypothesized (Lee et al., 2007b) that hydroxylated indoles may play a role in biofilm formation since many bacterial oxygenases such as dioxygenases from P. putida PpG7 (Ensley et al., 1983), Ralstonia pickettii PKO1 (Fishman et al., 2005), P. mendocina KR1 (Tao et al., 2004), and Burkholderia cepacia G4 (Rui et al., 2005) readily convert indole to oxidized compounds such as 2-hydroxyindole, 3-hydroxyindole, 4-hydroxyindole, isatin, indigo, isoindigo, and indirubin (Rui et al., 2005). In Luria-Bertani medium (LB) on polystyrene with quiescent conditions, 7-hydroxyindole (Fig. 3) decreased EHEC biofilm formation 27-fold and decreased K-12 biofilm formation eight-fold without affecting the growth of planktonic cells (Lee et al., 2007b). 5-Hydroxyindole also decreased biofilm formation 11-fold for EHEC and 6-fold for K-12. In contrast, isatin (indole-2,3-dione, Fig. 3) increased biofilm formation four-fold for EHEC while it had no effect on K-12. Whole transcriptome analysis revealed that isatin represses indole synthesis by repressing tnaABC 7- to 37-fold in EHEC, and extracellular indole levels were found to be 20-fold lower (Lee et al., 2007b). Furthermore, isatin repressed the AI-2 transporters lsrABCDFGKR, while significantly inducing the flagellar genes flgABCDEFGHIJK and fliAEFGILMNOPQ (which led to a 50% increase in motility). 7-Hydroxyindole induces the biofilm inhibitor/stress regulator bshA and represses cysADIJPU/fliC (which led to a 50% reduction in motility) and purBCDEFHKLMNRT. Isogenic mutants showed 7-hydroxyindole inhibits E. coli biofilm through cysteine metabolism. 7-Hydroxyindole (500 μM) also stimulates P. aeruginosa PAO1 biofilm formation two-fold; therefore, hydroxyindoles are interspecies bacterial signals, and 7-hydroxyindole is a potent EHEC biofilm inhibitor.
Similarly, the realization that uracil or a derivative is intertwined with the AI-2 and indole cell signaling pathways (Lee et al., 2008b) led to the discovery that the uracil analog, 5-fluorouracil inhibits E. coli biofilms; for example, 10 μM 5-fluorouracil inhibits biofilm formation five-fold while decreasing growth by 10% (author’s unpublished data). 5-Fluorouracil also affects P. aeruginosa PA14 by decreasing dramatically its quorum-sensing phenotypes, reducing biofilm formation, and reducing virulence (Ueda et al., 2008). Notably, 5-fluoruracil is already approved for treatment of human colon cancer (Wiebke et al., 2003); so it is relatively non-toxic to humans.
Given that AI-2 directly increases E. coli biofilm formation (González Barrios et al., 2006), compounds that mask AI-2 signaling should decrease biofilm formation. Indeed, (5Z)-4-Bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone (furanone, Fig. 3) of the alga Delisea pulchra inhibits E. coli biofilm formation by blocking AI-2 signaling (Ren et al., 2001) and the same genes induced by AI-2 are repressed by this furanone (Ren et al., 2004a). This organism makes more than twenty halogenated furanones (de Nys et al., 1993) to prevent biofilm formation so that it may conduct photosynthesis, and its effects with non-E. coli strains are well-studied (Rasmussen and Givskov, 2006). The mechanism by which furanones inhibit biofilm formation is by displacing AHL from LuxR (Manefield et al., 1999) and by decreasing the DNA-binding activity of LuxR which blocks all three quorum-sensing systems of V. harveyi (HAI-1 acylated homoserine lactone, AI-2, and CAI-1) (Defoirdt et al., 2007). By interfering with quorum sensing signaling and biofilm formation, these compounds have been shown to protect shrimp grown in aquaculture (Defoirdt et al., 2006) and to protect mice (Hentzer et al., 2003).
Like brominated furanones from algae, food ingredients have been found to inhibit the biofilm formation of E. coli. For example, the ground-beef fatty acids palmitic acid (Fig. 3), stearic acid, oleic acid, and linoleic acid inhibit AI-2 activity and decrease E. coli biofilm formation (Soni et al., 2008). Similarly, furocoumarins such as bergamottin from grapefruit inhibit both AHL and AI-2 activities as well as decrease biofilm formation by EHEC (Girennavar et al., 2008).
Note the shortage of structural information about the family of compounds that comprise AI-2 has slowed the development of AI-2-based, QS inhibitors (Lowery et al., 2008). However, propyl-DPD and butyl-DPD have been shown to inhibit DPD induction of lsr for S. typhimurium while the same compounds stimulate bioluminescence in V. harveyi (Lowery et al., 2008). In addition, two sulfone-based compounds were identified via virtual structure-based screening that inhibit bioluminescence in V. harveyi (Li et al., 2008).
Biofilm inhibitors not related to cell signaling
Screening 13,000 samples of compounds purified from plants resulted in the identification of another biofilm inhibitor, ursolic acid (Fig. 3) that is not toxic to E. coli and other bacteria as well as to hepatocytes (Ren et al., 2005). Whole transcriptome analysis showed ursolic acid induced genes related to chemotaxis and motility (cheA, tap, tar, and motAB), heat shock response (hslSTV and mopAB), and unknown functions (e.g., b1566 and yrfHI) and that ursolic acid repressed genes related to cysteine synthesis (cysK) and sulfur metabolism (cysD); however, the anti-biofilm effect of ursolic acid was not related to AI-2 (Ren et al., 2005). This manuscript and one by Rather and co-workers showed sulfur metabolism is related to biofilm formation as mutations in both cysB (Ren et al., 2005) and cysE (Sturgill et al., 2004) increase biofilm formation.
A related compound, asiatic acid (Fig. 3), was found to be even more effective than ursolic acid and whole transcriptome studies showed it also involves sulfur metabolism. Furthermore, aqueous fish muscle extract, composed primarily of fish muscle α-tropomyosin, was shown recently to significantly decrease attachment of a range of E. coli that cause urinary tract infections (Vejborg and Klemm, 2008).
Toxin/anti-toxin pairs
Toxin-antitoxin (T-AT) pairs consist of a stable toxin and a labile antitoxin. T-AT pairs appear to be involved in anti-phage defense (Pecota and Wood, 1996) and other possible functions include “genomic junk”, growth-rate control, programmed-cell death, and persister formation (Magnuson, 2007). The value of T-AT pairs to the cell has been questioned since after deleting five TAT pairs, it was shown the five best-studied E. coli T-AT pairs do not influence bacterial fitness of planktonic cells (Tsilibaris et al., 2007). However, MqsR is highly toxic since a deletion of the anti-toxin B3021 is lethal, and it appears MqsR is part of a T-AT pair that consists of MqsR (toxin) and B3021 (Shah et al., 2006). Since MqsR has been linked to biofilm formation via AI-2 (González Barrios et al., 2006) and via McbR (Zhang et al., 2008), T-AT pairs are clearly important for biofilm formation. In addition, deletion of the five best-studied E. coli T-AT pairs also alters biofilm formation (author’s unpublished results); this illustrates importance of studying biofilm cells along with planktonic cells.
Further evidence of this link between T-AT pairs and biofilm formation is provided by Hha and YbaJ (renamed TomB for toxin overexpression modulator in biofilms) (García-Contreras et al., 2008). Both Hha and TomB are highly induced in biofilms as found by whole-transcriptome profiling (Ren et al., 2004b), and Hha expression is toxic and TomB diminishes its toxicity (García-Contreras et al., 2008). Hha decreases biofilm formation by repressing type I fimbriae via fimA and ihfA and by inhibiting their translation via rare tRNAs (García-Contreras et al., 2008). Hha expression also induces ClpP and ClpX proteases that degrade many antitoxins, allowing free toxins to exert their inhibitory effects (García-Contreras et al., 2008). Note that decreases in translation efficiency activate toxins (Christensen et al., 2004). Hha also activates the prophage genes rzpD, yfjZ, alpA, and appY which actively lyse cells (García-Contreras et al., 2008). Hence, Hha is toxic indirectly by activating other toxins by changing translation efficiency (García-Contreras et al., 2008). Therefore, one of the most important roles of the nebulous T-AT pairs is to help control biofilm formation.
Small RNA and biofilm dispersal
Biofilm dispersal is important for disseminating the strain; however, for the bacterium to leave the solid matrix in which it is both protected and entrapped, it may be necessary to sacrifice part of the biofilm and have some cells undergo autolysis (Webb et al., 2003). Hence, programmed cell death may make sense for the biofilm and the primitive tissue that this collection of cells represents but not for planktonic cells (Webb et al., 2003). Biofilm dispersal for P. aeruginosa involves prophage (Webb et al., 2003) and in Pseudoalteromonas tunicata involves the autolytic protein AlpA (Mai-Prochnow et al., 2006). In E. coli, along with cell toxicity and biofilm formation, Hha appears to control biofilm dispersal. Initial evidence is that Hha leads to decreased biofilm in flow cells and to the formation of plaques (García-Contreras et al., 2008); cell lysis via Hha may aid biofilm dispersal by forming holes in the biofilm matrix.
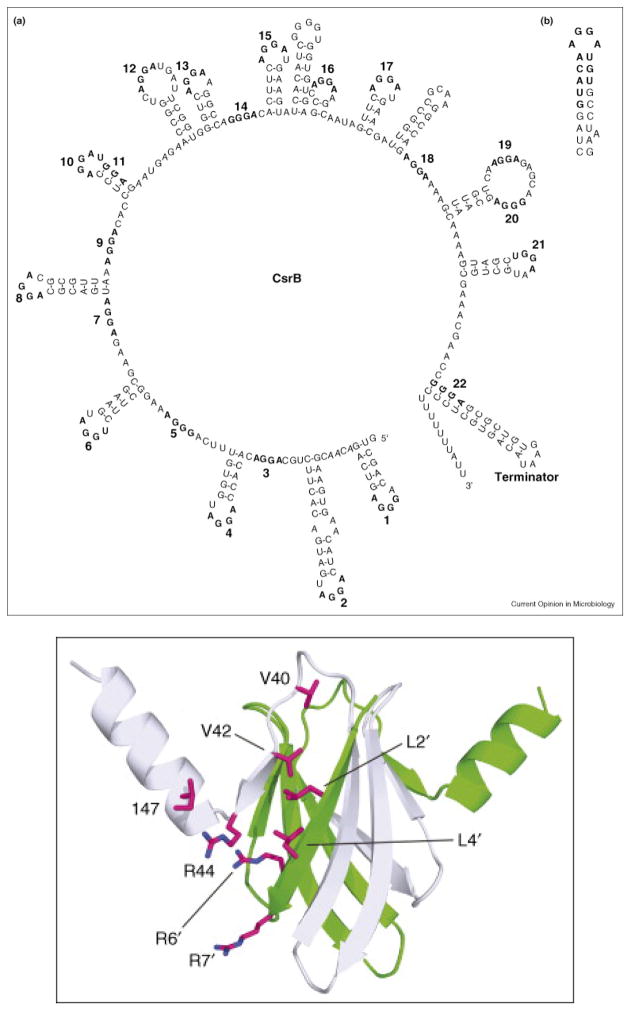
Some of the more than 60 sRNAs identified in E. coli (Kulesus et al., 2008) are related to biofilm dispersal. One of the first sRNA systems discovered in which sRNAs bind a regulator protein to control its activity is the carbon storage regulation system that consists of CsrA, the transcription regulator protein that binds specific mRNA to repress or activate transcription (Fig. 5), and the CsrA-binding sRNAs CsrB and CsrC that serve to titrate CsrA by binding nine CsrA dimers (Babitzke and Romeo, 2007). CsrA represses gluconeogenesis, glycogen metabolism, peptide transport, and production of the adhesion poly-β-1,6-_N-_acetyl-D-glucosamine while it activates glycolysis, acetate metabolism, and flagellum biosynthesis (Baker et al., 2007). CsrA also represses biofilm formation and increases biofilm dispersal (Jackson et al., 2002). In addition, this remarkable protein represses the global regulator Hfq that acts as a RNA chaperone by promoting sRNA-mRNA base pairing in E. coli (Baker et al., 2007). The Csr system is widespread in eubacteria and is also known as the Rsm (repressor of secondary metabolites) system (Babitzke and Romeo, 2007).
Fig. 5.
CsrA regulatory protein and sRNA CsrB which binds CsrA (Babitzke and Romeo, 2007). Secondary structure of sRNA CsrB showing 22 GGA regions for binding CsrA proteins (a). CsrA consensus binding sequence (b). Structure of CsrA dimer with possible CsrB binding residues shown.
CONCLUDING REMARKS
Whole transcriptome profiling has elucidated much in regard to E. coli biofilm formation. Differential gene expression between biofilm cells vs. planktonic cells initially identified important biofilm proteins (e.g., Hha as a toxin and regulator of translation efficiency, TomB as an anti-toxin, BhsA as a membrane mediator of stress, MqsR as an AI-2 mediator, BssR/BssS as signal-controlled biofilm regulators, and TqsA as the AI-2 exporter). Follow up approaches such as transcriptome profiling using isogenic mutants and in vivo DNA binding studies led to discoveries related to how these proteins affect biofilm formation (e.g., McbR/McbA as regulators of colanic acid, AriR as a new acid-resistance protein, the importance of rare tRNAs, and the importance toxin-anti toxin pairs).
E. coli as a reference system has also been important for discerning the role of small RNAs on biofilm formation (e.g., CsrB), and for discerning the role of toxins and anti-toxins for biofilm formation (e.g., Hha and MqsR). It is interesting that toxin/anti-toxin genes such as hok/sok that were postulated to protect E. coli cells from phage (Pecota and Wood, 1996) are now being related to biofilm dispersal and cell lysis (e.g., hha/tomB) (García-Contreras et al., 2008). It seems the cell is capable of taking the weapons of its enemy (phage) and using it to control its physiology (cell death) in a social manner (altruism).
E. coli has also been instrumental in discerning the role of both procaryotic and eucaryotic signals on biofilm formation. It is with this strain that the role of AI-2 on biofilm formation was clearly shown (by direct addition of enzymatically-synthesized compound), and EHEC has been a good model system for discerning the importance of interspecies and interkingdom signaling. To date, little research has been performed on the effect of plant signals on EHEC biofilm formation and this is important in regard to its pathogenicity.
Much research has been aimed at finding effective ways for the prevention, control, or eradication of biofilms (Labbate et al., 2004), and advances have also been made in E. coli biofilm inhibition and in anti-virulence measures. To date, there are few known anti-virulence compounds (Cegelski et al., 2008); anti-virulence compounds are an important way to fight infectious diseases because unlike antimicrobials, anti-virulence compounds do not affect growth and so there is less chance of developing resistance (Hentzer et al., 2002). Here, we have shown that several non-toxic anti-biofilm (anti-virulence) compounds exist for E. coli including brominated furanones (Ren et al., 2001), ursolic acid (Ren et al., 2005), indole derivatives (Lee et al., 2007b), and 5-fluorouracil. It is expected that there will be much activity in this area to find ever more potent compounds and that mixtures of these compounds will be required for efficacy in inhibiting biofilms.
Acknowledgments
This manuscript is written in loving memory of my father, Chester A. Wood, Jr. Funding for this review was supported by the NIH (5RO1EB003872-05) and ARO (W911NF-06-1-0408). I am grateful for the assistance with the figures provided by Dr. Jintae Lee and Qun Ma, and for the helpful comments provided by Dr. Jintae Lee, Dr. Xuesong Zhang, Dr. Rodolfo García-Contreras, Dr. Xiaoxue Wang, Tarun Bansal, Manjunath Hegde, Can Attila, and Qun Ma.
References
- An D, Parsek MR. The promise and peril of transcriptional profiling in biofilm communities. Current Opin Microbiol. 2007;10:292–296. doi: 10.1016/j.mib.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- Anyanful A, Dolan-Livengood JM, Lewis T, Sheth S, DeZalia MN, Sherman MA, et al. Paralysis and killing of Caenorhabditis elegans enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol Microbiol. 2005;57:988–1007. doi: 10.1111/j.1365-2958.2005.04739.x. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Current Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA Inhibits Translation Initiation of Escherichia coli hfq by Binding to a Single Site Overlapping the Shine-Dalgarno Sequence. J Bacteriol. 2007;189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T, Jesudhasan P, Pillai S, Wood T, Jayaraman A. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol. 2008;78:811–819. doi: 10.1007/s00253-008-1359-8. [DOI] [PubMed] [Google Scholar]
- Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A. Differential Effects of Epinephrine, Norepinephrine, and Indole on Escherichia coli O157:H7 Chemotaxis, Colonization, and Gene Expression. Infect Immun. 2007;75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008 doi: 10.1111/j.1462-2920.2008.01658.x. on-line. [DOI] [PubMed] [Google Scholar]
- Beloin C, Roux A, Ghigo J-M. In: Escherichia coli Biofilms. In Bacterial Biofilms. Romeo T, editor. Berlin: Springer-Verlag; 2008. pp. 249–289. [Google Scholar]
- Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, et al. Global Impact of Mature Biofilm Lifestyle on Escherichia coli K-12 Gene Expression. Mol Microbiol. 2004;51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant EL, Summers DK. Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol Microbiol. 2007;63:35–43. doi: 10.1111/j.1365-2958.2006.05481.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Brown DR, Xie Y, Green BT, Lyte M. Catecholamines Modulate Escherichia coli O157:H7 Adherence to Murine Cecal Mucosa. Shock. 2003;20:183–188. doi: 10.1097/01.shk.0000073867.66587.e0. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- Claret L, Hughes C. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J Mol Biol. 2002;321:185–199. doi: 10.1016/s0022-2836(02)00600-9. [DOI] [PubMed] [Google Scholar]
- Clarke MB, Sperandio V. Events at the Host-Microbial Interface of the Gastrointestinal Tract III. Cell-to-cell signaling among microbial flora, host, and pathogens: there is a whole lot of talking going on. Am J Physiol Gastrointest Liver Physiol. 2005a;288:G1105–1109. doi: 10.1152/ajpgi.00572.2004. [DOI] [PubMed] [Google Scholar]
- Clarke MB, Sperandio V. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2005b;57:1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- Collier-Hyams LS, Neish AS. Innate immune relationship between commensal flora and the mammalian intestine. Cell Mol Life Sci. 2005;62:1339–1348. doi: 10.1007/s00018-005-5038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell H, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Kolter R. Exopolysaccharide Production is Required for Development of Escherichia coli K-12 Biofilm Architecture. Journal Bacteriology. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- de Nys R, Wright AD, Konig GM, Sticher O. New halogenated furanones from the marine alga Delisea pulchra (cf fimbriata) Tetrahedron. 1993;49:11213–11220. [Google Scholar]
- Defoirdt T, Crab R, Wood TK, Sorgeloos P, Verstraete W, Bossier P. Quorum Sensing-Disrupting Brominated Furanones Protect the Gnotobiotic Brine Shrimp Artemia franciscana from Pathogenic Vibrio harveyi, Vibrio campbellii, and Vibrio parahaemolyticus Isolates. Appl Environ Microbiol. 2006;72:6419–6423. doi: 10.1128/AEM.00753-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt T, Miyamoto CM, Wood TK, Meighen EA, Sorgeloos P, Verstraete W, Bossier P. Brominated furanones disrupt quorum sensing-regulated gene expression in Vibrio harveyi by decreasing the concentration of the response regulator protein LuxR able to bind its target promoters. Environ Microbiol. 2007;9:2486–2495. doi: 10.1111/j.1462-2920.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- DeLisa MP, Wu C-f, Wang L, Valdes JJ, Bentley WE. DNA Microarray-Based Identification of Genes Controlled by Autoinducer 2 - Stimulated Quorum Sensing in Escherichia coli. J Bacteriol. 2001;183:5239–5247. doi: 10.1128/JB.183.18.5239-5247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino P, Fursy R, Bret L, Sundararaju B, Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol. 2003;49:443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- Domka J, Lee J, Wood TK. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol. 2006;72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T, Wood TK. Temporal Gene-Expression in Escherichia coli K-12 Biofilms. Environ Microbiol. 2007;9:322–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- Ensley BD, Ratzkin BJ, Osslund TD, Simon MJ, Wackett LP, Gibson DT. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983;222:167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- Fishman A, Tao Y, Rui L, Wood TK. Controlling the regiospecific oxidation of aromatics via active site engineering of toluene para-monooxygenase of Ralstonia pickettii PKO1. J Biol Chem. 2005;280:506–514. doi: 10.1074/jbc.M410320200. [DOI] [PubMed] [Google Scholar]
- Frenzen PD, Drake A, Angulo FJ. Economic cost of illness due to Escherichia coli O157 infections in the United States. J Food Prot. 2005;68:2623–2630. doi: 10.4315/0362-028x-68.12.2623. [DOI] [PubMed] [Google Scholar]
- García-Contreras R, Zhang XS, Kim Y, Wood TK. Protein Translation and Cell Death: The Role of Rare tRNAs in Biofilm Formation and in Activating Dormant Phage Killer Genes. PLoS ONE. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo JM. Natural Conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- Girennavar B, Cepeda ML, Soni KA, Vikram A, Jesudhasan P, Jayaprakasha GK, et al. Grapefruit juice and its furocoumarins inhibits autoinducer signaling and biofilm formation in bacteria. Int J Food Microbiol. 2008;125:204–208. doi: 10.1016/j.ijfoodmicro.2008.03.028. [DOI] [PubMed] [Google Scholar]
- González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 Controls Biofilm Formation in Escherichia coli Through a Novel Motility Quorum Sensing Regulator (MqsR, B3022) J Bacteriol. 2006;188:305–306. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BT, Lyte M, Chen C, Xie Y, Casey MA, Kulkarni-Narla A, et al. Adrenergic modulation of Escherichia coli O157:H7 adherence to the colonic mucosa. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1238–1246. doi: 10.1152/ajpgi.00471.2003. [DOI] [PubMed] [Google Scholar]
- Hancock V, Klemm P. Global Gene Expression Profiling of Asymptomatic Bacteriuria Escherichia coli during Biofilm Growth in Human Urine. Infect Immun. 2007;75:966–976. doi: 10.1128/IAI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie KR, Heurlier K. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat Rev Micro. 2008;6:635–643. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Wu H, Anderson JB, Riedel K, Rasmussen TB, Bagge N, et al. Attenuation of Pseudomonas aeruginosa Biofilm Virulence by Quorum Sensing Inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg M, Kaye IK, Peti W, Wood TK. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol. 2005;55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm Formation and Dispersal Under the Influence of the Global Regulator CsrA of Escherichia coli. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman A, Mansfeld FB, Wood TK. Inhibiting Sulfate-Reducing Bacteria in Biofilms by Expressing the Antimicrobial Peptides Indolicidin and Bactenecin. J Indust Microbiol Biotechnol. 1999;22:167–175. doi: 10.1007/s002530051520. [DOI] [PubMed] [Google Scholar]
- Jeon B, Itoh K, Misawa N, Ryu S. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol Immunol. 2003;47:833–839. doi: 10.1111/j.1348-0421.2003.tb03449.x. [DOI] [PubMed] [Google Scholar]
- Junker LM, Peters JE, Hay AG. Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology. 2006;152:2233–2245. doi: 10.1099/mic.0.28767-0. [DOI] [PubMed] [Google Scholar]
- Kawamura-Sato K, Shibayama K, Horii T, Iimuma Y, Arakawa Y, Ohta M. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol Lett. 1999;179:345–352. doi: 10.1111/j.1574-6968.1999.tb08748.x. [DOI] [PubMed] [Google Scholar]
- Kendall MM, Rasko DA, Sperandio V. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect Immun. 2007;75:4875–4884. doi: 10.1128/IAI.00550-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Oh S, Ahn EY, Imm JY, Oh S, Park S, Kim SH. Proteome Analysis of Virulence Factor Regulated by Autoinducer-2–like Activity in Escherichia coli O157:H7. Int J Food Protection. 2007;70:300–307. doi: 10.4315/0362-028x-70.2.300. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. Impact of the RNA Chaperone Hfq on the Fitness and Virulence Potential of Uropathogenic Escherichia coli. Infect Immun. 2008;76:3019–3026. doi: 10.1128/IAI.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbate M, Queck SY, Koh KS, Rice SA, Givskov M, Kjelleberg S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J Bacteriol. 2004;186:692–698. doi: 10.1128/JB.186.3.692-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jayaraman A, Wood TK. Indole Is an Inter-Species Biofilm Signal Mediated by SdiA. BMC Microbiol. 2007a;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol. 2007b;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Attila C, Cirillo SL, Cirillo JD, Wood TK. Indole and 7-hydoxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol. 2008a doi: 10.1111/j.1751-7915.2008.00061.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Zhang X-S, Hegde M, Bentley WE, Jayaraman A, Wood TK. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008b doi: 10.1038/ismej.2008.54. on-line. [DOI] [PubMed] [Google Scholar]
- Lee J, Page R, Garcia-Contreras R, Palermino JM, Zhang XS, Doshi O, et al. Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid Resistance. J Mol Biol. 2007c;373:11–26. doi: 10.1016/j.jmb.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Attila C, Wang L, Wood TK, Valdes JJ, Bentley WE. Quorum Sensing in Escherichia coli Is Signaled by AI-2/LsrR: Effects on Small RNA and Biofilm Architecture. J Bacteriol. 2007;189:6011–6020. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ni N, Chou HT, Lu CD, Tai Phang C, Wang B. Structure-Based Discovery and Experimental Verification of Novel AI-2 Quorum Sensing Inhibitors against Vibrio harveyi. ChemMedChem. 2008;3:1242–1249. doi: 10.1002/cmdc.200800076. [DOI] [PubMed] [Google Scholar]
- Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery CA, Park J, Kaufmann GF, Janda KD. An Unexpected Switch in the Modulation of AI-2-Based Quorum Sensing Discovered through Synthetic 4,5-Dihydroxy-2,3-pentanedione Analogues. J Am Chem Soc. 2008;130:9200–9201. doi: 10.1021/ja802353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Sanchez-Torres V, Wood TK. Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol. 2008;1:30–39. doi: 10.1111/j.1751-7915.2007.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson RD. Hypothetical Functions of Toxin-Antitoxin Systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai-Prochnow A, Webb JS, Ferrari BC, Kjelleberg S. Ecological Advantages of Autolysis during the Development and Dispersal of Pseudoalteromonas tunicata Biofilms. Appl Environ Microbiol. 2006;72:5414–5420. doi: 10.1128/AEM.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- Mendez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernandez J. Genome-wide Transcriptional Profile of Escherichia coli in Response to High Levels of the Second Messenger 3′,5′-Cyclic Diguanylic Acid. J Biol Chem. 2006;281:8090–8099. doi: 10.1074/jbc.M510701200. [DOI] [PubMed] [Google Scholar]
- Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira CG, Palmer K, Whiteley M, Sircili MP, Trabulsi LR, Castro AFP, Sperandio V. Bundle-Forming Pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J Bacteriol. 2006;188:3952–3961. doi: 10.1128/JB.00177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton WA, Snell EE. Formation and Interrelationships of Tryptophanase and Tryptophan Synthetases in Escherichia coli. J Bacteriol. 1965;89:355–364. doi: 10.1128/jb.89.2.355-364.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niba ETE, Naka Y, Nagase M, Mori H, Kitakawa M. A Genome-wide Approach to Identify the Genes Involved in Biofilm Formation in E. coli. DNA Res. 2007;14:237–246. doi: 10.1093/dnares/dsm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecota DC, Wood TK. Exclusion of T4 Phage by the hok/sok Locus of Plasmid R1. J Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Thi TTL, Vidal O, Lejeune P, Dorel C. Developmental Pathway for Biofilm Formation in Curli-Producing Escherichia coli Strains: Role of Flagella, Curli and Colanic Acid. Environmental Microbiology. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Prüβ BM, Besemann C, Denton A, Wolfe AJ. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J Bacteriol. 2006;188:3731–3739. doi: 10.1128/JB.01780-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The quorum sensing molecule AI-2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol. 2007;189:6109–6117. doi: 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TB, Givskov M. Quorum sensing inhibitors: a bargain of effects. Microbiology. 2006;152:895–904. doi: 10.1099/mic.0.28601-0. [DOI] [PubMed] [Google Scholar]
- Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol. 2003;48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- Ren D, Sims JJ, Wood TK. Inhibition of Biofilm Formation and Swarming of Escherichia coli by (5Z)-4-Bromo-5-(Bromomethylene)-3-Butyl-2(5H)-Furanone. Environ Microbiol. 2001;3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Differential Gene Expression Shows Natural Brominated Furanones Interfere with the Autoinducer-2 Bacterial Signaling System of Escherichia coli. Biotechnol Bioengr. 2004a;88:630–642. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene Expression in Escherichia coli Biofilms. Appl Microbiol Biotechnol. 2004b;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- Ren D, Zuo R, Gonzalez A, Bedzyk LA, Eldridge GR, Pasmore ME, Wood TK. Differential Gene Expression to Investigate Escherichia coli Biofilm Inhibition by Plant Extract Ursolic Acid. Appl Environ Microbiol. 2005;71:4022–4034. doi: 10.1128/AEM.71.7.4022-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard AH, Palmer RJ, Jr, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- Rui L, Reardon KF, Wood TK. Protein engineering of toluene ortho-monooxygenase of Burkholderia cepacia G4 for regiospecific hydroxylation of indole to form various indigoid compounds. Appl Microbiol Biotechnol. 2005;66:422–429. doi: 10.1007/s00253-004-1698-z. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Beuchat LR. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl Environ Microbiol. 2005;71:247–254. doi: 10.1128/AEM.71.1.247-254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Rickard AH, Davies DG. Biofilms and Biocomplexity. Microbe. 2007;2:347–353. [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Schembri MA, Kjærgaard K, Klemm P. Global Gene Expression in Escherichia coli Biofilms. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Lamont RJ, Demuth DR. Autoinducer-2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun. 2007;75:4211–4218. doi: 10.1128/IAI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JN, Dyszel JL, Soares JA, Ellermeier CD, Altier C, Lawhon SD, et al. SdiA, an N-Acylhomoserine Lactone Receptor, Becomes Active during the Transit of Salmonella enterica through the Gastrointestinal Tract of Turtles. PLoS ONE. 2008;3:e2826. doi: 10.1371/journal.pone.0002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni KA, Jesudhasan P, Cepeda M, Widmer K, Jayaprakasha GK, Patil BS, et al. Identification of Ground Beef–Derived Fatty Acid Inhibitors of Autoinducer-2–Based Cell Signaling. J Food Protection. 2008;71:134–138. doi: 10.4315/0362-028x-71.1.134. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Giron JA, Kaper JB. Quorum Sensing is a Global Regulatory Mechanism in Enterohemorrhagic Escherichia coli O157:H7. Journal of Bacteriology. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley NR, Lazazzera BA. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52:917–924. doi: 10.1111/j.1365-2958.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- Sturgill G, Toutain CM, Komperda J, O’Toole GA, Rather PN. Role of CysE in production of an extracellular signaling molecule in Providencia stuartii and Escherichia coli: loss of CysE enhances biofilm formation in Escherichia coli. J Bacteriol. 2004;186:7610–7617. doi: 10.1128/JB.186.22.7610-7617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga ME, Semmelhack JL, Bassler BL. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol Microbiol. 2001;42:777–793. doi: 10.1046/j.1365-2958.2001.02669.x. [DOI] [PubMed] [Google Scholar]
- Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50:1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- Tao Y, Fishman A, Bentley WE, Wood TK. Altering toluene 4-monooxygenase by active-site engineering for the synthesis of 3-methoxycatechol, methoxyhydroquinone, and methylhydroquinone. J Bacteriol. 2004;186:4705–4713. doi: 10.1128/JB.186.14.4705-4713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What Is the Benefit to Escherichia coli of Having Multiple Toxin-Antitoxin Systems in Its Genome? J Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Meth. 2008 doi: 10.1038/nmeth.1240. advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Attila C, Whiteley M, Wood TK. Uracil influences quorum sensing in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb Biotechnol. 2008 doi: 10.1111/j.1751-7915.2008.00060.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlich GA, Cooke PH, Solomon EB. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol. 2006;72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houdt R, Michiels CW. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol. 2005;156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- van Houdt R, Aertsen A, Moons P, Vanoirbeek K, Michiels CW. N-Acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol Letters. 2006;256:83–89. doi: 10.1111/j.1574-6968.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- Vejborg RM, Klemm P. Blocking of Bacterial Biofilm Formation by a Fish Protein Coating. Appl Environ Microbiol. 2008;74:3551–3558. doi: 10.1128/AEM.00279-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlisidou I, Lyte M, van Diemen PM, Hawes P, Monaghan P, Wallis TS, Stevens MP. The neuroendocrine stress hormone norepinephrine augments Escherichia coli O157:H7-induced enteritis and adherence in a bovine ligated ileal loop model of infection. Infect Immun. 2004;72:5446–5451. doi: 10.1128/IAI.72.9.5446-5451.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Ding X, Rather PN. Indole Can Act as an Extracellular Signal in Escherichia coli. Journal of Bacteriology. 2001a;183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Ding X, Rather PN. Indole can act as an extracellular signal in Escherichia coli. J Bacteriol. 2001b;183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. Cylclic AMP (cAMP) and cAMP Receptor Protein Influence Both Synthesis and Uptake of Extracellular Autoinducer 2 in Escherichia coli. J Bacteriol. 2005;187:2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, et al. Cell Death in Pseudomonas aeruginosa Biofilm Development. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. Cyclic-di-GMP-mediated signalling within the σS network of Escherichia coli. Mol Microbiol. 2006;62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- Wiebke EA, Grieshop NA, Loehrer PJ, Eckert GJ, Sidner RA. Antitumor effects of 5-fluorouracil on human colon cancer cell lines: antagonism by levamisole. J Surg Res. 2003;111:63–69. doi: 10.1016/s0022-4804(03)00053-2. [DOI] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can’t talk now - gone to lunch! Curr Opin Microbiol. 2002;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- Wong CS, Jelacic S, Habeeb RL, Watkins SL, Tarr PI. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;342:1930–1936. doi: 10.1056/NEJM200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK, Barrios AFG, Herzberg M, Lee J. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005a;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005b;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ma Q, Wood TK. The R1 Conjugative Plasmid Increases Escherichia coli Biofilm Formation through an Envelope Stress Response. Appl Environ Microbiol. 2008;74:2690–2699. doi: 10.1128/AEM.02809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C, Horn V, Gollnick P. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J Bacteriol. 1991;173:6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XS, García Contreras R, Wood TK. YcfR (BhsA) Influences Escherichia coli Biofilm Formation Through Stress Response and Surface Hydrophobicity. J Bacteriol. 2007;189:3051–3062. doi: 10.1128/JB.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XS, García Contreras R, Wood TK. Escherichia coli transcription factor YncC (McbR) regulates colanic acid and biofilm formation by repressing expression of periplasmic protein YbiM (McbA) ISME J. 2008;2:615–631. doi: 10.1038/ismej.2008.24. [DOI] [PubMed] [Google Scholar]