A genomic code for nucleosome positioning (original) (raw)
. Author manuscript; available in PMC: 2009 Jan 13.
Published in final edited form as: Nature. 2006 Jul 19;442(7104):772–778. doi: 10.1038/nature04979
Abstract
Eukaryotic genomes are packaged into nucleosome particles that occlude the DNA from interacting with most DNA binding proteins. Nucleosomes have higher affinity for particular DNA sequences, reflecting the ability of the sequence to bend sharply, as required by the nucleosome structure. However, it is not known whether these sequence preferences have a significant influence on nucleosome position in vivo, and thus regulate the access of other proteins to DNA. Here we isolated nucleosome-bound sequences at high resolution from yeast and used these sequences in a new computational approach to construct and validate experimentally a nucleosome-DNA interaction model, and to predict the genome-wide organization of nucleosomes. Our results demonstrate that genomes encode an intrinsic nucleosome organization and that this intrinsic organization can explain ∼50% of the in vivo nucleosome positions. This nucleosome positioning code may facilitate specific chromosome functions including transcription factor binding, transcription initiation, and even remodelling of the nucleosomes themselves.
Eukaryotic genomic DNA exists as highly compacted nucleosome arrays called chromatin. Each nucleosome contains a 147-base-pair (bp) stretch of DNA, which is sharply bent and tightly wrapped around a histone protein octamer1. This sharp bending occurs at every DNA helical repeat (∼10 bp), when the major groove of the DNA faces inwards towards the histone octamer, and again ∼5 bp away, with opposite direction, when the major groove faces outward. Bends of each direction are facilitated by specific dinucleotides2,3. Neighbouring nucleosomes are separated from each other by 10-50-bp-long stretches of unwrapped linker DNA4; thus, 75-90% of genomic DNA is wrapped in nucleosomes. Access to DNA wrapped in a nucleosome is occluded1 for polymerase, regulatory, repair and recombination complexes, yet nucleosomes also recruit other proteins through interactions with their histone tail domains5. Thus, the detailed locations of nucleosomes along the DNA may have important inhibitory or facilitatory roles6,7 in regulating gene expression.
DNA sequences differ greatly in their ability to bend sharply2,3,8. Consequently, the ability of the histone octamer to wrap differing DNA sequences into nucleosomes is highly dependent on the specific DNA sequence9,10. In vitro studies show this range of affinities to be 1,000-fold or greater11. Thus, nucleosomes have substantial DNA sequence preferences. A key question is whether genomes use these sequence preferences to control the distribution of nucleosomes in vivo in a way that strongly impacts on the ability of DNA binding proteins to access particular binding sites. By controlling binding site accessibility in this way, genomes could, for example, target the binding of transcription factors towards appropriate sites and away from irrelevant, non-functional sites9.
One view is that the sequence preferences of nucleosomes might not be meaningful. Nucleosome positions might be regulated in cells in trans by the abundant12 ATP-dependent nucleosome remodelling complexes13, which might over-ride the sequence preferences of nucleosomes and move them to new locations whenever needed. Another view, however, is that remodelling factors do not themselves determine the destinations of the nucleosomes that they mobilize. Rather, the remodelling complexes may allow nucleosomes to sample alternative positions rapidly, resulting in a thermodynamic equilibrium between the nucleosomes and the site-specific DNA binding proteins that compete with nucleosomes for occupancy along the genome. In this view, nucleosome positions are regulated in cis by their intrinsic sequence preferences, which would then have significant regulatory roles. In this cis regulation model, we expect the genome to encode a nucleosome organization, intrinsic to the DNA sequence alone, comprising sequences with both low and high affinity for nucleosomes. Many of the high-affinity sequences should then be occupied by nucleosomes in vivo. Moreover, the detailed distribution of nucleosome positions encoded by the genome should significantly influence chromosome functions genome-wide.
Here we report the results of a combined experimental and computational approach to detect the DNA sequence preferences of nucleosomes and the intrinsic nucleosome organization of the genome that these preferences dictate. Our findings demonstrate that eukaryotic genomes use a nucleosome positioning code, and link the resulting nucleosome positions to specific chromosome functions.
Validating a nucleosome-DNA interaction model
To construct a model for nucleosome-DNA interactions in yeast (Fig. 1a), we used a genome-wide assay to isolate DNA regions that were stably wrapped in nucleosomes. Our experimental method maps nucleosomes on the yeast genome with greater accuracy than previous approaches, resulting in a set of 199 mononucleosome DNA sequences of length 142-152 bp (Supplementary Fig. 1). We used this collection of sequences to construct a probabilistic model that represents the DNA sequence preferences of yeast nucleosomes (Supplementary Fig. 2). Our approach resembles that used for representing the binding specificities of transcription factors from a collection of known sites, but with two main distinctions: first, in contrast to the mononucleotide probability distributions used for transcription factors, we use dinucleotide probability distributions (as dinucleotides are the simplest sequence elements to capture the sequence-dependent mechanics of DNA bending14 that are essential for histone-DNA association3); second, when constructing the model we represent the two-fold symmetry axis of the nucleosome structure1 by including the reverse complement of each sequence in the nucleosome collection. More sophisticated nucleosome-DNA interaction models based on mixture models15 or on the expectation-maximization algorithm16 yielded equivalent results.
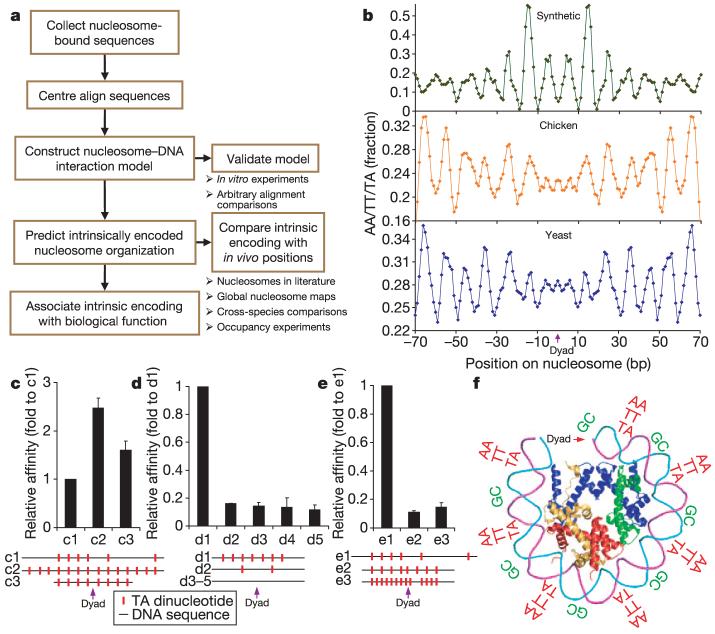
Figure 1. Probabilistic nucleosome-DNA interaction model.
a, Flow chart illustrating our approach. b, Fraction (3-bp moving average) of AA/TT/TA dinucleotides at each position of centre-aligned yeast, chicken2 or random chemically synthesized17 nucleosome-bound DNA sequences, showing ∼10-bp periodicity of these dinucleotides. c-e, In vitro experiments. Positions of the key AA/TT/TA dinucleotides on the tested sequences are indicated. Error bars are s.e.m. c, Nucleosome binding affinities of sequences c2 and c3 (ref. 44), which include additional dinucleotide motifs at key positions, relative to the affinity of c1. d, Sequences d2-d5 have dinucleotide motifs removed from key positions in e1. e, Sequences e2 and e3 have disrupted spacing between the key dinucleotide motifs. f, Key dinucleotides inferred from the alignments are shown relative to the three-dimensional structure of one-half of the symmetric nucleosome.
As expected for a nucleosome-DNA interaction model, the resulting model exhibits distinctive sequence motifs that recur periodically at the DNA helical repeat and are known to facilitate the sharp bending of DNA around the nucleosome3. These include ∼10-bp periodic AA/TT/TA dinucleotides that oscillate in phase with each other (Fig. 1b) and out of phase with ∼10-bp periodic GC dinucleotides. Moreover, the same periodicities and phase relationships were derived independently from a collection of 177 natural nucleosomes from chicken2, and they arose again in three independent in vitro experiments that selected for stable nucleosomes. These in vitro selection experiments include one on chemically synthesized random DNA17, one on mouse genomic DNA18, and a new experiment that we performed on yeast genomic DNA (see Methods). The similarities among these independently derived nucleosome patterns are striking and quantitatively significant (Fig. 1b and Supplementary Figs 3-5), for example, P < 10-50 for yeast-chicken in vivo similarity.
We experimentally validated the importance of these periodic sequence motifs for nucleosome-DNA interactions in vitro. Improving the agreement of a sequence with these motifs increased its binding affinity to the nucleosome, whereas changing the periodicity or deleting the key motifs decreased that affinity (Fig. 1c-e and Supplementary Fig. 6). In addition, these periodic motifs did not arise in alignments of randomly chosen regions in the yeast or chicken genomes (Supplementary Fig. 7). Together, these results establish that the distinctive motifs in our model represent DNA sequence preferences of nucleosomes (Fig. 1f).
If genomes use these sequence preferences, then high-affinity sequences should be prevalent in the genome. Indeed, we found that intergenic and coding regions in the yeast genome contain many more high-affinity DNA sequences than expected by chance (P < 10-200 for both intergenic and coding regions; Supplementary Fig. 8), and that scores at positions separated by 10 bp are strongly correlated (Supplementary Fig. 9). Together with the distinctive features of the yeast in vivo nucleosome collection, these results show that sequence motifs for positioning nucleosomes are abundantly encoded in the yeast genome and that nucleosomes occupy these sequences in vivo.
Predicting nucleosome organization in genomic DNA sequence
We next sought to understand how the encoded nucleosome preferences integrate to specify the intrinsic genome-wide positioning of nucleosomes. This task is non-trivial because encoded nucleosome positions are correlated through steric hindrance. We designed a thermodynamic model that defines an apparent free energy for every organization of nucleosomes on the DNA, taking steric hindrance and competition between nucleosomes into account (see Methods). A dynamic programming method19 evaluated efficiently all sterically allowed organizations, yielding both the probability that each base pair is occupied by any nucleosome (average nucleosome occupancy) and the genomic locations of the sites at which nucleosomes have a high probability of starting (stably positioned nucleosomes).
The resulting intrinsic nucleosome organization differs qualitatively at different genomic locations. In some cases, several mutually exclusive organizations dominate (Supplementary Fig. 10a, b); in others, a single organization dominates (Supplementary Fig. 10c); and yet in others no particular organization dominates (Supplementary Fig. 10d). Comparing these diverse intrinsic organizations to known transcription factor binding sites20 reveals the potential regulatory role of nucleosomes: nucleosomes may have a strong affinity to occupy transcription factor binding sites (rendering them inaccessible) in some genomic locations (Supplementary Fig. 10a), but a weak affinity to occupy sites (thereby increasing their accessibility) in other locations (Supplementary Fig. 10b).
Predicted nucleosome organization reflects in vivo data
By comparing actual in vivo nucleosome positions to our predicted or experimentally measured intrinsically encoded positions, we can test whether in vivo positions are dictated by the genomic sequence. To this end, we used five different approaches. First, we measured the distance between our predicted stable nucleosome positions (stability probability ≥0.2; see Methods) and 99 experimentally mapped nucleosome positions at 11 loci21-28 (Supplementary Fig. 11). There is some disagreement between different experimental measurements of nucleosome positions (Fig. 2b and Supplementary Fig. 12), hence discrepancies between our predictions and literature reports are attributable to inaccuracies both in our model and in the literature. Even so, six loci showed substantial correspondence (Fig. 2 and Supplementary Figs 13-22). Overall, 54% of our predicted stable nucleosomes were within 35 bp of the literature positions, significantly more than the 39 ± 1% expected by chance (P < 10-16).
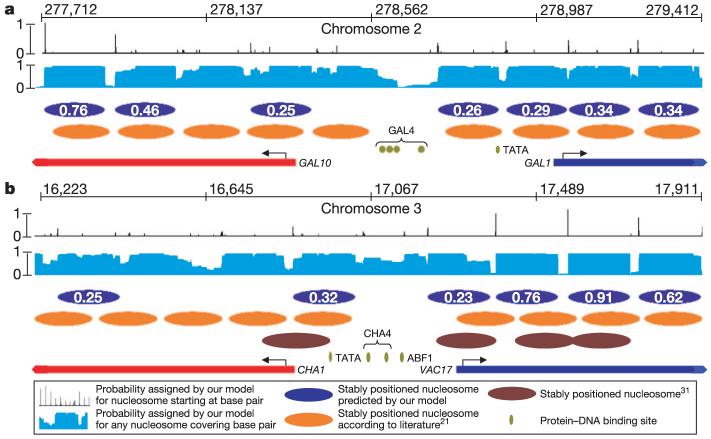
Figure 2. Genome-wide prediction of intrinsic nucleosome organization and comparison to literature-reported, experimentally identified31 nucleosome positions.
a, Detailed view of the GAL1-10 locus, with literature-reported nucleosome positions21 (orange ovals). Black trace, probability of a nucleosome starting at each base pair; blue ovals, high probability nucleosomes predicted from our model (probability is indicated); light-blue trace, average occupancy by any nucleosome at each base pair; red and blue bars, protein-coding regions; green ovals, conserved and bound DNA-binding sites20. b, Same as in a, but for the CHA1 locus24; brown ovals, nucleosomes reported from other experiments31. The discrepancies between the two sets of literature-reported nucleosome positions highlight the uncertainty in such measurements.
Second, we compared our predictions to three genome-wide measurements of nucleosome positions at low29,30 or higher31 resolution. Our model showed significant correspondence to these experiments, predicting lower occupancy at nucleosome-depleted (low nucleosome abundance) coding or intergenic regions29,30 (Supplementary Figs 23-25; 68% of 57 depleted coding regions and 76% of 294 depleted intergenic regions had predicted low occupancy compared with 30% (P < 10-6) and 56% (P < 10-9), respectively, expected by chance). The model also showed strong correspondence with the higher resolution nucleosome map31: 45% of our predicted stable nucleosomes were within 35 bp of experimentally determined nucleosome positions31 compared with 32 ± 1% expected by chance, P < 10-15 (Supplementary Figs 26 and 27). Notably, our predictions also match closely the stereotyped chromatin organization at Pol II promoters as revealed by the higher resolution nucleosome map31, and the most stable nucleosome predicted by our model at promoters is located precisely (within 8 bp) where stable nucleosomes containing the histone variant H2A.Z are located in vivo32 (Fig. 5a).
Figure 5. Genomes encode unstable nucleosomes at transcriptional start sites.
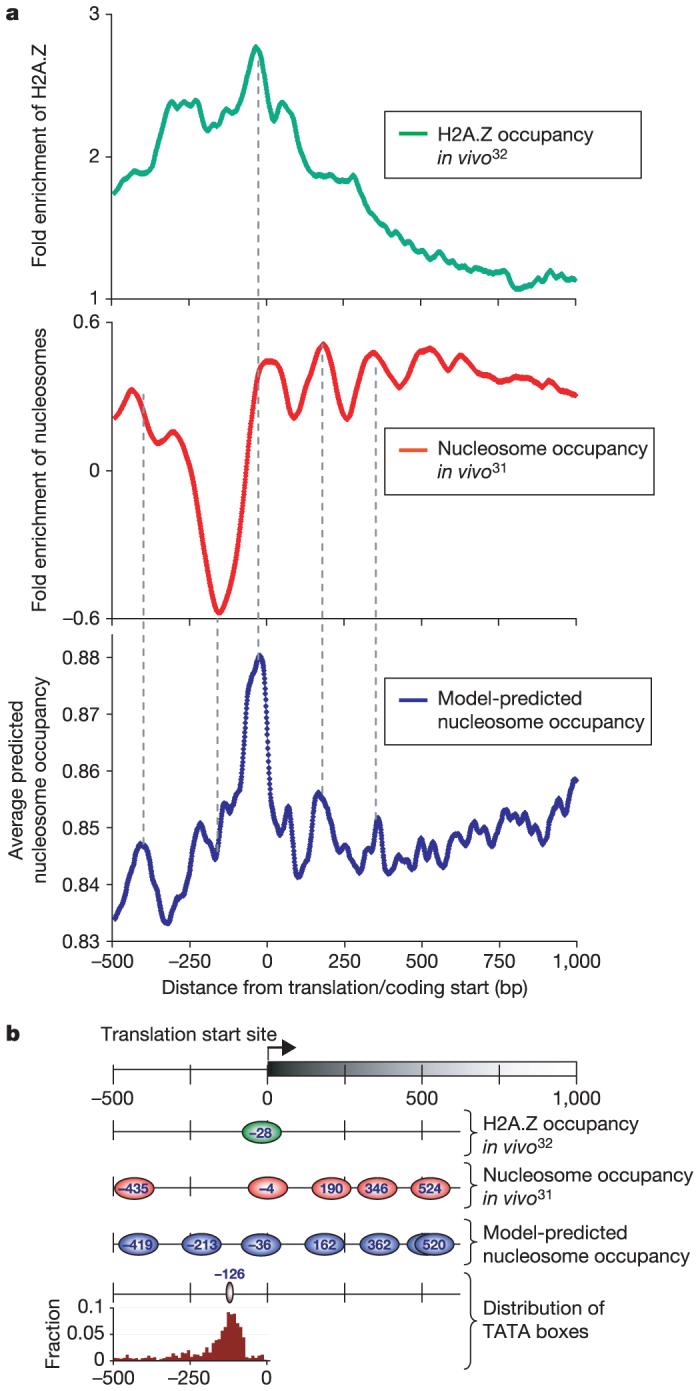
a, Average across all yeast genes of the in vivo occupancy of nucleosomes containing the histone variant H2A.Z32 (green) or of canonical nucleosomes31 (red), compared to the nucleosome occupancy predicted by our yeast nucleosome model (blue); all versus distance from the translation open reading frame (ORF) start site. The ORF-proximal peak of our model is statistically significant (Supplementary Fig. 37). b, The most probable nucleosome organization, based on a. Each nucleosome (ovals; labels represent nucleosome centres) is centred to a corresponding peak in a. Bottom graph shows distribution of TATA boxes42 relative to ORF start sites; brown oval is median TATA box location.
Third, we compared the yeast model predictions to those of a model constructed independently using only nucleosome-bound sequences from chicken. The predictions of the chicken model when applied to the yeast genome correlated strongly with those of the yeast model (Supplementary Fig. 28) and with the genome-wide experimental measurements of nucleosome occupancy at yeast coding and intergenic regions29-31: 35% of 57 depleted coding regions and 72% of 294 depleted intergenic regions had predicted low occupancy compared with 4% (P < 10-4) and 53% (P < 10-8) expected by chance.
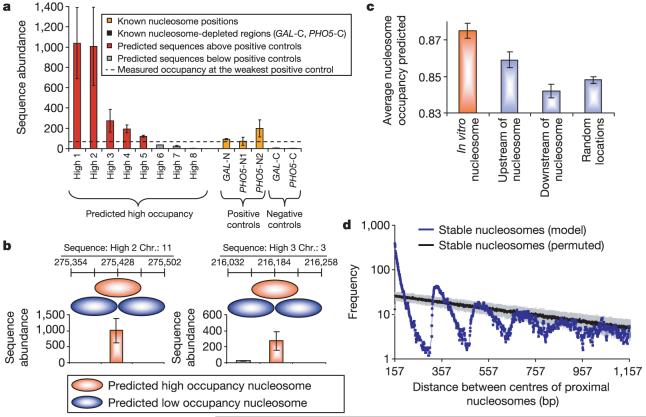
Fourth, we carried out a new selection for nucleosome formation on yeast genomic DNA in vitro. This experiment directly reveals intrinsically encoded, individual high-affinity nucleosome positions. These in vitro nucleosome locations overlap significantly with our in vivo yeast nucleosome collection: 32% of 339 selected in vitro nucleosomes overlapping the in vivo bound sequences compared with 5% (P < 10-5) expected by chance. The in vitro selected nucleosomes are particularly enriched in intergenic regions that have a high predicted nucleosome occupancy, compared with random genomic locations and to locations immediately upstream or downstream of the selected nucleosomes (P < 10-3; Fig. 3c and Supplementary Figs 29 and 30).
Figure 3. Higher-order features of intrinsic nucleosome organization and comparison with in vivo occupancy experiments.
a, Experimentally measured nucleosome occupancy in vivo for eight high-occupancy predictions, compared with high- and low-occupied locations in the GAL1-10 and PHO5 promoters. Error bars are s.d. b, In vivo nucleosome occupancy measured at predicted low-occupancy regions that are one-half nucleosome distance upstream and downstream (light blue) from the high-occupancy (orange) predictions of a. See Supplementary Fig. 31 for additional measurements. Results of a and b were consistent when normalized for the sequence specificity of micrococcalnuclease (Supplementary Fig. 32). Error bars are s.d. c, Predicted nucleosome occupancy in intergenic regions for nucleosomes obtained from an in vitro selection experiment (orange) compared with predicted nucleosome occupancy in immediately upstream or downstream locations, or to random genomic locations (light blue). Error bars are s.e.m. d, Number of all pairs of proximal stable nucleosomes per centre-to-centre nucleosome distance, compared to the mean (black) and standard deviation (grey) in 100 permutations. Blue, yeast model (stability probability ≥0.5).
Finally, we experimentally tested whether our highest occupancy predictions are highly occupied by nucleosomes in vivo, by measuring their in vivo nucleosome occupancies and comparing them to the occupancies at three nucleosome sites flanking the GAL1-10 and PHO5 promoters for which the nucleosome positions are known. Five of the eight predictions tested yielded in vivo occupancies comparable to or greater than those of the known nucleosome positions (Fig. 3a), indicating that ∼60% of the intrinsically high-occupancy nucleosome sites on the DNA sequence are strongly occupied in vivo. In 10 out of 11 cases, these predicted nucleosome positions also had higher occupancy than regions 73 bp (one-half the length of a nucleosome) upstream or downstream from the predicted position (Fig. 3b and Supplementary Figs 31 and 32).
Taken together, these results show that ∼50% of the in vivo nucleosome organization can be explained solely by the sequence preferences of nucleosomes. Moreover, these results indicate that the nucleosome depletions observed at coding and intergenic regions29-31 are attributable in part to unstable nucleosomes (that is, positions on the DNA sequence that nucleosomes have a low probability of occupying) encoded in these regions.
Global features of intrinsic nucleosome organization in yeast
We next studied global properties of the intrinsic nucleosome organization in yeast. First, we examined the predicted stability of all 11,802,267 possible genome-wide nucleosome positions; 15,777 were highly stable (stability probability ≥0.5), significantly more than the 10,940 ± 339 (P < 10-20) expected by chance. This result may indicate the existence of many genomic locations that encode highly stable nucleosomes, together covering 20% of the genome.
Second, we asked whether individual nucleosomes are organized into higher-ordered nucleosome arrays. The distribution of pairwise distances between positions of the highly stable nucleosomes revealed significant correlations persisting over at least six adjacent nucleosomes, with an average nucleosome repeat length of 177 bp (Fig. 3d). We found similar strong correlations when considering the average nucleosome occupancy predictions (Supplementary Fig. 33). We conclude that the yeast genome not only encodes the preferred positions of individual nucleosomes, but also directly encodes higher structural levels of chromatin organization.
Nucleosome organization varies by type of genomic region
We next asked whether the genome’s intrinsic encoding of nucleosome occupancy varies across different types of chromosomal regions, including centromeres, telomeres, intergenic and coding regions, and specific gene classes (Fig. 4a and Supplementary Fig. 34). Indeed, several types of regions had markedly high or low predicted occupancy. The highest predicted occupancy was over centromeres, indicating that centromere function requires enhanced stability of histone-DNA interactions that are encoded in the genomic sequence.
Figure 4. Intrinsic nucleosome occupancy varies with genomic location type and is low at functional transcription factor binding sites.
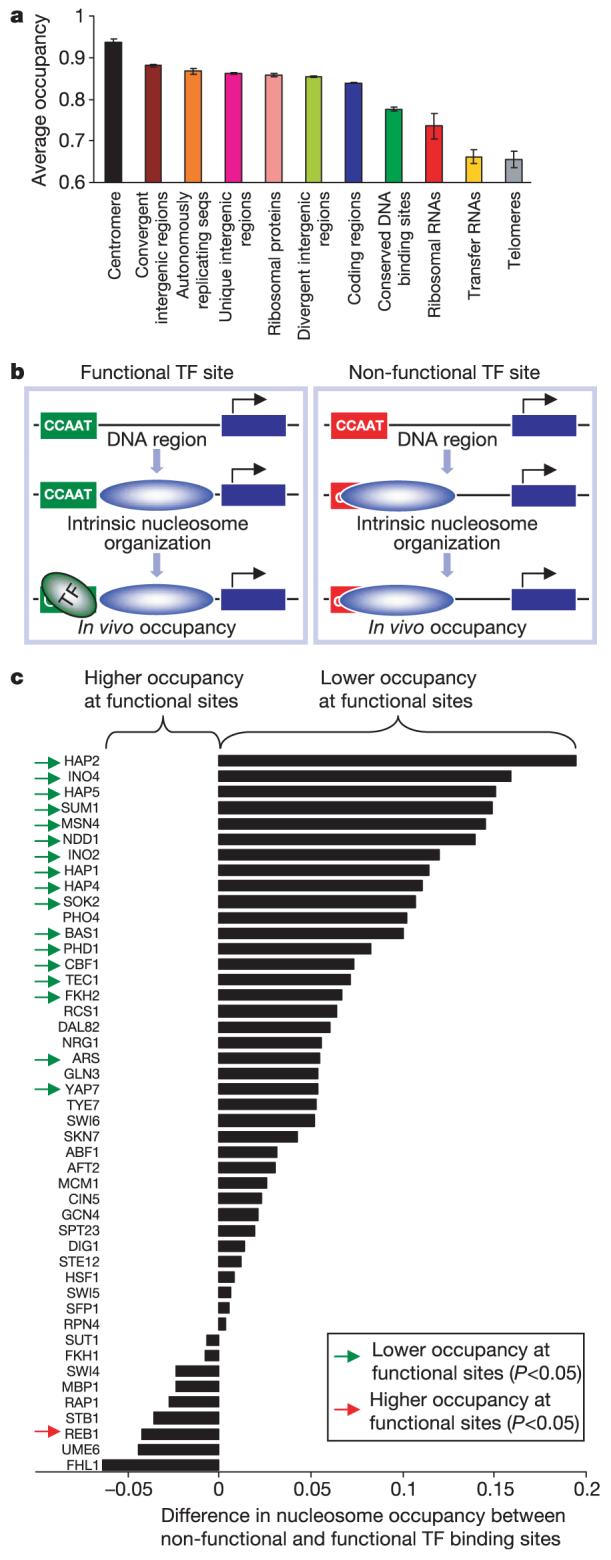
a, Average occupancies and standard errors for different types of genomic regions. b, Schematic illustrating how the intrinsic nucleosome organization may facilitate binding of transcription factors (TF) at functional sites, while disfavouring binding at identical non-functional sites that occur by chance. c, Difference in predicted nucleosome occupancy between non-functional and functional transcription factor binding sites (absolute occupancy levels are shown in Supplementary Fig. 36). Green arrows, 17 factors having significantly lower nucleosome occupancy at functional sites compared with non-functional sites20; red arrow, 1 factor having significantly higher nucleosome occupancy at non-functional sites compared with functional sites.
One might think that genomes would facilitate high gene expression levels by encoding unstable nucleosomes over highly expressed genes. Consistent with this expectation, the highly expressed ribosomal RNA and transfer RNA genes stood out as having markedly low predicted nucleosome occupancy.
In contrast to the ubiquitously expressed tRNAs, many other genes vary their expression between high and low levels in different conditions. However, as the genome sequence is static, it cannot simultaneously encode a nucleosome organization that would facilitate both high and low expression levels. Ribosomal proteins are one such example. Our model predicts high nucleosome occupancy encoded over these genes. Thus, the genome sequence does not facilitate the nucleosome depletion29 and high expression of ribosomal proteins observed during normal growth, which therefore must be governed by other factors. Instead, the genome facilitates the rapid nucleosome reassembly29 and strong repression of these genes observed under stress33,34. These results show how the genome’s statically encoded nucleosome organization may contribute to the dynamic process of gene regulation.
Nucleosomes facilitate their own remodelling
We tested whether the variation of nucleosome occupancy that we observed at different types of chromosomal region also extended to other sets of functionally related genes. We collected 1,949 different sets of yeast genes from a functional gene annotation database35 and from a wide range of genomic studies20,36-40, and found that indeed many gene sets showed a significant association with either high or low predicted nucleosome occupancy (Supplementary Fig. 35). Notably, of all gene sets tested, the most significant association predicted low occupancy at regions bound by the chromatin remodelling complex RSC40 (P < 10-34). This implies that genomes facilitate their own chromatin remodelling by encoding intrinsically low nucleosome occupancy at sites destined for remodelling.
Low nucleosome occupancy encoded at functional binding sites
For any given transcription factor, some of its canonical target sites in the genome are occupied by a nucleosome, whereas others are not. Many of the unoccupied sites are thought to occur at random and to be functionally irrelevant20,41, but the mechanism by which they are kept unoccupied is not known. An intriguing hypothesis is that genomes use their intrinsic nucleosome organization for this task by encoding stable nucleosomes over non-functional sites, thereby decreasing their accessibility to transcription factors (Fig. 4b). We tested this hypothesis by examining our predictions at binding sites for 46 transcription factors. Notably, for 17 (37%) transcription factors the predicted nucleosome occupancy at their functional and conserved DNA binding sites20 was significantly lower compared with predicted occupancy at their other canonical (but presumed non-functional) sites (Fig. 4c). Only one (2%) factor exhibited significantly higher predicted occupancy at its functional binding sites. These results illustrate how the intrinsic nucleosome organization may help in directing transcription factors towards the appropriate subset of their target sites while excluding them from irrelevant sites.
Low nucleosome occupancy encoded at transcription start sites
Recent nucleosome maps indicate that nucleosomes are depleted from transcriptional start sites31 (TSSs), but the mechanism for this depletion is not known. For two promoter regions, this depletion was shown experimentally to be intrinsically encoded in the DNA sequence9. We asked whether this intrinsically encoded depletion occurs globally by examining the encoded nucleosome organization at all TSSs in yeast (Fig. 5a). We found that the most probable location for TATA elements42 places them in areas of the genomic sequence that remain unoccupied by nucleosomes; that is, just outside a stably positioned nucleosome (Fig. 5b). Strikingly, the location of the stably positioned nucleosome is conserved across all fungal species (Supplementary Fig. 38). We obtained all of the above results independently, applying both the chicken and yeast models to the yeast genomes. Together, these results may indicate that eukaryotic genomes direct the transcriptional machinery to functional sites by encoding unstable nucleosomes over these elements, thereby enhancing their accessibility.
Conclusions and prospects
Our results establish that nucleosome organization is encoded in eukaryotic genomes. This newly characterized genetic information occurs chromosome-wide, explains ∼50% of the in vivo nucleosome organization, and may facilitate specific chromosome functions. The consistency between the predictions on the yeast genome using models derived independently from information concerning only yeast or chicken nucleosomes implies that the genomic signals for nucleosome positioning are strong.
Despite its successes, our approach has several limitations and represents only a first step towards understanding the DNA preferences of nucleosomes and the biological implications. First, additional experiments are needed to derive a more accurate nucleosome-DNA interaction model. Second, our representation of nucleosome-nucleosome interactions derived from a thermodynamic model does not yet account for favourable interactions43, or for the steric hindrance constraints implied by the three-dimensional nucleosome structure. Finally, we examined the intrinsic nucleosome organization without regard for the collection of DNA binding proteins that influence nucleosome positioning by competing for DNA occupancy. At equilibrium, this competition would depend on the concentrations and sequence specificities of both the DNA binding proteins and nucleosomes. The DNA binding proteins have high binding specificity but are present at low concentrations, whereas the nucleosomes have lower binding specificity but are present at high concentrations, covering 75-90% of the DNA. Thus, both are expected to make important contributions to the outcome (Supplementary Figs 39 and 40).
Overall, our results establish that genomes encode the positioning and stability of nucleosomes in regions that are critical for gene regulation and for other specific chromosome functions, and establish that this nucleosome positioning code can be successfully decoded. The genome-wide predictions of nucleosome occupancy and stability that we generated should facilitate the understanding of specific natural gene regulatory phenomena, such as the mechanism by which transcription factors bind preferentially to appropriate sites in promoters rather than to the excess of irrelevant sites in the genome. Our approach may also be useful for improving the performance of engineered transgenes. Our model and results provide a concrete framework for quantitatively integrating chromatin structure into models of gene regulation, and thus represent an essential step towards the goal of developing a quantitative, predictive understanding of transcriptional regulation in all eukaryotes.
Supplementary Material
Supplement
Acknowledgements
We thank A. Travers for providing the chicken nucleosome core DNA sequences; M. Kubista for providing selected mouse DNA sequences; O. Rando for providing access to their nucleosome data before publication; J. Lieb, E. Nili and P. Jones for sharing their respective unpublished data; Y. Lubling for creating the supplementary website; and H. Chang, N. Friedman, U. Gaul, A. Matouschek, B. Meyer, M. Ptashne, E. Siggia and A. Tanay for useful comments on the manuscript. E.S. was supported by a fellowship from the Center for Studies in Physics and Biology at Rockefeller University and by an NIH grant. J.W. thanks the Center for their hospitality during a sabbatical. J.-P.Z.W. acknowledges support from an NIH grant and J.W. acknowledges support from two NIH grants. E.S. is the incumbent of the Soretta and Henry Shapiro career development chair.
Appendix
METHODS
See Supplementary Information for a more detailed description of the methods.
Molecular biology methods
Mononucleosomes were extracted from log-phase yeast (Saccharomyces cerevisiae) cells using standard methods. The DNA was extracted, and protected fragments of length ∼147 bp were cloned and sequenced. An in vitro selection for nucleosome formation on the yeast genome was performed using purified yeast genomic DNA and substoichiometric purified histone octamer by salt gradient dialysis44. The resulting chromatin was treated as for the in vivo selection. In vitro affinity measurements for core histone H32H42 tetramers were performed as described44. In vivo nucleosome occupancies were measured as described9.
Probabilistic nucleosome-DNA interaction model
Given a collection of nucleosome DNA sequences, we aligned all sequences and their reverse complements about their centres, and associated a dinucleotide distribution with each position i, estimated from the combined dinucleotide counts at three neighbouring positions, such that the probability assigned by the model to a 147-bp sequence S is:
P(S)=P1(S1)∏i=2147Pi(Si∣Si−1)
Thermodynamic model for predicting nucleosome positions genome-wide
We used the above probabilistic nucleosome-DNA model within a statistical mechanics framework to compute the nucleosome organization intrinsic to the genomic DNA sequence. We took the partition function to be all ‘legal configurations’ of nucleosomes on a sequence S, where a legal configuration specifies start positions for a set of non-overlapping 147-bp nucleosomes on S, thus respecting steric hindrance effects between nucleosomes. Using our probabilistic model and an apparent nucleosome concentration parameter, we assigned a statistical weight to each configuration and used the Boltzmann distribution to compute the probability of every configuration. A dynamic programming method19 was used to efficiently compute the probability that each base pair of S starts a nucleosome or is occupied by a nucleosome.
Additional methods and URLs
For our data, model and genome-wide occupancy predictions, see http://genie.weizmann.ac.il/pubs/nucleosomes06. Our results are also viewable in Genomica (http://Genomica.weizmann.ac.il).
Footnotes
The authors declare no competing financial interests.
References
- 1.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 3.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 4.van Holde KE. Chromatin. Springer; New York: 1989. [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 7.Wyrick JJ, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 8.Trifonov EN. Sequence-dependent deformational anisotropy of chromatin DNA. Nucleic Acids Res. 1980;8:4041–4053. doi: 10.1093/nar/8.17.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thåström A, et al. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 12.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 13.Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JP, Widom J. Improved alignment of nucleosome DNA sequences using a mixture model. Nucleic Acids Res. 2005;33:6743–6755. doi: 10.1093/nar/gki977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B. 1977;39:1–39. [Google Scholar]
- 17.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 18.Widlund HR, et al. Identification and characterization of genomic nucleosome-positioning sequences. J. Mol. Biol. 1997;267:807–817. doi: 10.1006/jmbi.1997.0916. [DOI] [PubMed] [Google Scholar]
- 19.Rabiner LR. A tutorial on Hidden Markov Models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- 20.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 22.Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss K, Simpson RT. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J. 1997;16:4352–4360. doi: 10.1093/emboj/16.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreira JM, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu M, Roth SY, Szent-Gyorgyi C, Simpson RT. Nucleosomes are positioned with base pair precision adjacent to thea 2 operator in Saccharomyces cerevisiae. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent NA, Tsang JS, Crowther DJ, Mellor J. Chromatin structure modulation in Saccharomyces cerevisiae by centromere and promoter factor 1. Mol. Cell Biol. 1994;14:5229–5241. doi: 10.1128/mcb.14.8.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdone L, Camilloni G, Di Mauro E, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 32.Guillemette B, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Iyer VR. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol. Cell Biol. 2004;24:8104–8112. doi: 10.1128/MCB.24.18.8104-8112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 37.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 38.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casolari JM, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 40.Robert F, et al. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Rhodius V, Gross C, Siggia ED. Identification of the binding sites of regulatory proteins in bacterial genomes. Proc. Natl Acad. Sci. USA. 2002;99:11772–11777. doi: 10.1073/pnas.112341999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 43.Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thåström A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement