Genome-wide Association Study of Vitamin B6, Vitamin B12, Folate, and Homocysteine Blood Concentrations (original) (raw)
Abstract
The B vitamins are components of one-carbon metabolism (OCM) that contribute to DNA synthesis and methylation. Homocysteine, a by-product of OCM, has been associated with coronary heart disease, stroke and neurological disease. To investigate genetic factors that affect circulating vitamin B6, vitamin B12, folate and homocysteine, a genome-wide association analysis was conducted in the InCHIANTI (N = 1175), SardiNIA (N = 1115), and BLSA (N = 640) studies. The top loci were replicated in an independent sample of 687 participants in the Progetto Nutrizione study. Polymorphisms in the ALPL gene (rs4654748, p = 8.30 × 10−18) were associated with vitamin B6 and FUT2 (rs6022662, p = 2.83 × 10−20) with vitamin B12 serum levels. The association of MTHFR, a gene consistently associated with homocysteine, was confirmed in this meta-analysis. The ALPL gene likely influences the catabolism of vitamin B6 while FUT2 interferes with absorption of vitamin B12. These findings highlight mechanisms that affect vitamin B6, vitamin B12 and homocysteine serum levels.
Main Text
One-carbon metabolism (OCM) is a process whereby folate transfers one-carbon groups in a range of biological processes including DNA synthesis, methylation, and homocysteine metabolism.1,2 Water-soluble B vitamins folate, vitamin B6, and vitamin B12 play key roles as enzyme cofactors or substrates in OCM. Patients with deficiencies in these vitamins can develop anemia (MIM 170900 and 261100) and, in the case of vitamin B12 deficiency, neurological problems. Subclinical deficiencies during pregnancy have been linked with neural tube defects (NTD [MIM 601634]).3,4 In adults, subclinical B vitamin deficiency has been associated with increased risk of coronary artery disease (CAD [MIM 607339])5–8 and some cancers such as colorectal cancer (CRC [MIM 114500]).9,10
The exact mechanism that links B vitamins with human health is unknown, but is thought to involve the OCM metabolic pathways.1,11 One of the main hypotheses for the protective effect of B vitamins on CAD is that folate, vitamin B12, and vitamin B6 deficiencies increase the production and decrease the catabolism of homocysteine, which is an independent risk factor for CAD.12–14 Homocysteine affects various proatherogenic processes including inflammation, thrombosis, endothelial dysfunction, and vascular smooth muscle cell proliferation.15,16 Folate insufficiency alters gene expression through changes in methylation pattern of DNA and histones.17–19
The study of genetic variants that affect circulating levels of B vitamins is important for understanding the interplay of diet, genetics, and human health. Circulating levels of B vitamins and homocysteine are at least in part genetically determined. The most-studied polymorphism is the 677T←C (rs1801133) in exon 5 of the 5,10-methylenetetrahydrofolate reductase (MTHFR [MIM 607093]) gene. The 677T variant results in an thermolabile enzyme that is less effective in the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.20 Accordingly, individuals with the 677T variant have higher homocysteine concentrations as well as lower genomic DNA methylation.21,22 In addition to MTHFR, genetic variations in other genes have been linked to B vitamin and homocysteine concentrations, although results have been inconsistent.1,23 The goal of the present study was to conduct a genome-wide investigation to identify SNPs associated with differential concentrations of folate, vitamin B12, vitamin B6, and homocysteine in the InCHIANTI study from the Chianti region in Tuscany, Italy;24,25 SardiNIA study from Ogliastra province of Sardinia, Italy;26 and Baltimore Longitudinal Study of Aging (BLSA) based in the Baltimore-Washington DC area.27 The results from the genome-wide association (GWA) were examined in the Progetto Nutrizione study based in Tuscany, Italy.28
With the exception of folic acid, the ranges of B vitamin concentrations were similar in the SardiNIA, InCHIANTI, BLSA, and Progetto Nutrizione studies (Table 1). Folic acid concentration was higher in the BLSA (which is U.S. based) compared to the Italian studies, reflecting fortification policies in the United States. For the same reason, homocysteine concentration was lowest in the BLSA even though the individuals in this study, on average, were the oldest in the four studies.
Table 1.
Descriptive Characteristics of InCHIANTI, SardiNIA, BLSA, and Progetto Nutrizione Study
| Studies | InCHIANTI | BLSA | SardiNIA | Progetto Nutrizione | ||||
|---|---|---|---|---|---|---|---|---|
| N | 1178 | 641 | 1115 | 686 | ||||
| Vitamin B6 (ng/mL) | 7.5 | (7.8) | – | – | 12.7 | (10.2) | ||
| Vitamin B12 (pg/mL) | 467.4 | (330.4) | 487.2 | (240.5) | 404.1 | (278.2) | 387.8 | (194.8) |
| Folic acid (ng/mL) | 3.3 | (2.0) | 15.8 | (8.6) | 3.4 | (1.7) | 5.9 | (2.9) |
| Homocysteine (μmol/L) | 15.2 | (6.9) | 10.2 | (5.3) | 11.7 | (5.1) | 11.0 | (5.5) |
| Age (years) | 68.1 | (15.5) | 68.3 | (15.5) | 44.7 | (18.4) | 47.0 | (12.7) |
| BMI (kg/m2) | 27.2 | (4.2) | 27.2 | (4.2) | 25.5 | (4.7) | 24.9 | (4.3) |
| % Males (N) | 44.4% | (523) | 54.0% | (346) | 42.2% | (470) | 40.0% | (276) |
| % Smokers (N) | 18.9% | (222) | 3.7% | (24) | 19.4% | (216) | 40.2% | (276) |
Genome-wide significant associations were observed for vitamin B6, B12, and homocysteine concentrations. Vitamin B6 was only measured in the InCHIANTI study, thus the GWA of 484,115 autosomal SNPs was assessed with a Bonferroni corrected p value threshold of 1 × 10−7 (Figure 1A and Figure S1 available online). Three SNPs that met the threshold were located in a region with high linkage disequilibrium (LD) including the neuroblastoma breakpoint family, member 3 (NBPF3 [MIM 612367]), and ∼12–50 kb upstream of the tissue nonspecific alkaline phosphatase (ALPL [MIM 171760]) gene (Figure S2). The most significant SNP was rs4654748 (p = 1.21 × 10−8), in which the presence of the C allele was associated with 1.38 ng/mL lower vitamin B6 concentrations (Table 2). This association was replicated in the Progetto Nutrizione study (rs4654748; ALPL p = 2.08 × 10−11). A sample-size-weighted meta-analysis across the two studies resulted in a p value of 8.3 × 10−18 and 1.45 ng/mL lower vitamin B6 per copy of the C allele.
Figure 1.
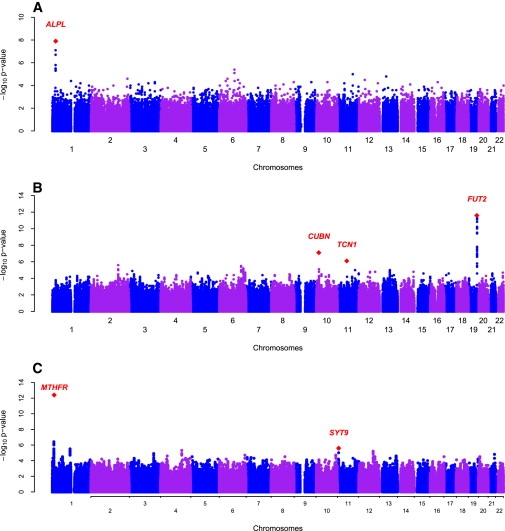
Genome-wide Scans of Plasma Vitamin B12 and B6 in the InCHIANTI Study on Aging
(A) Genome-wide associations of plasma vitamin B6 graphed by chromosome position and −log10 p value. The most significant variant was in the ALPL and NBPF3 genes on chromosome 1.
(B) Genome-wide associations of plasma vitamin B12 graphed by chromosome position and −log10 p value. The most significant variant was in the FUT2 gene on chromosome 19. The next genes of interested included CUBN gene on chromosome 10 and TCN1 on chromosome 11.
(C) Genome-wide associations of plasma homocysteine graphed by chromosome position and −log10 p value. The most significant variant was in the MTHFR gene on chromosome 1. The second significant region was in the SYT9 gene on chromosome 11.
Table 2.
Top SNPs for Vitamin B6, Vitamin B12, Folate, and Homocysteine Concentrations
| GWAS Meta-analysis | Replication | Four Cohort Meta-analysisc | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentrations | Chr | SNP | Position (bp) | Genea | Gene Region | Alleles (+/−) | Freq (+)b | N | Effect | SE | p value | Effect | SE | P value | N | Effect | SE | p value |
| Vitamin B6 (ng/mL) | ||||||||||||||||||
| 1 | rs4654748 | 21531374 | ALPL | 5′UTR | C/T | 0.50 | 1178 | −1.38 | 0.32 | 1.21 × 10−8 | −1.668 | 0.56 | 2.08 × 10−11 | 1864 | −1.45 | 0.28 | 8.30 × 10−18 | |
| Vitamin B12 (pg/mL) | ||||||||||||||||||
| 19 | rs602662 | 53898797 | FUT2 | exon2 | A/G | 0.53 | 2927 | 44.20 | 8.26 | 2.43 × 10−12 | 58.65 | 10.43 | 2.19 × 10−10 | 3613 | 49.77 | 6.47 | 2.83 × 10−20 | |
| 10 | rs11254363 | 17170699 | CUBN | intron52 | A/G | 0.70 | 2927 | −39.16 | 9.18 | 7.24 × 10−8 | 3.62 | 10.94 | 0.815 | 3613 | −21.49 | 7.03 | 1.11 × 10−6 | |
| 11 | rs526934 | 59390069 | TCN1 | intron8 | A/G | 0.67 | 2927 | 36.76 | 10.35 | 8.33 × 10−7 | 12.83 | 13.24 | 0.519 | 3613 | 27.62 | 8.15 | 1.51 × 10−6 | |
| Homocysteine (μmol/L) | ||||||||||||||||||
| 1 | rs1801133d | 11790644 | MTHFR | exon8 | A/G | 0.47 | 2965 | 1.31 | 0.16 | 4.36 × 10−13 | - | - | - | - | - | - | - | |
| 11 | rs11041321 | 7310445 | SYT9 | intron3 | T/C | 0.10 | 2965 | −0.97 | 0.25 | 2.42 × 10−6 | 0.30 | 0.49 | 0.280 | 3651 | −0.70 | 0.22 | 1.11 × 10−4 | |
| Folate (ng/mL) | ||||||||||||||||||
| 1 | rs1999594 | 11893482 | MTHFRd | - | A/G | 0.34 | 2931 | 0.30 | 0.07 | 1.12 × 10−7 | - | - | - | - | - | - | - | |
| 3 | rs153734 | 64053049 | PRICKLE2 | 3′UTR | T/C | 0.84 | 2931 | 0.35 | 0.08 | 1.01 × 10−6 | −0.01 | 0.19 | 0.913 | 3617 | 0.29 | 0.08 | 7.20 × 10−6 |
Mutations in the ALPL gene cause hypophosphotasia (MIM 241510, 231500, and 146300), an inborn error of metabolism characterized by low or complete absence of alkaline phosphatase (ALP) activity. The clinical manifestation of hypophosphatasia is highly variable, ranging from a lethal perinatal form to a more moderate adult form presenting with bone abnormalities.29,30 One characteristic of hypophosphatasia is the accumulation of phospho compounds including vitamin B6. All of the hypophophatasia mutations identified, many of which are missense mutations, lie within the ALPL gene.30 In a recent meta-analysis that included the InCHIANTI study, rs1780324 within the ALPL gene region was the top signal associated with ALP concentration.31 This SNP was one of the top-three SNPs in the vitamin B6 analysis in the current study and is in LD with the most significant SNP, rs4654748 (r2 = 0.6). When the regression model was conditioned on rs4654748, rs1780324 was no longer significantly associated with vitamin B6 (p = 0.358), indicating that these two SNPs represent the same locus. In addition, when ALP levels are included in the regression model, the associations of both rs4654748 (p = 0.0331) and rs1780324 (p = 0.019) with vitamin B6 were no longer significant. This indicates that the association of the two ALPL gene SNPs with vitamin B6 is mediated by ALP. Mechanistically, ALP is the major enzyme involved in the clearance of vitamin B6,32 and therefore the lower vitamin B6 in C allele carriers most likely results from more efficient clearance of the vitamin.
Vitamin B12, folate, and homocysteine concentrations were available for GWA in InCHIANTI, SardiNIA, and BLSA. To conduct a meta-analysis of the results from these three studies, which were genotyped with different platforms (Affymetrix and Illumina), we imputed ∼2.5 million SNPs with MACH using the HapMap CEPH sample as reference.33 The SNPs that passed quality control (MAF > 1%, r2hat > 0.3) were used to conduct a GWA analysis in each study. For the meta-analysis, an arbitrary reference allele is selected and a z statistic summarizing the magnitude and direction of effect relative to the reference allele is weighted by the square root of the sample size of each study. A fixed-effects inverse-variance method was used for calculating the effect sizes. For the meta-analysis, a Bonferroni corrected p value threshold of 5 × 10−8 was considered to be genome-wide significant.
One locus reached genome-wide significance in the meta-analysis of vitamin B12 concentrations (Figure 1B; Figure S1; Table 2). The top SNP in this locus, rs6022662 (pmeta = 2.43 × 10−12), mapped to exon 2 of the fucosyltransferase 2 (FUT2) gene (Figure S2A). The presence of the A allele was associated with 44.2 pg/mL higher vitamin B12 concentrations. This is the same SNP reported in a recent GWA analysis in 2717 women in the Cancer Genetic Markers of Susceptibility projects and the Nurses' Health Study.34 Polymorphisms in FUT2 determine the human secretor (Se) blood group through the expression of α1,2-fucosyltransferase that mediates the fucosylation of oligosaccharides to form H type 1 and 2 antigens.35,36 These H antigens mediate the adhesion of various gastric pathogens such as Helicobactor pylori to the gastric and duodenal mucosal.36–38 Overgrowth of gastric bacteria, such as with H. pylori, has been associated with vitamin B12 deficiency.39,40 Interestingly, rs602662 has been identified in nonsecretor status, or the absence of H antigens, in individuals from Northern Portugal.41 The reduced activity of the FUT2 enzyme with the A allele may decrease susceptibility to bacterial infection and indirectly lower the risk of vitamin B12 malabsorption, thereby resulting in higher vitamin B12 concentrations in A allele carriers.
The second- and third-top SNPs for vitamin B12 analysis did not reach genome-wide significance but were mapped to genes involved in the absorption and transport of vitamin B12. These were rs11254363 in intron 52 of the intrinsic factor—cobalamin receptor, cubilin (CUBN [MIM 602997]) gene (pmeta = 7.24 × 10−8; Table 2, Figure S2) and rs526934 in intron 8 of the transcobalamin I (TCN1 [MIM 189905]) gene (pmeta = 8.33 × 10−7; Table 2, Figure S2). Mutations in CUBN cause a rare autosomal-recessive disorder termed megaloblastic anemia 1 (MGA [OMIM 261100]), characterized by juvenile pernicious anemia.42,43 Mutations in TCN1 result in transcobalamin I deficiency (OMIM 193090), characterized by low vitamin B12. These SNPs, however, were not replicated in the Progetto nutrizione study. There is evidence for heterogeneity among studies for rs11254363 (_χ_2 = 9.24, p = 0.03) but not rs526934 (_X_2 = 5.37, p = 0.15). Subjects in the Progetto nutrizione study come from a similar source population as the InCHIANTI study; thus it is difficult to identify any factor contributing to the observed heterogeneity and warrants further investigation.
In the meta-analyses of homocysteine concentrations, rs1801133 reached genome-wide significance (pmeta = 4.36 × 10−13; Figure 1C; Figure S1). This SNP is the C677T polymorphism in the MTHFR gene (Figure S4). Although this polymorphism was highly associated with homocysteine concentrations in both InCHIANTI (β = 4.2, p = 7.24 × 10−12) and SardiNIA (β = 2.2, p = 7.24 × 10−6), there was no evidence of association in BLSA (β = 0.36, p = 0.819). There was significant evidence of heterogeneity (χ2 = 18.11, p = 1.2 × 10−4) between the studies. The most likely source of these differences is the higher folate status in the United States due to food fortification. This compensation of 677C→T effect by folate status has been described in other studies.44,45
The second and third SNPs in the homocysteine analyses were rs11041321 (Synaptotagmin IX (SYT9 [GeneID 143425]) p = 2.4 × 10−6; Figure S4) and rs2713280 (no nearby genes; p = 5.41 × 10−6), respectively. Synaptotagmins are calcium sensors that regulate exocytosis and have important roles in neurotransmission.46,47 Homocysteine has been implicated in the pathogenesis of neurological conditions such as dementia (MIM 600274). In light of the potential role of SYT9 in neurotransmission, rs11041321 was tested for replication but the association was not confirmed in Progetto nutrizione study.
No SNPs reached genome-wide significance in the meta-analysis of folate concentrations (Figure S1). The most significant rs1999594 (pmeta = 1.06 × 10−7) is located ∼100 kb from MTHFR (Figure S5). Because this SNP is in partial LD rs1801133 (r2 = 0.2 in the HapMAP CEU sample) and did not reach genome-wide significance, it was tested for replication. The second SNP was rs153734 in the 3′ untranslated region of the prickle-like homolog 2 gene, (PRICKLE2 [MIM 608501], Figure S5). In mice, PRICKEL2 homolog is continuously expressed in postmitotic neurons in early embryogenesis and has been implicated in neuron formation during brain development.48 For this observation, PRICKLE2 was a good candidate gene in the context of folate deficiency and NTD. This SNP was tested for replication in the Progetto nutrizione study, but the association was not confirmed.
Several studies have investigated the associations of genes other than MTHFR in the OCM pathway and circulating B vitamins and homocysteine concentrations. We examine the associations of polymorphisms located in ten candidate genes (MTHFR and MTR [MIM 156570], MTRR [MIM 602568], MTHFD1 [MIM 172460], BHMT [MIM 602888], CBS [MIM 236200], TCN2 [MIM 275350], PON1 [MIM 168820], DHFR [MIM 126060], and FOLH1 [MIM 600934]) in the current analyses. None of these genes were associated with vitamin B6 or vitamin B12 (data not shown). Variants in BHMT (rs651852, p = 0.003), FOLH1 (rs202700, p = 0.0008), and CBS (rs2124459, p = 0.0034) showed the greatest evidence for significance association with folate concentrations (Table S1). For homocysteine, MTR (rs12060264, p = 0.0005), MTRR (rs7703033, p = 0.005), and CBS (rs6586282, p = 0.0002) showed the greatest evidence of association (Table S1).
In summary, we report two variants associated with vitamin B6 and vitamin B12 concentrations. In addition to MTHFR, several genes in the OCM pathway were associated with circulating folate and homocysteine. These SNPs may be important markers to identify people at risk for life-long low vitamin and high homocysteine levels. In particular, this may be relevant to persons at risk for vitamin deficiencies such as the elderly in nursing homes. Future studies may investigate the potentially additive nature of multiple polymorphisms of genes in the OCM pathway on homocysteine concentrations and test the hypothesis that polymorphisms in these genes are risk factors for subclinical B vitamin deficiency and its consequences.
Acknowledgments
The InCHIANTI study baseline (1998-2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336); the InCHIANTI Follow-up 1 (2001–2003) was funded by the U.S. National Institute on Aging (Contracts: N.1-AG-1-1 and N.1-AG-1-2111); the InCHIANTI Follow-ups 2 and 3 studies (2004-2010) were financed by the U.S. National Institute on Aging (Contract: N01-AG-5-0002). BLSA and InCHIANTI were supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health (NIH), Baltimore, Maryland. A portion of that support was through a R&D contract with MedStar Research Institute. The SardiNIA team was supported by Contract NO1-AG-1-2109 from the NIA. The replication experiments were supported by a grant from Genopolis government FIRB project (RBLA038RMA_008).
Supplemental Data
Document S1. Five Figures and One Table
Web Resources
The URLs for data presented herein are as follows:
- EIGENSTRAT software, http://genepath.med.harvard.edu/∼reich/EIGENSTRAT.htm
- GAINQC Package, http://www.sph.umich.edu/csg/abecasis/GainQC/
- Genome browser, http://genome.ucsc.edu/
- International HapMap Project, http://www.hapmap.org/
- Markov Chain Haplotyping Package, http://www.sph.umich.edu/csg/abecasis/mach/ index.html
- MERLIN Package, http://www.sph.umich.edu/csg/abecasis/Merlin/index.html
- METAL Package, http://www.sph.umich.edu/csg/abecasis/metal/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/sites/Omim
- The R project for Statistical Computing, http://www.r-project.org/
References
- 1.Haggarty P. B-vitamins, genotype and disease causality. Proc. Nutr. Soc. 2007;66:539–547. doi: 10.1017/S0029665107005861. [DOI] [PubMed] [Google Scholar]
- 2.Mason J.B. Biomarkers of nutrient exposure and status in one-carbon (methyl) metabolism. J. Nutr. 2003;133(Suppl 3):941S–947S. doi: 10.1093/jn/133.3.941S. [DOI] [PubMed] [Google Scholar]
- 3.Botto L.D., Moore C.A., Khoury M.J., Erickson J.D. Neural-tube defects. N. Engl. J. Med. 1999;341:1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- 4.Daly L.E., Kirke P.N., Molloy A., Weir D.G., Scott J.M. Folate levels and neural tube defects. Implications for prevention. JAMA. 1995;274:1698–1702. doi: 10.1001/jama.1995.03530210052030. [DOI] [PubMed] [Google Scholar]
- 5.Rimm E.B., Willett W.C., Hu F.B., Sampson L., Colditz G.A., Manson J.E., Hennekens C., Stampfer M.J. Folate and vitamin B6 from diet and supplements in relation to risk of coronary heart disease among women. JAMA. 1998;279:359–364. doi: 10.1001/jama.279.5.359. [DOI] [PubMed] [Google Scholar]
- 6.Tavani A., Pelucchi C., Parpinel M., Negri E., La Vecchia C. Folate and vitamin B(6) intake and risk of acute myocardial infarction in Italy. Eur. J. Clin. Nutr. 2004;58:1266–1272. doi: 10.1038/sj.ejcn.1601960. [DOI] [PubMed] [Google Scholar]
- 7.de Bree A., Verschuren W.M., Blom H.J., Nadeau M., Trijbels F.J., Kromhout D. Coronary heart disease mortality, plasma homocysteine, and B-vitamins: A prospective study. Atherosclerosis. 2003;166:369–377. doi: 10.1016/s0021-9150(02)00373-8. [DOI] [PubMed] [Google Scholar]
- 8.Friso S., Girelli D., Martinelli N., Olivieri O., Lotto V., Bozzini C., Pizzolo F., Faccini G., Beltrame F., Corrocher R. Low plasma vitamin B-6 concentrations and modulation of coronary artery disease risk. Am. J. Clin. Nutr. 2004;79:992–998. doi: 10.1093/ajcn/79.6.992. [DOI] [PubMed] [Google Scholar]
- 9.Giovannucci E., Stampfer M.J., Colditz G.A., Hunter D.J., Fuchs C., Rosner B.A., Speizer F.E., Willett W.C. Multivitamin use, folate, and colon cancer in women in the Nurses' Health Study. Ann. Intern. Med. 1998;129:517–524. doi: 10.7326/0003-4819-129-7-199810010-00002. [DOI] [PubMed] [Google Scholar]
- 10.Sharp L., Little J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: A HuGE review. Am. J. Epidemiol. 2004;159:423–443. doi: 10.1093/aje/kwh066. [DOI] [PubMed] [Google Scholar]
- 11.Beaudin A.E., Stover P.J. Folate-mediated one-carbon metabolism and neural tube defects: Balancing genome synthesis and gene expression. Birth Defects Res. C Embryo Today. 2007;81:183–203. doi: 10.1002/bdrc.20100. [DOI] [PubMed] [Google Scholar]
- 12.Lowering blood homocysteine with folic acid based supplements: Meta-analysis of randomised trials. Homocysteine Lowering Trialists' Collaboration. BMJ. 1998;316:894–898. [PMC free article] [PubMed] [Google Scholar]
- 13.Selhub J., Jacques P.F., Wilson P.W., Rush D., Rosenberg I.H. Vitamin status and intake as primary determinants of homocysteinemia in an elderly population. JAMA. 1993;270:2693–2698. doi: 10.1001/jama.1993.03510220049033. [DOI] [PubMed] [Google Scholar]
- 14.Wald D.S., Law M., Morris J.K. Homocysteine and cardiovascular disease: Evidence on causality from a meta-analysis. BMJ. 2002;325:1202. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Splaver A., Lamas G.A., Hennekens C.H. Homocysteine and cardiovascular disease: Biological mechanisms, observational epidemiology, and the need for randomized trials. Am. Heart J. 2004;148:34–40. doi: 10.1016/j.ahj.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 16.McCully K.S. Homocysteine, vitamins, and vascular disease prevention. Am. J. Clin. Nutr. 2007;86:1563S–1568S. doi: 10.1093/ajcn/86.5.1563S. [DOI] [PubMed] [Google Scholar]
- 17.Choi S.W., Friso S., Dolnikowski G.G., Bagley P.J., Edmondson A.N., Smith D.E., Mason J.B. Biochemical and molecular aberrations in the rat colon due to folate depletion are age-specific. J. Nutr. 2003;133:1206–1212. doi: 10.1093/jn/133.4.1206. [DOI] [PubMed] [Google Scholar]
- 18.Choi S.W., Friso S., Keyes M.K., Mason J.B. Folate supplementation increases genomic DNA methylation in the liver of elder rats. Br. J. Nutr. 2005;93:31–35. doi: 10.1079/bjn20041283. [DOI] [PubMed] [Google Scholar]
- 19.Jang H., Mason J.B., Choi S.W. Genetic and epigenetic interactions between folate and aging in carcinogenesis. J. Nutr. 2005;135:2967S–2971S. doi: 10.1093/jn/135.12.2967S. [DOI] [PubMed] [Google Scholar]
- 20.Frosst P., Blom H.J., Milos R., Goyette P., Sheppard C.A., Matthews R.G., Boers G.J., den Heijer M., Kluijtmans L.A., van den Heuvel L.P. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 21.Friso S., Choi S.W., Girelli D., Mason J.B., Dolnikowski G.G., Bagley P.J., Olivieri O., Jacques P.F., Rosenberg I.H., Corrocher R. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klerk M., Verhoef P., Clarke R., Blom H.J., Kok F.J., Schouten E.G. MTHFR 677C→T polymorphism and risk of coronary heart disease: A meta-analysis. JAMA. 2002;288:2023–2031. doi: 10.1001/jama.288.16.2023. [DOI] [PubMed] [Google Scholar]
- 23.Fredriksen A., Meyer K., Ueland P.M., Vollset S.E., Grotmol T., Schneede J. Large-scale population-based metabolic phenotyping of thirteen genetic polymorphisms related to one-carbon metabolism. Hum. Mutat. 2007;28:856–865. doi: 10.1002/humu.20522. [DOI] [PubMed] [Google Scholar]
- 24.Ferrucci L., Bandinelli S., Benvenuti E., Di Iorio A., Macchi C., Harris T.B., Guralnik J.M. Subsystems contributing to the decline in ability to walk: Bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J. Am. Geriatr. Soc. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 25.Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilia G., Chen W.M., Scuteri A., Orru M., Albai G., Dei M., Lai S., Usala G., Lai M., Loi P. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shock N.W., Greulich R.C., Costa P.T., Andres R., Lakatta E.G., Arenberg D., Tobin J.D. US Government Printing Office; Washington, DC: 1984. Normal Human Aging: The Baltimore Study of Aging. [Google Scholar]
- 28.Sofi F., Vecchio S., Giuliani G., Martinelli F., Marcucci R., Gori A.M., Fedi S., Casini A., Surrenti C., Abbate R. Dietary habits, lifestyle and cardiovascular risk factors in a clinically healthy Italian population: The ‘Florence’ diet is not Mediterranean. Eur. J. Clin. Nutr. 2005;59:584–591. doi: 10.1038/sj.ejcn.1602112. [DOI] [PubMed] [Google Scholar]
- 29.Henthorn P.S., Raducha M., Fedde K.N., Lafferty M.A., Whyte M.P. Different missense mutations at the tissue-nonspecific alkaline phosphatase gene locus in autosomal recessively inherited forms of mild and severe hypophosphatasia. Proc. Natl. Acad. Sci. USA. 1992;89:9924–9928. doi: 10.1073/pnas.89.20.9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mornet E. Hypophosphatasia: The mutations in the tissue-nonspecific alkaline phosphatase gene. Hum. Mutat. 2000;15:309–315. doi: 10.1002/(SICI)1098-1004(200004)15:4<309::AID-HUMU2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 31.Yuan X., Waterworth D., Perry J.R., Lim N., Song K., Chambers J.C., Zhang W., Vollenweider P., Stirnadel H., Johnson T. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson B.B., O'Brien H., Griffin G.E., Mollin D.L. Hydrolysis of pyridoxal-5′-phosphate in plasma in conditions with raised alkaline phosphate. Gut. 1980;21:192–194. doi: 10.1136/gut.21.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Abecasis G.R. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 34.Hazra A., Kraft P., Selhub J., Giovannucci E.L., Thomas G., Hoover R.N., Chanock S.J., Hunter D.J. Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat. Genet. 2008;40:1160–1162. doi: 10.1038/ng.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly R.J., Rouquier S., Giorgi D., Lennon G.G., Lowe J.B. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J. Biol. Chem. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 36.Watkins W.M. Biochemistry and genetics of the ABO, Lewis, and P blood group systems. Adv. Hum. Genet. 1980;10:1–136. doi: 10.1007/978-1-4615-8288-5_1. 379–185. [DOI] [PubMed] [Google Scholar]
- 37.Boren T., Falk P., Roth K.A., Larson G., Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 38.Ikehara Y., Nishihara S., Yasutomi H., Kitamura T., Matsuo K., Shimizu N., Inada K., Kodera Y., Yamamura Y., Narimatsu H. Polymorphisms of two fucosyltransferase genes (Lewis and Secretor genes) involving type I Lewis antigens are associated with the presence of anti-Helicobacter pylori IgG antibody. Cancer Epidemiol. Biomarkers Prev. 2001;10:971–977. [PubMed] [Google Scholar]
- 39.Carmel R., Perez-Perez G.I., Blaser M.J. Helicobacter pylori infection and food-cobalamin malabsorption. Dig. Dis. Sci. 1994;39:309–314. doi: 10.1007/BF02090202. [DOI] [PubMed] [Google Scholar]
- 40.Kaptan K., Beyan C., Ural A.U., Cetin T., Avcu F., Gulsen M., Finci R., Yalcin A. Helicobacter pylori–is it a novel causative agent in Vitamin B12 deficiency? Arch. Intern. Med. 2000;160:1349–1353. doi: 10.1001/archinte.160.9.1349. [DOI] [PubMed] [Google Scholar]
- 41.Serpa J., Mendes N., Reis C.A., Santos Silva L.F., Almeida R., Le Pendu J., David L. Two new FUT2 (fucosyltransferase 2 gene) missense polymorphisms, 739G→A and 839T→C, are partly responsible for non-secretor status in a Caucasian population from Northern Portugal. Biochem. J. 2004;383:469–474. doi: 10.1042/BJ20040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanner S.M., Li Z., Bisson R., Acar C., Oner C., Oner R., Cetin M., Abdelaal M.A., Ismail E.A., Lissens W. Genetically heterogeneous selective intestinal malabsorption of vitamin B12: Founder effects, consanguinity, and high clinical awareness explain aggregations in Scandinavia and the Middle East. Hum. Mutat. 2004;23:327–333. doi: 10.1002/humu.20014. [DOI] [PubMed] [Google Scholar]
- 43.Tanner S.M., Li Z., Perko J.D., Oner C., Cetin M., Altay C., Yurtsever Z., David K.L., Faivre L., Ismail E.A. Hereditary juvenile cobalamin deficiency caused by mutations in the intrinsic factor gene. Proc. Natl. Acad. Sci. USA. 2005;102:4130–4133. doi: 10.1073/pnas.0500517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis S.J., Ebrahim S., Davey Smith G. Meta-analysis of MTHFR 677C->T polymorphism and coronary heart disease: Does totality of evidence support causal role for homocysteine and preventive potential of folate? BMJ. 2005;331:1053. doi: 10.1136/bmj.38611.658947.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q.H., Botto L.D., Gallagher M., Friedman J.M., Sanders C.L., Koontz D., Nikolova S., Erickson J.D., Steinberg K. Prevalence and effects of gene-gene and gene-nutrient interactions on serum folate and serum total homocysteine concentrations in the United States: Findings from the third National Health and Nutrition Examination Survey DNA Bank. Am. J. Clin. Nutr. 2008;88:232–246. doi: 10.1093/ajcn/88.1.232. [DOI] [PubMed] [Google Scholar]
- 46.Zhu D., Zhou W., Liang T., Yang F., Zhang R.Y., Wu Z.X., Xu T. Synaptotagmin I and IX function redundantly in controlling fusion pore of large dense core vesicles. Biochem. Biophys. Res. Commun. 2007;361:922–927. doi: 10.1016/j.bbrc.2007.07.083. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-Chacon R., Konigstorfer A., Gerber S.H., Garcia J., Matos M.F., Stevens C.F., Brose N., Rizo J., Rosenmund C., Sudhof T.C. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 48.Okuda H., Miyata S., Mori Y., Tohyama M. Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth. FEBS Lett. 2007;581:4754–4760. doi: 10.1016/j.febslet.2007.08.075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Five Figures and One Table