Use of Spoligotyping and Large Sequence Polymorphisms To Study the Population Structure of the Mycobacterium tuberculosis Complex in a Cohort Study of Consecutive Smear-Positive Tuberculosis Cases in The Gambia (original) (raw)
Abstract
Mycobacterium africanum, first described in Senegal in 1968, causes up to half of the smear-positive pulmonary tuberculosis cases in West Africa, but it has not been found in other geographical areas except among recent West African migrants. The reasons for the geographic restriction of M. africanum are unknown. We used molecular tools to determine the population structure of the Mycobacterium tuberculosis complex in a cohort study of consecutive smear-positive tuberculosis cases in The Gambia. We collected and genotyped 386 clinical isolates using spoligotype analysis and PCRs for large sequence polymorphisms (LSPs) and compared the genotype patterns to the patterns in an international database. The results of spoligotyping and LSP analysis for the study population were also compared to determine the correlation between them. The main lineages within the Mycobacterium tuberculosis complex identified in The Gambia included M. africanum type I (38.4%), characterized by an LSP in region of difference 702 (RD702; West African type 2). Among the M. tuberculosis sensu stricto isolates, lineages characterized by RD182 and by RD174 were the most common. We also detected a gradient in the prevalence of M. africanum that extended from neighboring Guinea-Bissau. The genotypic diversity of the spoligotype patterns was greater among the isolates of M. africanum than among the isolates of M. tuberculosis. We postulate that M. africanum became endemic in West Africa first, before the introduction of different lineages within M. tuberculosis sensu stricto.
Molecular epidemiologic techniques have been used to compare isolates within the Mycobacterium tuberculosis complex and have clarified many aspects of tuberculosis (TB) transmission, such as the importance of ongoing transmission in low-incidence countries (21) and the limited importance of within-household transmission in high-incidence settings (26). In recent years, genotyping techniques have also been applied to infer the phylogenetic relationships between lineages within the M. tuberculosis complex (2, 17). Different genetic markers have different rates of evolutionary change. Genetic markers such as large sequence polymorphisms (LSPs) that change slowly are essentially limited to analysis by phylogenetic studies (11, 24). On the other hand, spoligotyping, which is based on the polymorphism of the direct repeat region and which has the ability to detect changes with a faster molecular clock, has been used for both phylogeny and molecular epidemiological studies of M. tuberculosis (10).
The population structure of the M. tuberculosis complex in The Gambia or Senegal has not been described since biochemical methods were used to demonstrate that M. africanum caused approximately 20% of the TB cases in Senegal during the 1970s (9). Since the advent of molecular methods, the strains previously classified as East African M. africanum type II have been reclassified as M. tuberculosis sensu stricto (22). West African M. africanum type I is now subdivided into West African M. africanum type 1, characterized by region of difference (RD) 711 (RD711) (10) and reported mostly around the Gulf of Guinea (19), and West African M. africanum type 2, characterized by RD702 and prevalent mostly in western West Africa (10, 18).
We used molecular methods to determine the genetic population structure of the M. tuberculosis complex in The Gambia, a country with an incidence of sputum smear-positive TB of approximately 80/100,000 population per year (1) and rising. We compared spoligotyping with LSP analysis to determine if the two methods gave concordant results and then combined spoligotyping with LSP analysis to infer phylogenetic relationships and to compare the genetic diversity between different lineages of the M. tuberculosis complex.
MATERIALS AND METHODS
Collection of mycobacterial isolates.
Consecutive sputum smear-positive TB patients from the Medical Research Council (MRC) Outpatient Department and the major Gambian government TB clinics in the greater Banjul area in The Gambia, were enrolled in the study if they provided informed consent. Patients were eligible if they were diagnosed with smear-positive TB, were over 15 years of age, and had not been previously treated for TB. The enrolled patients received voluntary counseling and testing for human immunodeficiency virus (HIV) infection. Those who tested HIV seropositive were referred for HIV care, including free antiretroviral therapy, which became available in The Gambia in 2004. The study was approved by the joint Gambian Government-MRC ethics committee and the Institutional Review Board at Stanford University.
Mycobacteriology.
The sputum samples were stained with auramine and by the Ziehl-Neelsen method. Sputum was decontaminated by using _N_-acetyl cysteine-NaOH before culture in Bactec vials (Becton Dickinson) and on paired Lowenstein-Jensen slopes. Positive cultures were confirmed by the Ziehl-Neelsen smear method and were stored in glycerol at −70°C.
Mycobacterial genotyping.
Stored isolates were grown in Middlebrook 7H9 broth with oleic acid-dextrose-albumin-catalase supplement for DNA extraction by previously published methods (25). After spectrophotometry, we used 10 ng of DNA for spoligotype analysis (16) with commercially available membranes (Isogen Biosciences, Maarssen, The Netherlands). Isolates that repeatedly failed to yield a spoligotype pattern were analyzed, as described previously (13), for classification as atypical mycobacteria versus members of the M. tuberculosis complex. Spoligotype films were scanned and classified by using software designed in the Matlab program (Mathworks, Natick, MA) (D. Jeffries, unpublished data), followed by manual editing and confirmation. Each spoligotype pattern was classified into a binary code, and the result was entered in a Microsoft (Redmond, WA) Access database.
The presence or absence of lineage-defining LSPs was assessed by using the methods and the flanking primers described for 100 clinical isolates in San Francisco, CA (10, 24). We used a hierarchical screening sequence, first testing one isolate representative of each of the different spoligotype patterns to detect different lineage-defining LSPs. Isolates with a certain lineage-defining LSP were not tested for further LSPs. We subsequently screened all isolates with a similar spoligotype pattern to determine whether they had identical lineage-defining LSPs. If spoligotype analysis did not result in a clear pattern of spacers, the isolate underwent repeat spoligotyping. If a spoligotype pattern did not fit the prior association of that pattern with a certain RD, both the spoligotype analysis and the PCR for the RD were repeated.
Data analysis.
The phylogenetic tree was constructed by first classifying isolates by their lineage-defining deletion on the basis of the LSP results, followed by the construction of subbranches within each lineage by using the minimum-spanning-trees method with the spoligotype data in GelCompar software (Bionumerics; Applied Maths, Kortrijk, Belgium). The following rules were applied to tree construction: first, link the maximum number of single-locus variants, and second, use the maximum number with double (or more)-locus variants. The third rule was to include the maximum number of entries, and the fourth rule was that no hypothetical types were allowed. Spacers commonly missing from the spoligotype patterns by lineage were identified by visual comparison and consensus (3). The Hunter-Gaston discriminatory index was used to calculate the level of discrimination of each typing method (14).
The individual spoligotype patterns were compared with an updated in-house proprietary version of the SpolDB4 database at the Institut Pasteur de Guadeloupe, named SITVIT2 (the initial version is available at http://www.pasteur-guadeloupe.fr:8081/SITVITDemo). At the time of this comparison, the updated in-house proprietary version contained a total of 2,808 shared types (or spoligotype international types [SITs]) corresponding to 63,473 clinical isolates from 122 countries of isolation and 160 countries of origin. A shared type, or SIT, is defined as a spoligotype pattern common to two or more M. tuberculosis isolates. Major phylogenetic clades were assigned according to the signatures provided in SpolDB4 (5), which defines 62 genetic lineages or sublineages. The reader is referred to the original paper for a detailed description. Some major lineages include M. bovis (BOV) and its 4 sublineages, M. africanum (AFRI) and its 3 sublineages, the Central Asian (CAS) clade and its 2 sublineages, the East African-Indian (EAI) clade and its 9 sublineages, the Haarlem (H) clade and its 4 sublineages, the Latin American-Mediterranean (LAM) clade and its 12 sublineages, the Manu family and its 3 sublineages, the IS_6110_-low banding X clade and its 3 sublineages, and an ill-defined T clade and its 13 sublineages.
Within the SITVIT2 database, we compared the spoligotype patterns of M. africanum (missing spacers 7 to 9 and 39) with the patterns of the Euro-American lineage within M. tuberculosis sensu stricto (missing spacers 33 to 36). We compared both the number of isolates and the number of different spoligotype patterns between these groups.
We compared the demographic variables and the HIV test results for the TB cases by the mycobacterial lineage of the patient's isolate using the χ2 test of proportions. We compared several groups of patients, according to the lineage of mycobacteria in their isolate, if there was information on at least 10 patients in a group.
RESULTS
Sampling frame and mycobacterial genotyping.
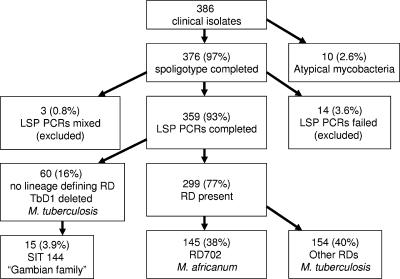
The isolates were collected from 386 consecutive smear-positive TB patients residing in the greater Banjul area, which is a semiurban coastal area of The Gambia and which is where 60% of Gambians live. Among the 386 isolates, 10 did not yield a spoligotype pattern and were confirmed to be atypical mycobacteria, as 16S RNA but not any of the M. tuberculosis complex-specific regions could be amplified from them (Fig. 1). Among the remaining 376 isolates, 93 spoligotype patterns were identified. For LSP analysis, 14 isolates for which amplification failed were excluded, as were 3 isolates with PCR results suggestive of mixed DNA, leaving a total of 359 isolates for LSP analysis. These were grouped by LSPs into 11 lineages (Fig. 2), with the exception of 60 strains that could not be classified in a precise lineage by LSP analysis alone; TbD1 was concomitantly deleted from all strains (i.e., they were modern strains), and the strains were tentatively categorized as “H37Rv-like.”
FIG. 1.
Genotyping results for clinical TB isolates in The Gambia. TbD1, region present in ancestral strains of the M. tuberculosis complex.
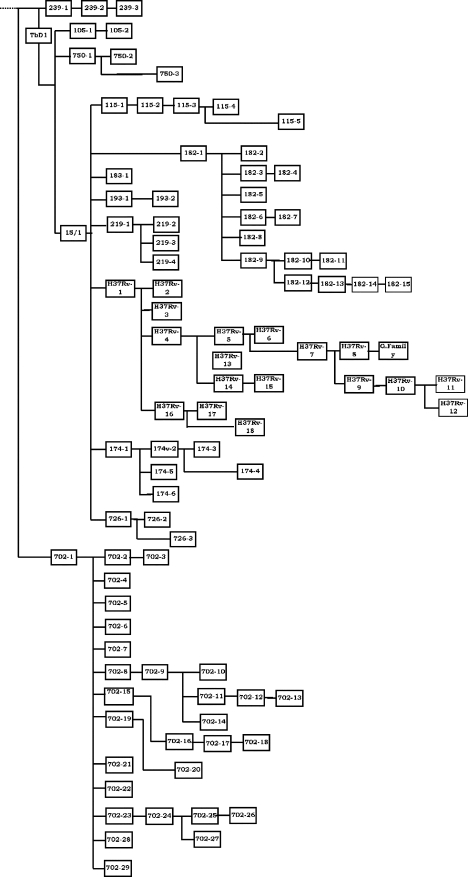
FIG. 2.
Population structure of the M. tuberculosis complex in The Gambia determined by combining the results of LSP analysis and spoligotyping.
Prevalence of the different lineages within the M. tuberculosis complex.
The main lineages identified in The Gambia include RD702 (M. africanum type I, West African type 2) (10, 18), which accounted for 38.6% of all smear-positive TB cases; and within M. tuberculosis sensu stricto, the main lineages identified were RD182, RD174, and RD219. We did not detect any M. bovis, M. canettii, or other subspecies of the M. tuberculosis complex. We refer to M. africanum type I, West African type 2, when using “M. africanum,” unless otherwise specified. Table 1 lists the different spoligotype patterns within each lineage, their classification in SITVIT2, their prevalence, and their spoligotype signatures. M. africanum was the most prevalent genotype and also showed the most spoligotype diversity, with 29 different patterns. RD105, the Beijing family lineage which has a worldwide distribution, caused 3% of the smear-positive TB cases in The Gambia. RD750 was found to have two subtypes, one with a PCR product larger than the intact region.
TABLE 1.
Spoligotype patterns and SITs for clusters of M. tuberculosis complex isolates defined by RDs
Concordance between LSPs and spoligotype analysis.
Figure S4 in the supplemental material shows the phylogenetic tree in Fig. 2 (see Figure S4a in the supplemental material) in comparison with a tree prepared by using the same minimum-spanning method but based solely on spoligotype patterns, without LSPs (see Figure S4b in the supplemental material). Tree 4b is markedly different from tree 4a because several LSPs cluster in succession, on the basis of the spoligotype patterns. This finding confirms that spoligotype analysis by itself does not provide a robust phylogeny. In contrast, the subspecies determination by spoligotype pattern between the two distinct members of the M. tuberculosis complex identified in The Gambia, M. africanum (lacking spacers 7 to 9 and 39) and M. tuberculosis, corresponds 100% with the phylogeny defined by LSP RD702.
Some LSPs could not be predicted from the spoligotype patterns (Table 1), whereas some LSPs were consistently correlated with a particular spoligotype pattern, such as RD182, which always lacked spacers 31 and 33 to 36 in the spoligotype pattern. Some LSPs occurred in only a fraction of the isolates with a given spoligotype pattern; e.g., RD219 was identified in 52% of the isolates whose spoligotype pattern lacked spacers 33 to 36, whereas no lineage-defining LSPs were identified in the remainder of the isolates with the same spoligotype pattern. Similarly, some isolates had spoligotype patterns that could be categorized into multiple, different LSP lineages; e.g., isolates with the spoligotype pattern missing spacers 21 to 24 and 33 to 36 (LAM according to SpolDB4) can belong to lineage RD115 or RD174 (Euro-American lineage) or do not belong to either lineage. LSP RD724, which is adjacent to an IS_6110_ insertion element (10), was not a unique event/lineage-defining polymorphism. It occurred in isolates from different phylogenetic branches.
The Hunter-Gaston discriminatory indices were 0.78 for typing by LSPs and 0.94 for spoligotype analysis, confirming the higher discriminatory power of spoligotype analysis (Fig. 2).
Diversity of spoligotype patterns.
A total of 92 distinct spoligotype patterns were obtained from 359 isolates. By comparison with the patterns in the SITVIT2 international spoligotype database, 338/359 (94.1%) of the isolates had a spoligotype pattern that matched a spoligotype pattern that had previously been placed in the database. The spoligotype patterns of 313 (87.2%) isolates were clustered in 46 clusters, each of which contained between 2 and 80 isolates. Of the 46 (12.8%) isolates with unclustered spoligotype patterns, 21 were novel (i.e., they were not represented in the SITVIT2 database) and 25 were unique spoligotype patterns in our study from The Gambia that matched a spoligotype pattern in the database. In total, addition of the spoligotype patterns of these 359 isolates resulted in 14 new shared-type patterns from 32 different isolates added to the international database. A group of 15 isolates with the same spoligotype pattern had sporadically been identified in the SITVIT2 database. We labeled this group of isolates, which lacked TbD1 but which had no other lineage-defining deletions, the “Gambian family.”
Gradient of M. africanum prevalence in West Africa.
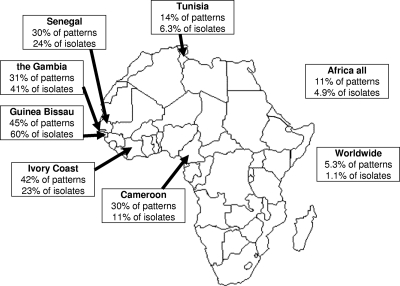
The high prevalence and the high degree of genetic diversity of M. africanum suggest the hypothesis that M. africanum has been evolving in Africa for quite some time. Consistent with this hypothesis, only a subset of the Euro-American lineages of the M. tuberculosis complex occur at high frequencies in The Gambia (i.e., RD174 or LAM and RD182 or Haarlem) and were most likely introduced since the beginning of European exploration and colonization. To test this hypothesis more formally, we compared the distribution and genetic diversity of the M. africanum and the Euro-American lineages within M. tuberculosis sensu stricto (as defined earlier) in our strain collection and in the international SITVIT2 database. We compared genotyped isolates from The Gambia and genotyped isolates from the rest of the world at three different levels: individual countries, the African continent as a whole, and worldwide. We compared both the variability in spoligotype patterns (each pattern was counted once) and the variability in the total number of isolates within each subspecies (Fig. 3). A statistical analysis by use of the χ2 test showed that the proportion of M. africanum spoligotype patterns within the _M. africanum_-M. tuberculosis Euro-American group in The Gambia was 31%, whereas the proportions were 45.2% in Guinea-Bissau (P < 0.08), 42.1% in Ivory Coast (P < 0.3), 30.3% in Senegal (P < 0.9), 30% in Cameroon (P < 0.9), 13.8% in Tunisia (P < 0.001), 10.9% for the African continent (P < 0.001), and 5.3% at the worldwide level (P < 0.001).
FIG. 3.
Distribution of M. africanum type I, West African type 2, relative to that of the Euro-American lineage of M. tuberculosis sensu stricto in the SITVIT2 database. Patterns, the proportion of different spoligotype patterns within the lineage M. africanum (with signature spacers 7 to 9 and 39 absent and at least one of the spacers from spacers 33 to 36 present) relative to the pattern of the Euro-American lineage of M. tuberculosis (with spacers 33 to 36 absent and at least one of the spacers from spacers 7 to 9 and 39 present); isolates, the proportion of isolates with the signature for M. africanum described above versus the signature for the Euro-American lineage of M. tuberculosis. (Map courtesy of http://www.theodora.com/maps, used with permission.)
The proportion of M. africanum isolates within the _M. africanum_-M. tuberculosis Euro-American group in The Gambia was 41.02%, whereas the proportions were 59.91% in Guinea-Bissau (P < 0.001), 24.36% in Senegal (P < 0.01), 22.91% in Ivory Coast (P < 0.02), 10.95% in Cameroon (P < 0.001), 6.29% in Tunisia (P < 0.001), 4.89% for the African continent (P < 0.001), and 1.12% at the worldwide level (P < 0.001). Taken together, these data show that M. africanum is particularly frequent and much more genetically diverse than the Euro-American lineage in The Gambia and elsewhere in West Africa compared to its frequency of occurrence and diversity in other regions of the world.
Molecular epidemiological associations.
Finally, we looked for clinical and epidemiological associations with the different strain lineages. By univariate analysis, M. africanum (RD702)-infected patients were more likely to be older, male, and HIV infected than those infected with other lineages (Table 2). However, none of the associations were statistically significant.
TABLE 2.
Epidemiological associations by lineage
| Lineage (no. of isolates) | Median age (yr) | Sex (% male) | HIV (% positive) |
|---|---|---|---|
| RD105 (10) | 24.5 | 70 | 0 |
| RD174 (44) | 28 | 59 | 9.3 |
| RD182 (53) | 28 | 66 | 0 |
| RD219 (18) | 26.5 | 83 | 6.7 |
| RD702 (138) | 30 | 72 | 13 |
| RD726 (13) | 27 | 77 | 0 |
| H37Rv-like (45) | 29 | 69 | 10 |
| Gambian family (15) | 26 | 67 | 0 |
| Other (23) | 32 | 61 | 5.9 |
| Total (359) | 29 | 69 | 8.4 |
| Pa | 0.76 | 0.66 | 0.17 |
DISCUSSION
This study identified M. africanum as the cause of 39% of the smear-positive pulmonary TB cases in The Gambia, with the remainder (61%) being caused by lineages within M. tuberculosis sensu stricto. DNA from both M. africanum and M. tuberculosis sensu stricto was recovered from Egyptian mummies (28), yet today M. africanum isolates are rarely encountered outside of West Africa, except among first-generation immigrants (8). This lack of spread of M. africanum, despite large migrations, such as through the slave trade to the New World, which lasted several centuries, suggests that M. africanum has established a specific geographic niche in West Africa. Our findings confirm the specificity of the M. africanum lineage to the African continent, with a decreasing gradient of prevalence from West to East. The focal point of endemicity is the region around Guinea-Bissau, The Gambia, and Senegal. Moreover, the greater genetic variability in this geographic region relative to the genetic variability of different lineages within M. tuberculosis sensu stricto supports the hypothesis that M. africanum established itself in West Africa before the Euro-American M. tuberculosis lineage was introduced during European exploration and colonization (12, 23). However, there are alternative explanations for the high degree of genetic diversity among M. africanum strains. For example, M. africanum might not be primarily a human pathogen and as yet unknown animal or environmental reservoirs exist. If this explanation is true, M. africanum could be under very different selective pressures, which could partially explain the observed greater spoligotype diversity. However, M. africanum is a very successful pathogen of humans in West Africa, causing up to 45% of the disease cases and having a clear capacity to transmit from human to human (6). We previously found that M. africanum was more strongly associated with HIV infection than M. tuberculosis, all lineages combined (7), but this association was not significant in the present study when we compared the HIV prevalence across the different lineages within the M. tuberculosis complex (Table 2). We found the clinical and radiographic presentations and the outcome of disease for patients infected with M. africanum to be indistinguishable from those for patients infected with M. tuberculosis, all lineages combined (5), although _M. africanum_-infected patients and their contacts were less likely to mount a response to the _M. tuberculosis-_specific protein ESAT-6 (6-kDa early secreted antigenic target) (6). Further studies will be needed to understand the striking geographical restriction in the prevalence of M. africanum. In addition, DNA sequence-based sequencing approaches will provide a more robust measure of the relative genetic diversity between different mycobacterial lineages.
In addition to M. africanum, many other global lineages within M. tuberculosis sensu stricto were encountered in The Gambia, predominantly those with genomic deletions in RD174, RD182, and RD219. Strains with the RD174 deletion were found to have an increased secondary case rate ratio in San Francisco, CA (K. DeRiemer, unpublished data). RD174 is located within the dormancy regulon of the M. tuberculosis genome (27) and has a spoligotype signature of the LAM clade (Table 1), previously identified in Guinea-Bissau (15). The spoligotype pattern that correlates with an LSP in RD726 has previously been described in Cameroon (20). In Table 1, it corresponded to spoligotype pattern SIT61 (absence of spacers 23 to 25 and 33 to 36) in 9/13 strains, which is the signature of the Cameroon family (20), also termed LAM10-CAM in SpolDB4 (3). By comparing the genotypes of recent TB cases to the genotypes of historical specimens in Cameroon by biochemical methods, TB caused by isolates with this specific spoligotype pattern appear to have replaced M. africanum type I, West African type 1. One spoligotype pattern within lineage RD174 was found in lineage RD115 and in a different isolate that lacked a specific lineage-defining LSP. These shared patterns likely correspond to the pattern of the ancestral strain common to both RD115 and RD174 or reflect homoplasy, i.e., convergence to the same spoligotype pattern through different evolutionary pathways (3). Thus, the spoligotype method is best complemented by a genotyping method with a slower molecular clock, such as LSPs.
The LSPs used for this study were identified in a set of clinical isolates in San Francisco, CA, and isolates of African origin were underrepresented. As a result, a number of Gambian strains, such as the Gambian family, clustered without a lineage-defining deletion in the H37Rv-like group. The genomewide identification of large and small polymorphisms in African strains would likely identify novel genotypes circulating on the African continent. The identification of lineages within the M. tuberculosis complex that are particularly well adapted to HIV-infected hosts, such as M. africanum (7), may prove important in African countries, where the incidence of coinfection with tuberculosis and HIV is increasing (4). The genomewide identification of polymorphisms in strains commonly isolated in Africa will require investments to improve genotyping facilities in the countries where outbreaks of M. tuberculosis and M. africanum prevail (3). To our knowledge, our work in The Gambia is the first effort in West Africa to perform the laboratory work involved with molecular genotyping in the country of origin of the isolates.
In conclusion, the current description of the population structure of the M. tuberculosis complex in The Gambia confirms the high prevalence of M. africanum in West Africa, which accounts for 38.4% of the sputum smear-positive TB cases. The higher degree of diversity of the spoligotype patterns within the subspecies M. africanum supports the hypothesis that M. africanum became endemic in West Africa first, after which several lineages within subspecies M. tuberculosis sensu stricto were introduced.
Supplementary Material
[Supplemental material]
Acknowledgments
This work was supported by MRC core funds and by NIH grant TW006083 awarded to B.C.D.J. The work performed in Nalin Rastogi's laboratory was partially funded by the European Regional Development Fund, European Commission (ERDF/FEDER, A34-05).
We thank the participants and TB diagnostics staff, in particular, Neneh Sallah and Ebou Mbowe. We also thank colleagues at the Institut Pasteur de Guadeloupe for various aspects regarding genotyping analysis, Christophe Sola for an initial comparison of preliminary results using the SpolDB4 database, and Christophe Demay for the entry of new data from the updated data set into the SITVIT2 database.
Footnotes
▿
Published ahead of print on 4 February 2009.
REFERENCES
- 1.Anonymous. 2004. National Leprosy and Tuberculosis Programme. Annual Report. The Gambia. KNCV Tuberculosis Foundation, The Hague, The Netherlands.
- 2.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 993684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 1631009-1021. [DOI] [PubMed] [Google Scholar]
- 5.de Jong, B. C., P. C. Hill, A. Aiken, D. J. Jeffries, A. Onipede, P. M. Small, R. A. Adegbola, and T. P. Corrah. 2007. Clinical presentation and outcome of tuberculosis patients infected by M. africanum versus M. tuberculosis. Int. J. Tuberc. Lung Dis. 11450-456. [PubMed] [Google Scholar]
- 6.de Jong, B. C., P. C. Hill, R. H. Brookes, S. Gagneux, D. J. Jeffries, J. K. Otu, S. A. Donkor, A. Fox, K. P. McAdam, P. M. Small, and R. A. Adegbola. 2006. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J. Infect. Dis. 1931279-1286. [DOI] [PubMed] [Google Scholar]
- 7.de Jong, B. C., P. C. Hill, R. H. Brookes, J. K. Otu, K. L. Peterson, P. M. Small, and R. A. Adegbola. 2005. Mycobacterium africanum: a new opportunistic pathogen in HIV infection? AIDS 191714-1715. [DOI] [PubMed] [Google Scholar]
- 8.Desmond, E., A. T. Ahmed, W. S. Probert, J. Ely, Y. Jang, C. A. Sanders, S. Y. Lin, and J. Flood. 2004. Mycobacterium africanum cases, California. Emerg. Infect. Dis. 10921-923. [DOI] [PubMed] [Google Scholar]
- 9.Diop, S., D. de Medeiros, G. de Medeiros, R. Baylet, and M. Sankale. 1976. Incidence and geographic distribution of Mycobacterium africanum in Senegal. Bull. Soc. Med. Afr. Noire Lang Fr. 2150-56. (In French.) [PubMed] [Google Scholar]
- 10.Gagneux, S., K. DeRiemer, T. Van, M. Kato-Maeda, B. C. de Jong, S. Narayanan, M. Nicol, S. Niemann, K. Kremer, M. C. Gutierrez, M. Hilty, P. C. Hopewell, and P. M. Small. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1032869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagneux, S., and P. M. Small. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7328-337. [DOI] [PubMed] [Google Scholar]
- 12.Hershberg, R., M. Lipatov, P. M. Small, H. Sheffer, S. Niemann, S. Homolka, J. C. Roach, K. Kremer, D. A. Petrov, M. W. Feldman, and S. Gagneux. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huard, R. C., L. C. de Oliveira Lazzarini, W. R. Butler, D. van Soolingen, and J. L. Ho. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 411637-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E. Svensson, F. Dias, B. I. Marklund, and S. B. Svenson. 1999. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin. Microbiol. 373872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150483-496. [DOI] [PubMed] [Google Scholar]
- 18.Mostowy, S., A. Onipede, S. Gagneux, S. Niemann, K. Kremer, E. P. Desmond, M. Kato-Maeda, and M. Behr. 2004. Genomic analysis distinguishes Mycobacterium africanum. J. Clin. Microbiol. 423594-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, P. Cunin, J. Thonnon, C. Sola, N. Rastogi, V. Vincent, and M. C. Gutierrez. 2003. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 412547-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, J. Thonnon, V. Vincent, and M. C. Gutierrez. 2004. Molecular characteristics of strains of the Cameroon family, the major group of Mycobacterium tuberculosis in a country with a high prevalence of tuberculosis. J. Clin. Microbiol. 425029-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 3301703-1709. [DOI] [PubMed] [Google Scholar]
- 22.Sola, C., N. Rastogi, M. C. Gutierrez, V. Vincent, R. Brosch, and L. Parsons. 2003. Is Mycobacterium africanum subtype II (Uganda I and Uganda II) a genetically well-defined subspecies of the Mycobacterium tuberculosis complex? J. Clin. Microbiol. 411345-1346. (Author reply, 41:1346-1348.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka, M. M., and A. R. Francis. 2006. Detecting emerging strains of tuberculosis by using spoligotypes. Proc. Natl. Acad. Sci. USA 10315266-15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 1014865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verver, S., R. M. Warren, Z. Munch, M. Richardson, G. D. van der Spuy, M. W. Borgdorff, M. A. Behr, N. Beyers, and P. D. van Helden. 2004. Proportion of tuberculosis transmission that takes place in households in a high-incidence area. Lancet 363212-214. [DOI] [PubMed] [Google Scholar]
- 27.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zink, A. R., C. Sola, U. Reischl, W. Grabner, N. Rastogi, H. Wolf, and A. G. Nerlich. 2003. Characterization of Mycobacterium tuberculosis complex DNAs from Egyptian mummies by spoligotyping. J. Clin. Microbiol. 41359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]