DIVERSITY-BASED, MODEL-GUIDED CONSTRUCTION OF SYNTHETIC GENE NETWORKS WITH PREDICTED FUNCTIONS (original) (raw)
. Author manuscript; available in PMC: 2009 Nov 1.
Published in final edited form as: Nat Biotechnol. 2009 Apr 19;27(5):465–471. doi: 10.1038/nbt.1536
SUMMARY
Engineering artificial gene networks from modular components is one of the major goals of synthetic biology. However, the construction of gene networks with predictable functions remains hampered by a lack of suitable components and the fact that assembled networks often require extensive, iterative retrofitting to work as intended. Here we present an approach that couples libraries of diversified components (synthesized with randomized non-essential sequence) with in silico modeling to guide predictable gene network construction without the need for post-hoc tweaking. We demonstrate our approach in S. cerevisiae by synthesizing regulatory promoter libraries and using them to construct feedforward loop networks with different predicted input-output characteristics. We then expand our method to produce a synthetic gene network acting as a predictable timer, modifiable by component choice. We utilize this network to control the timing of yeast sedimentation, illustrating how the plug-and-play nature of our design can be readily applied to biotechnology.
INTRODUCTION
Synthetic biology promises to revolutionize biotechnology by applying engineering principles to biological systems1. In less than a decade this field has already yielded technological applications, providing new avenues for drug manufacture2, 3, biofabrication4 and therapeutics5, 6, while also offering promises in alternative energy7. A major focus of the field is the synthesis of gene networks with predictable behavior8-10, either to endow cells with novel functions11-15 or provide study for analogous natural systems8, 16-19. Despite a booming community and notable successes, the basic task of assembling a predictable gene network from biomolecular parts remains a significant challenge and often takes many months before a desired network is realized20. To advance synthetic biology, it is essential to identify techniques that increase the predictability of gene network engineering and decrease the amount of hands-on molecular biology required to get a functional network up-and-running.
Current approaches of gene network construction typically use a small set of components plundered from different natural systems, which are then assembled and tested in vivo, often without guidance from a priori mathematical modeling13, 21. Networks rarely behave as intended the first time, usually because chosen parts have the correct function but lack the specific quantitative properties required. Even for those few synthetic biology studies which do utilize computational assistance22-25, in silico results have been mainly used for data interpretation, not for guiding design and assembly. Instead, in most projects an initial failed network is usually resolved over months of iterative retrofitting20, often by fine-tuning imperfect parts by mutation, by identifying alternative parts, or by adding on extra features to counterbalance the problems. Directed evolution has been shown to provide a short-cut through this phase21, but is complicated by the additional work needed to couple networks to selective pressures.
This time-consuming post-hoc tweaking phase stems, in part, from having to work with a limited set of imperfect components. Although this lack of reliable parts is being addressed by community efforts26, it remains an acute problem due to there being a limited number of components and the fact that the majority are inadequately characterized, e.g., many promoters are simply characterized as being ‘weak’ or ‘strong’. What is needed to resolve this problem and fast-track synthetic biology is a new approach that creates libraries of components ahead of any assembly; and then, by starting with a finer granular range of choices for each component, modeling can be used to quickly pick out the correct part needed to generate the intended network function. This approach offers the added attraction of allowing substantially different network outcomes to be chosen in advance, simply by selecting functionally-equivalent components with slightly different properties. This exploits a feature common to many types of finely-balanced networks, where small changes to one component can dramatically impact the behavior of the entire system.
Using regulated promoters as our example, we describe here how a simple synthesis technique can be used to rapidly create and parallel-characterize component libraries for synthetic biology. Working in S. cerevisiae, we demonstrate how such libraries can be teamed with predictive modeling to rationally guide the construction of gene networks that have diverse outputs. We also illustrate a plug-and-play application for one of our network designs by using it to control the timing of yeast sedimentation.
RESULTS
Parallel Synthesis and Characterization of Regulatory Promoter Libraries
To demonstrate our library-modeling approach, we focused on regulated promoters, as they typically control gene network logic and modulate responses to stimuli. Promoter libraries have been created using DNA-shuffling/combinatorial approaches27-29 and mutation-selection techniques30-33. We modified an efficient synthesis-with-degenerate-sequence method32 to yield libraries of regulatory promoters that have a range of inputs and outputs. In this technique, promoters are constructed with runs of unspecified (‘N’) sequence separating key motifs32; the fixed motif sequences ensure promoter function, and the random bases surrounding them modulate their efficiency, presumably by subtly altering local DNA conformation34.
Our first library was designed to yield yeast promoters repressed by TetR (Tn_10_.B Tetracycline repressor35) and inducible with the TetR-inhibitor anhydrotetracycline (ATc). Jensen and Hammer’s Klenow_-based synthesis method using inexpensive oligonucleotides31, 34 was utilized to build promoters containing the TATA box and start site from the commonly-used GAL1 promoter36. To introduce controlled regulation, we placed two tandem TetR operators (Tn_10 operator _tet_O2) into the promoter at positions previously shown to provide tight repression29.
A schematic of our library synthesis technique is shown in Figure 1 and detailed in the Methods section. To permit screening, libraries were cloned in a vector with the strong constitutive TEF1 promoter37 directing TetR expression and the GAL1 upstream activation signal (UAS) region placed upstream of the engineered promoter to direct the removal of nucleosomes from the promoter in the presence of galactose38. We arbitrarily chose to build a library of 20 promoters (T1-T20) covering a wide range of expression and inhibition levels. We used flow cytometry detection of yeast-enhanced green fluorescent protein (yEGFP39) to quantitatively measure the promoters for minimum repressed output (Smin) and maximum unrepressed output (Smax) (Table 1). The approach was designed to yield interchangeable promoters that are identical except for Smin, Smax, and inter-motif sequences. This was confirmed by comparison to TX, a control promoter retaining the GAL1 wild-type sequence between defined motifs, as well by DNA sequencing (Supplementary Fig. 1) and dose-response curves (Supplementary Fig. 3).
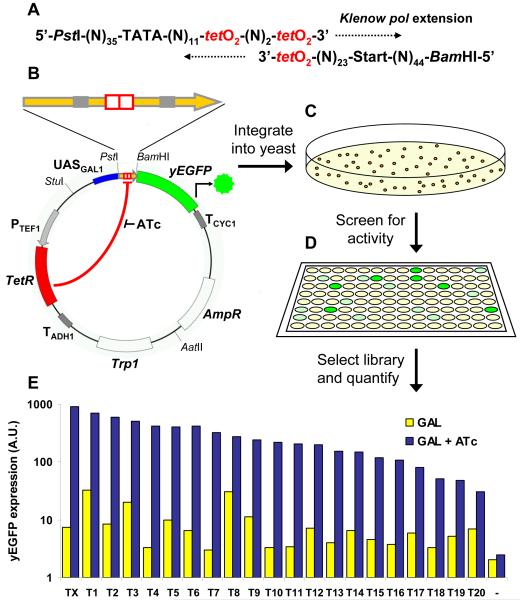
Figure 1. Regulatory promoter library synthesis, screening and characterization.
Schematic design for TetR-regulated promoter synthesis. (A) Promoters are constructed by Klenow pol extension from two overlapping 110mer oligonucleotides synthesized with unspecified nucleotides (N) between defined motifs. (B) Promoters are ligated between the GAL1 upstream activation signal (UASGAL1) and the yEGFP coding sequence in the characterization vector, which also expresses TetR from the TEF1 promoter (PTEF1). (C) Transformation of yeast yields thousands of colonies with the genome-integrated vector. (D) Individual colonies are screened in 96-well format by measuring fluorescence in induction conditions after 22 hours growth (media supplemented with 2% galactose + 250 ng/ml ATc). (E) A library of 20 regulated promoters covering a range of expression levels is selected from screening data and quantitatively characterized using flow cytometry measurement of yEGFP expression after 22 hours growth in media supplemented with 2% galactose (GAL) and with 250 ng/ml ATc (GAL + ATc). TX = control promoter, T1-T20 = library promoters, and - = null promoter.
Table 1. Maximum and minimum output values of members of the promoter libraries.
Cultures grown in 2% galactose-supplemented media for 22 hours were measured for median yEGFP expression by flow cytometry; values are the means of three biological repeats with standard error of the mean given. For characterization purposes, the repressor of interest (TetR or LacI) was constitutively expressed from the TEF1 promoter, which was also measured in this study and found to remain constant in all growth conditions used. Maximum output (Smax) is measured with saturating concentrations of repressor inhibitor (250 ng/ml ATc for TetR, 10 mM IPTG for LacI); minimum output (Smin) is measured without inhibitors
| PLibT TetR-regulated promoters | PLibL LacI-regulated promoters | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Promoter | Smax | error | Smin | error | Promoter | Smax | error | Smin | error |
| TX | 918.00 | 33.83 | 7.46 | 0.46 | LX | 717.38 | 21.06 | 13.06 | 0.77 |
| T1 | 694.23 | 19.89 | 32.79 | 2.58 | L1 | 399.90 | 25.02 | 11.11 | 0.60 |
| T2 | 595.79 | 17.07 | 8.38 | 0.50 | L2 | 372.59 | 16.87 | 9.71 | 0.11 |
| T3 | 506.31 | 27.48 | 20.22 | 2.16 | L3 | 292.11 | 11.60 | 83.05 | 1.09 |
| T4 | 421.78 | 5.83 | 3.26 | 0.16 | L4 | 282.01 | 13.61 | 50.55 | 1.92 |
| T5 | 408.04 | 22.91 | 9.87 | 0.41 | L5 | 246.73 | 6.42 | 151.75 | 2.77 |
| T6 | 418.60 | 16.63 | 6.46 | 1.68 | L6 | 228.45 | 15.37 | 23.79 | 0.31 |
| T7 | 319.66 | 13.41 | 3.04 | 0.15 | L7 | 139.99 | 8.43 | 5.40 | 0.35 |
| T8 | 277.75 | 12.94 | 30.88 | 1.75 | L8 | 141.86 | 6.23 | 7.67 | 0.35 |
| T9 | 244.21 | 11.79 | 11.34 | 0.62 | L9 | 134.04 | 9.73 | 23.54 | 1.55 |
| T10 | 216.99 | 7.34 | 3.27 | 0.18 | L10 | 129.13 | 8.04 | 4.96 | 0.30 |
| T11 | 203.14 | 6.90 | 3.41 | 0.18 | L11 | 108.27 | 4.18 | 5.74 | 0.45 |
| T12 | 201.76 | 3.75 | 7.08 | 0.53 | L12 | 107.35 | 4.73 | 5.07 | 0.36 |
| T13 | 154.46 | 12.15 | 4.01 | 0.23 | L13 | 103.58 | 9.54 | 4.37 | 0.29 |
| T14 | 151.03 | 10.36 | 6.42 | 0.19 | L14 | 82.32 | 1.50 | 4.15 | 0.23 |
| T15 | 118.93 | 5.85 | 4.62 | 0.19 | L15 | 70.91 | 4.42 | 20.83 | 0.96 |
| T16 | 108.22 | 3.40 | 3.71 | 0.13 | L16 | 72.03 | 3.05 | 4.28 | 0.23 |
| T17 | 81.70 | 3.39 | 5.91 | 0.27 | L17 | 56.97 | 1.77 | 5.15 | 0.36 |
| T18 | 51.75 | 3.27 | 3.26 | 0.25 | L18 | 47.16 | 1.33 | 3.91 | 0.28 |
| T19 | 48.29 | 1.10 | 5.13 | 0.89 | L19 | 44.10 | 2.25 | 4.25 | 0.20 |
| T20 | 30.69 | 0.40 | 6.95 | 0.45 | L20 | 37.08 | 2.12 | 9.41 | 0.69 |
| TEF1 | 287.38 | 14.38 |
Feedforward Loop Networks
To demonstrate how our approach can be applied in a gene network, we used the TetR-regulated promoter library with in silico modeling to investigate the incoherent type II negative feedforward loop network. This is a genetic motif found in S. cerevisiae and mammalian cells that consists of an output gene regulated by two repressor genes, one of which is also inhibited by the other40.
For our network, we used TetR and a eukaryotic-optimized version of the E. coli Lac inhibitor41 (LacI), as repressors (Fig. 2A). Each controlled yEGFP expression by regulating a hybrid _GAL1_-based promoter (POR-LT) containing both the K-12 E. coli O1 Lac Operator (OLac) and a _tet_O2 site, thus acting as an OR-gate promoter. TetR expression was constitutive from the TEF1 promoter, while LacI expression was driven by TetR-regulated promoters selected from our library (PLibT). By varying the concentrations of two inputs - ATc and the LacI inhibitor Isopropyl β-D-1-thiogalactopyranoside (IPTG) - the repressive effects of TetR and LacI could be tuned, modulating yEGFP expression output.
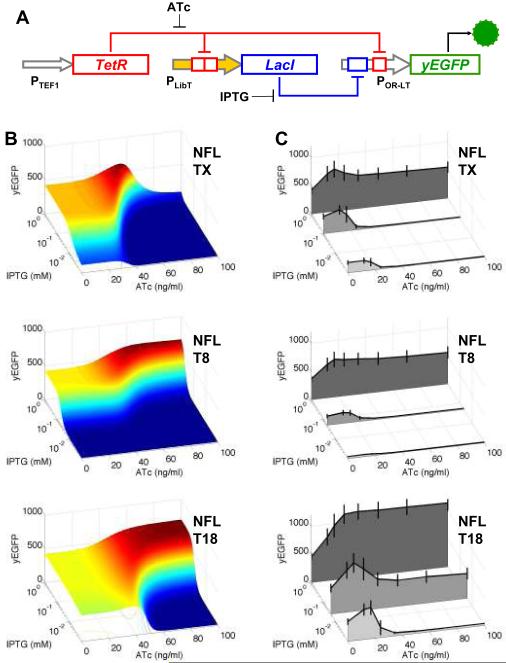
Figure 2. Modeling and synthesis of feedforward loop networks using a promoter library.
TetR-regulated promoter library data were used in conjunction with in silico modeling to construct negative feedforward loop (NFL) gene networks with different predicted input-output functions. (A) Schematic of the network, where PTEF1 = TEF1 promoter, PLibT = TetR-regulated promoter library, and POR-LT = LacI-TetR dual-regulated promoter. (B) In silico modeling of the network from component properties predicts yEGFP expression (output) in response to varied concentrations of ATc and IPTG (inputs) when three different TetR-regulated promoters are used. (C) The three networks were assembled in S. cerevisiae, and median yEGFP expression was measured by flow cytometry after 22 hours growth of cells in media supplemented with 2% galactose plus varying concentrations of ATc and IPTG. Error bars show the standard deviation of the gated cell population.
Before any network assembly, we used component properties from our experimental characterization steps to build a mathematical model to predict how network output would change when input levels (ATc/IPTG) and promoter properties were varied. The model served as a guide, predicting which components from the library could be selected to yield different network outcomes, and what dosage of induction would be most experimentally informative. In the model, the experimentally determined Smax and Smin values for the promoters (Table 1) were utilized; generic values were assumed for other parameters (see Supplementary Information for modeling details).
A simulation with PLibT = TX (control promoter: Smax 918.0, Smin 7.46) leads to an interesting non-monotonic expression landscape with an output peak at intermediate inputs (Fig. 2B). This occurs because TetR has simultaneously opposing effects on yEGFP output - inhibiting the production of yEGFP by binding to POR-LT, while also relieving LacI inhibition of yEGFP by binding to PLibT and repressing LacI production. This is consistent with previous in situ studies of naturally occurring negative feedforward loops42. Our synthetic library-modeling approach enables the investigation of this motif without the hindrance of inter-connected regulatory networks42-44.
By changing the Smin and Smax values of PLibT in the model, we can examine computationally how different promoters from our library affect this response landscape. The model predicts significant changes in output, and two examples most divergent from the TX simulation are shown in Figure 2B. Increasing the Smin value of the TetR-regulated promoter removes almost all expression in low concentrations of IPTG (PLibT = T8: Smin 30.88), whereas decreasing the Smax value shifts peak expression to occur only at higher concentrations of ATc (PLibT = T18: Smax 51.75). This demonstrates quantitatively that the same external induction (ATc or IPTG) can elicit very different responses from the motif simply due to small changes in promoter strength.
To test these in silico predictions, we assembled the three negative feedforward loop networks shown in Figure 2B using corresponding components from our libraries, and quantified their output responses to varied ATc/IPTG inputs using flow cytometry measurement of yEGFP. The experimental data (Fig. 2C) correlated very well with our computational predictions, particularly considering the many non-fitted generic parameters used in the model. These results demonstrate how small changes in promoter strength can have dramatic consequences on network responses. These findings also show that a model built from just component data and generic parameters can offer insights into a network response landscape, and such a model, when teamed with component libraries, can serve as a useful, rapid guide for producing networks with different predictable characteristics.
Genetic Timer Networks
Having demonstrated our approach in a relatively simple network using one promoter library, we next utilized two promoter libraries in a more complex network with a richer set of dynamics. Using the mutual-repression motif of the genetic toggle switch14, we set out to produce predictable genetic ‘timers’. These timers exploit the finely-balanced nature of a mutual inhibitory network14, where changes in opposing repressor levels can disrupt bistability, and memory of induction can be lost as the system resets back to its original default state. These timers are effectively genetic toggle switches operating in a monostable regime, and their rate of resetting is directly related to relative expression levels of the two repressors - the further they are from the balanced values required for bistability14, the more rapidly memory of induction is lost.
For yeast timers, we used LacI and TetR as the two mutually-repressive genes (Fig. 3A). LacI is expressed from a TetR-regulated promoter (PLibT) selected from our component library described above, and TetR is expressed from a LacI-regulated promoter (PLibL) taken from the second component library. This second library of promoters (L1-L20 plus control LX) was synthesized and characterized as before, but with the Lac operator (OLac) in place of the tandem Tet operators (promoter data are shown in Table 1 and sequences in Supplementary Fig. 2). To follow the expression state of the timers, yEGFP was placed under the control of the LX promoter, giving an expression read-out directly correlated to TetR expression (Fig. 3A).
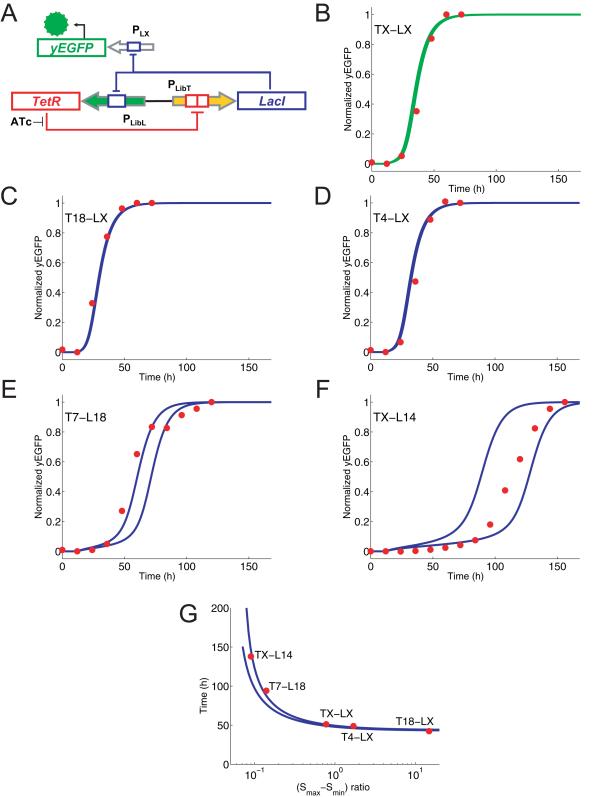
Figure 3. Predictable genetic timer networks constructed from two promoter libraries.
TetR- and LacI-regulated promoter libraries were used to construct a mutually-repressive gene network that acts as a predictable timer. (A) Schematic of the timer network; PLibL = LacI-regulated promoter library, PLX = LacI-regulated control promoter, and PLibT = TetR-regulated promoter library. (B) In silico model of the network fitted to TX-LX experimental data (Supplementary Fig. 4) shows yEGFP expression changing over time after ATc induction is removed at time 0. Yeast cells with the TX-LX network genomically-integrated were grown for 36 hours with 250 ng/ml ATc to induce the network, washed three times and monitored starting from time 0 until the expression state reset to a maximum. Normalized yEGFP output (red circles), which was calculated from flow cytometry measurements taken every 12 hours, matches the model output (green lines) well. Both upper and lower bound of model fittings are plotted (details in Supplementary Information). (C,D,E,F) Median yEGFP expression (red circles) was measured by flow cytometry every 12 hours for four different promoter combinations (T18-LX, T4-LX, T7-L18 and TX-L14). Cultures were induced and treated the same as in (B). Blue lines are model predictions based on parameters inferred from (B). Lower and upper bounds use the parameters corresponding to lower and upper bounds in (B). (G) The relationship between the reset time and the ratio for all of its values is plotted. Lower and upper bounds use the parameters corresponding to lower and upper bounds in (B). The reset time can be approximated as T = C1 + C2/√(|r-C3|), where T = reset time, C1 = basal reset time, C2 = scale factor and C3 = bifurcation ratio (details in Supplementary Information).
An initial model built with component properties from both libraries gave us qualitative insights into how timers can be set via imbalanced mutual inhibition (see Supplementary Information). However, a model built solely from component data cannot in this case capture important quantitative features of the network. The initial model with generic parameters revealed that changing the ratio of expression from the two opposing promoters affects the reset time, but it was not able to predict accurately by how much the reset time would change. The temporal dynamics of this system cannot be quantitatively predicted without first seeing a system in action to dissect some of the lumped parameters that remain fixed for all possible timers (see Supplementary Information for details). To address this, we assembled and tested a single example timer using the two control promoters (TX-LX); we then used the experimental data (Fig. 3B) from this system to calibrate a quantitatively predictive model for the other 440 possible timers afforded by our libraries.
The quantitative model gave us predictions as to how reset time could be varied by promoter selection, specifically by adjusting the ratio of relative expression from opposing promoters (using Smax-Smin to determine relative expression for each). We used the model to quantitatively predict the reset behavior of two timers (T18-LX and T4-LX) with ratios greater than that for TX-LX, and two timers (T7-L18 and TX-L14) with smaller ratios. These timers were assembled and tested in yeast. The experimental data for all four networks fell within the upper and lower bounds of the model predictions, validating our approach and ability to make quantitative predictions (Fig. 3C,D,E,F).
The model with calibrated parameters provides us with a “confidence interval” of reset times for all ratios, shown as blue lines in Figure. 3G. Closer inspection of the model reveals that reset times of different timer networks with low Smin values are approximately proportional to the reciprocal of the square root of the distance between the Smax-Smin ratio and the bifurcation ratio (the ideal Smax-Smin ratio for bistability). Mathematically, this is due to a temporal lag in resetting caused by the network passing through a ‘bottleneck’ as it leaves bistability45, 46. This direct relationship, shown in Figure. 3G, allows timers with any reset time between 50 and 150 hours to be chosen simply based on the strengths of the two promoters selected from the respective libraries.
Control of Yeast Sedimentation Timing
To demonstrate how our approach can be readily applied in a biotechnology scenario, we used the plug-and-play nature of our timer networks to control the flocculation of yeast. Flocculation occurs when yeast cells express FLO1, which functions as yeast-specific adhesin that causes cells to clump together and sediment from the medium47-49. The phenotype is crucial in industrial beer, wine and bioethanol fermentation, as it allows for easy removal of yeast sediment after all sugars have been converted to ethanol48.
Since the reset times of T4-LX and T18-LX are very close to that of TX-LX, we chose TX-LX, T7-L18 and TX-L14 to test the application of genetic timers. Using these three networks, we controlled the timing of sedimentation by replacing yEGFP with the FLO1 gene (Fig. 4A). In our laboratory yeast strain, FLO1 is not expressed, but replacing its native promoter with a strong promoter re-activates flocculation, causing sedimentation to occur when a threshold of FLO1 expression is passed. The timing of sedimentation can therefore be tied to the resetting of each timer network by choosing an appropriate regulated promoter from the libraries. With the 441 possible timers, and a choice from two library sets of promoters for controlling FLO1 expression, we had the potential to produce >17,000 different flocculating networks. We selected the L7 promoter, which has relatively high Smax and very low Smin, to give a wide dynamic range. When LacI was abundant, the minimal expression from this promoter did not elicit sedimentation, allowing yeast to grow in suspension.
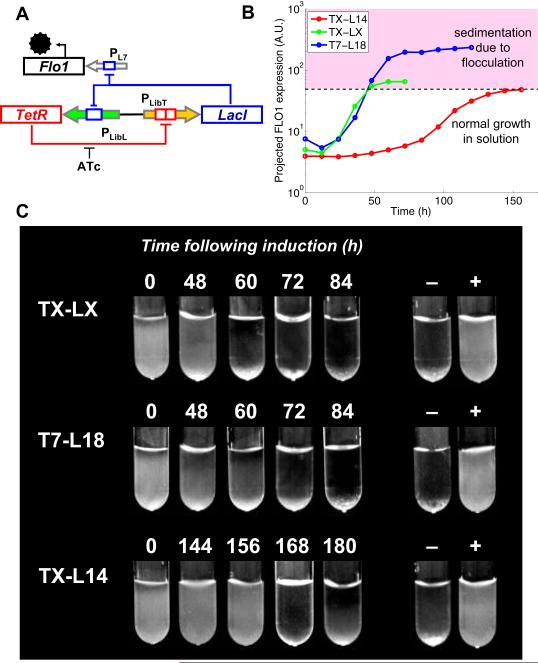
Figure 4. Controlling the timing of yeast sedimentation using a predictable gene network.
The synthetic networks tested in Figure 3B,E,F were used to control the timing of yeast sedimentation caused by flocculation. (A) Schematic of flocculation gene networks. Flocculation is regulated by replacing yEGFP and PLX in the gene network shown in Figure 3A with FLO1 under the control of the L7 promoter (PL7). (B) Rescaled yEGFP data from Figure 3B,E,F were used to project temporal FLO1 expression levels and predict the timing of cell sedimentation due to flocculation (details in Supplementary Information). (C) The timing of sedimentation from the three synthetic networks. Cultures induced by growth with 250 ng/ml ATc for 36 hours were washed twice and grown at high OD600 with shaking and diluted into fresh media every 12 hours, until sedimentation cleared the suspension. Images shown here are 1ml cultures at 12 hour intervals, 10 minutes after brief vortexing. Controls: - = growth in 10 mM IPTG, + = growth in 250 ng/ml ATc.
We experimentally determined the threshold of FLO1 expression that causes sedimentation (see Supplementary Information). Rescaling the timer data in Figure 3B,E,F to match the Smax and Smin values for the L7 promoter allowed us to plot estimates as to when this threshold would be passed for our networks (Fig. 4B). We tested these predictions by assembling the networks in yeast, and then growing the yeast cultures until sedimentation, days after the initial induction (ATc) was removed (Fig. 4C). For the TX-LX, T7-L18 and TX-L14 networks, sedimentation was seen at 60, 60 and 168 hours, respectively. These findings closely matched the predicted times assuming a 12-hour lag, presumably due to a longer phenotype maturation for flocculation compared to yEGFP fluorescence. This experiment demonstrates that we can quickly apply predictable networks built with our approach to control an industrially-relevant phenotype. Such accurate control of flocculation timing provides a wide window of opportunity to harvest fermentation product from cells and could be applied to improve biomass recycling in the biofuels industry.
DISCUSSION
This work establishes a new approach that can be used to rapidly increase the number of network components as well as decrease the time and effort required to engineer gene networks with desired functions. Our approach is compatible with plug-and-play synthetic biology and provides a platform to fast-track gene network construction. Here we focused on generating, characterizing and utilizing component libraries of promoters, but our approach is also applicable to other biomolecular components, as diversity in non-essential sequence also affects functional efficiency in proteins and RNA.
Although screening of mutated parts is not a new technique, our approach represents an advance over previous methods by coupling qualitative and quantitative modeling with library diversity to guide the construction of synthetic gene networks with predictable functions. In robust networks like our feedforward loop, models built entirely from component property sets are sufficient to guide the choice of parts required to elicit specific network phenotypes, such as high sensitivity to inputs or low maximum output. Although previous studies have shown that it is possible, in some cases, to accurately predict network behaviors based solely on component properties8, 50, we found that this conclusion cannot be generalized to more complex, finely-balanced networks, such as the timers described here. Instead we found that one must first assemble and experimentally characterize a single exemplary network of interest, in order to create a generalizable model with quantitative predictive capabilities. The experimental work needed to construct and test this one network is quickly offset by yielding accurate quantitative predictions for all other possibilities, and the benefit of this is especially significant when one considers that each additional component library incorporated increases the number of potential networks exponentially.
Our approach effectively moves component ‘tweaking’ to the front-end of gene network engineering. This arrangement is instinctively more rational than network retrofitting and made feasible by the coupling with mathematical modeling. As component libraries are produced in parallel at the same point in the process that individual parts are typically characterized for modeling, they require little extra effort in return for significant reward. Projects undertaken with this approach will help accelerate synthetic biology by yielding many more components for the community, and as library-synthesized components are designed to show variation only in intended properties, the need for extensive characterization of each component is eliminated or substantially reduced. Our work also provides an accessible method for introducing predictable, controlled variability to networks, a feature that is increasingly becoming desirable as synthetic biology enters its second decade18, 19. With advances in modern DNA synthesis technologies, the range of our approach will expand as synthesis becomes faster and cheaper, and as longer regions of biomolecules are able to be specifically varied in a systematic fashion.
METHODS
Strains and media
S. cerevisiae strain YPH500 (α, ura3-52, lys2-801, ade2-101, trp1Δ63, his3Δ200, leu2Δ1) (Stratagene, La Jolla, CA) was used in all experiments, and all genomic integrations were specifically targeted to the redundant ura3-52 locus. Culturing, genetic transformation and verification of transformation were done as previously described29, using either the TRP1, HIS3 or LEU2 genes as selectable markers.
Plasmid construction
The TetR-regulated promoter library characterization vector (pTVGI, Fig. 1B) was adapted from the previously described yeast integrative plasmid pRS4D129, removing the GAL1/GAL10 promoter region and replacing it with the S. cerevisiae TEF1 promoter directing TetR expression and the GAL1 UAS region plus a synthesized library promoter directing yEGFP expression. A 489 bp span of arbitrary sequence from the ura3 gene was included between these promoters to buffer any cross-talk between them and to allow the vector to site-specifically integrate into the ura3-52 locus. For the LacI-regulated promoter library characterization vector (pLVGI), TetR was replaced by a synthetic codon-optimized LacI41 that had been altered to remove both an internal _Pst_I restriction site (without changing the codon sequence or efficiency) and the hyper-strong SV40 nuclear localization signal. The control TX promoter was amplified directly from pRS4D1, whereas the LX promoter and OR-LT promoters were generated, as previously described, by standard oligonucleotide PCR mutation methods from the TX promoter and T123 pRS4D1 promoter, respectively29. All plasmids were constructed and transformed into E. coli to harvest DNA for yeast transformations, as previously described29.
Promoter library synthesis and screening
Promoters were created from partially-overlapping pairs of 110mer PAGE-purified oligonucleotides (listed in Supplementary Information), which were custom synthesized by Sigma-Genosys (The Woodlands, TX). Second strand DNA synthesis by Klenow polymerase was followed by agarose gel electrophoresis purification to obtain fragments ready for insertion into characterization vectors34. Upon ligation, DH5α E. coli (NEB, Beverly MA) were transformed with the plasmid vectors and clones were selected by ampicilin resistance. 104-105 colonies were pooled directly from LB agar plates and harvested for plasmid using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). The promoter plasmid library was then used to transform yeast as previously described29, scaling up by a factor of 10 and plating on 250mm × 250mm plates to yield ∼3500 individual colonies. 192 of these colonies were transferred to two 96-well plates, and grown for 22 hours in 300 μl of media supplemented with 2% galactose and 250 ng/ml ATc. Cell fluorescence was measured at 450nm using a SpectraFluor Plate Reader (Tecan, Durham NC). Approximately one quarter of clones produced a detectable level of expression when TetR was inhibited by ATc, and roughly three quarters of these responded to the removal of ATc with a drop in expression, indicating controlled regulation. Expression was undetectable in glucose. Colonies selected to create our 20-member library were PCR-tested for single genomic integration, and then characterized by flow cytometry in the presence and absence of 250 ng/ml ATc. For LacI-regulated libraries, ATc was replaced with 10 mM IPTG in the screening and characterization stages.
Flow cytometry and data analysis
Flow cytometry measurements were carried out as previously described8, running samples on a medium flow rate until 20,000 cells had been collected within a small forward and side scatter gate to reduce extrinsic noise. Data files were analyzed using MatLab (The MathWorks, Natick, MA), linearizing log-binned fluorescence intensity values and then calculating the median and standard deviation of the gated population. For both promoter library data (Table 1) and control ATc and IPTG induction curves, 3 ml cultures were grown for 20 hours to an optical density at 600 nm (OD600) of 1.00 at 30°C with orbital shaking before measurement. For the negative feedforward loop and genetic timer data, 300 μl of cells were grown to OD600 of 1.00 at 30°C in 96-well format. For the negative feedforward loop data, cells were grown for 22 hours before measurement. For the genetic timer data, cells were grown for 12 hours, a sample was taken for measurement, and then a fraction of the remaining cells was diluted into fresh media for the next 12 hours of growth.
Flocculation
To obtain flocculating strains, the L7 library promoter was inserted into pFA6a-KanMX6-pGAL1 in place of the GAL1 promoter sequences, and PCR amplification from primer pairs FL1 and FL2 was used to integrate these in place of the wild-type FLO1 promoter as described previously47. To measure flocculation over time, 1.2 ml cultures were grown for 12 hours to OD600 = 1.50 at 30°C with orbital shaking. 1 ml was removed for measurement and replaced with 1 ml of fresh culture media to continue growth. For measurement, 1 ml of culture was vortexed for 5 seconds before sitting for 10 minutes in a clear 3 ml culture tube. Cultures were photographed with a light box behind, and the image inverted and auto-contrasted using Picasa imaging software (Google, Mountain View, CA).
Supplementary Material
1
ACKNOWLEDGMENTS
We thank Peter R. Jensen for advice on promoter library synthesis methods, Kevin Verstrepen for guidance and materials relating to yeast flocculation, and Henry H. Lee for valuable ideas in genetic device construction. This work was supported by the National Institutes of Health through the NIH Director’s Pioneer Award Program, grant number DP1 OD003644, the NSF FIBR program, and the Howard Hughes Medical Institute.
References
- 1.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100073. 2006 0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Withers ST, Keasling JD. Biosynthesis and engineering of isoprenoid small molecules. Appl Microbiol Biotechnol. 2007;73:980–990. doi: 10.1007/s00253-006-0593-1. [DOI] [PubMed] [Google Scholar]
- 3.Weber W, et al. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc Natl Acad Sci U S A. 2008;105:9994–9998. doi: 10.1073/pnas.0800663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 5.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106:4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol. 2008 doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Guido NJ, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 9.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 10.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 11.Levskaya A, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 12.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 13.Kramer BP, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 14.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 15.Weber W, et al. A synthetic time-delay circuit in mammalian cells and mice. Proc Natl Acad Sci U S A. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grilly C, Stricker J, Pang WL, Bennett MR, Hasty J. A synthetic gene network for tuning protein degradation in Saccharomyces cerevisiae. Mol Syst Biol. 2007;3:127. doi: 10.1038/msb4100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 18.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 20.Marguet P, Balagadde F, Tan C, You L. Biology by design: reduction and synthesis of cellular components and behaviour. J R Soc Interface. 2007;4:607–623. doi: 10.1098/rsif.2006.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi H, et al. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 24.Blake WJ, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 26.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 27.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy KF, Balazsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci U S A. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 31.Jensen PR, Hammer K. Artificial promoters for metabolic optimization. Biotechnol Bioeng. 1998;58:191–195. [PubMed] [Google Scholar]
- 32.Hammer K, Mijakovic I, Jensen PR. Synthetic promoter libraries--tuning of gene expression. Trends Biotechnol. 2006;24:53–55. doi: 10.1016/j.tibtech.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 36.Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schirmaier F, Philippsen P. Identification of two genes coding for the translation elongation factor EF-1 alpha of S. cerevisiae. Embo J. 1984;3:3311–3315. doi: 10.1002/j.1460-2075.1984.tb02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci U S A. 2008;105:2975–2980. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cormack BP, et al. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143(Pt 2):303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 40.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scrable H, Stambrook PJ. Activation of the lac repressor in the transgenic mouse. Genetics. 1997;147:297–304. doi: 10.1093/genetics/147.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan S, Bren A, Dekel E, Alon U. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol Syst Biol. 2008;4:203. doi: 10.1038/msb.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Entus R, Aufderheide B, Herbert M, Sauro MH. Design and implementation of three incoherent feed-forward motif based biological concentration sensors. Systems and Synthetic Biology. 2007;1:119–128. doi: 10.1007/s11693-007-9008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Hao N, Dohlman HG, Elston TC. Bistability, stochasticity, and oscillations in the mitogen-activated protein kinase cascade. Biophys J. 2006;90:1961–1978. doi: 10.1529/biophysj.105.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strogatz SH. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry, and Engineering. Addison-Wesley Pub.; Reading, Mass: 1994. [Google Scholar]
- 47.Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- 49.Smukalla S, et al. FLO1 Is a Variable Green Beard Gene that Drives Biofilm-like Cooperation in Budding Yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Accurate prediction of gene feedback circuit behavior from component properties. Mol Syst Biol. 2007;3:143. doi: 10.1038/msb4100185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1