Mutations in the Nonstructural Protein 3A Confer Resistance to the Novel Enterovirus Replication Inhibitor TTP-8307 (original) (raw)
Abstract
A novel compound, TTP-8307, was identified as a potent inhibitor of the replication of several rhino- and enteroviruses. TTP-8307 inhibits viral RNA synthesis in a dose-dependent manner, without affecting polyprotein synthesis and/or processing. Drug-resistant variants of coxsackievirus B3 were all shown to carry at least one amino acid mutation in the nonstructural protein 3A. In particular, three mutations located in a nonstructured region preceding the hydrophobic domain (V45A, I54F, and H57Y) appeared to contribute to the drug-resistant phenotype. This region has previously been identified as a hot sport for mutations that resulted in resistance to enviroxime, the sole 3A-targeting enterovirus inhibitor reported thus far. This was corroborated by the fact that TTP-8307 and enviroxime proved cross-resistant. It is hypothesized that TTP-8307 and enviroxime disrupt proper interactions of 3A(B) with other viral or cellular proteins that are required for efficient replication.
Enteroviruses comprise several pathogens that are implicated in an large variety of clinical manifestations that range from mild illnesses to more serious or even life-threatening diseases, such as meningitis, encephalitis, myocarditis, pancreatitis, acute paralysis, or neonatal sepsis (30, 40). Enteroviruses are small, nonenveloped, and spherical in shape, with a diameter of about 30 nm. The icosahedrally shaped capsids are assembled from 60 protomers, each composed of four structural proteins, designated VP1 (for viral protein 1), VP2, VP3, and VP4 (38, 39). The enteroviral genome consists of a single-stranded, positive-sense RNA of approximately 7,500 bases in length. The coding region of the viral genome is divided into three primary precursor molecules which contain the four structural (derived from P1) and 10 nonstructural viral proteins (derived from P2 and P3).
The nonstructural protein 3A and its precursor 3AB are derived from P3 and are indispensable for viral replication. A feature of 3A that has been the subject of many studies is its ability to serve as a membrane anchor through the presence of a 22-residue hydrophobic domain that forms an amphipathic helix near its C terminus (25). In infected cells, both 3A and 3AB are found in association with membranes (17). In the context of the viral replication complex, 3AB serves to deliver the basic protein VPg (3B) at the 5′ ends of plus- and minus-strand RNA during replication (18, 41) and, hence, to recruit the other proteins of the replication complex to the cellular membranes, the site of viral replication. The 3B protein then serves as a primer for the initiation of RNA synthesis, probably only after it has been cleaved from the 3A portion and not when it is still in the 3AB membrane-bound state (17). Cleavage of 3AB is mediated by 3Cpro/3CDpro and can only occur when the protein is membrane bound (26). Moreover, this proteolysis was shown to be enhanced in the presence of purified 3AB but not 3A (26, 31, 49). Stimulation of catalytic activity by 3AB has also been observed for the 3Dpol, both when 3AB is membrane bound and purified, and this has been suggested to occur via stabilization of the primer-template/3Dpol complex (26, 28, 35-37). 3AB but not 3A shows nonspecific RNA-binding activity but binds specifically to the 5′ RNA cloverleaf of the viral RNA genome when it is complexed with 3CDpro (20, 28, 48). The protein also binds to the 3′ untranslated region but, in contrast to binding at the 5′ cloverleaf, binding at the 3′ untranslated region can also occur in the absence of other proteins (20). Moreover, 3AB has been shown to induce membrane permeability in bacterial but not mammalian cells (1, 24, 25, 29), to induce membrane alterations in the endoplasmic reticulum (ER) (15), to form homodimers (26, 42, 47), and to be involved in host range pathogenicity (2, 27, 33, 34).
Apart from these features that are directly or indirectly associated with viral replication, 3A and 3AB are also involved in processes that specifically affect the host cell. It is well documented that protein 3A is able to interfere with cellular protein secretion via inhibition of ER-to-Golgi transport, causing accumulation of proteins otherwise destined for export (4, 13, 14). Determinants for this feature are located at the N terminus (4, 13, 44-46). This inhibition of ER-to Golgi transport has been shown to reduce or inhibit the secretion of antiviral cytokines such as interleukin-6, interleukin-8, and beta interferon (12), the concentration of tumor necrosis factor receptor on the surfaces of infected cells (32), and the presentation of antigen in the context of major histocompatibility complex class I molecules (7). In doing so, ER-to-Golgi transport inhibition in infected cells might help in evading the host cell's immune response and, hence, promote viral replication in an indirect way, although this inhibition is not required for efficient viral replication per se (12, 13).
The indispensable presence of 3A(B) during viral replication makes this protein an attractive candidate as a target for inhibition of viral replication. Thus far, only one compound (enviroxime) has been reported to target protein 3A (8, 22, 23). Despite its potent antiviral activity, however, the development of this compound was halted, mainly because of toxicity and an unfavorable pharmacokinetic profile (6). We here report on a novel compound (TTP-8307) that was identified in a screening campaign for inhibitors of the replication of enteroviruses. The compound appeared to be a potent inhibitor of the replication of several rhino- and enteroviruses.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (ATCC CCL-81), Buffalo green monkey (BGM) cells (ECACC 90092601), and HeLa cells (ATCC CCL-2) were grown in minimal essential medium (Gibco, Merelbeke, Belgium) supplemented with 10% heat-inactivated fetal bovine serum (Integro, Leuvenheim, The Netherlands), 5% bicarbonate (Gibco), and 5% l-glutamine (Gibco). Cells were grown at 37°C in a 5% CO2 incubator. Coxsackievirus B3 (CVB3) was derived from plasmid pCB53/T7, which contains a full-length cDNA of CVB3 strain Nancy behind a T7 RNA promoter (45). For assays involving virus growth, 2% fetal bovine serum was used instead of 10% fetal bovine serum. Rhinoviruses were provided by K. Andries, and the poliovirus Sabin 1, 2, and 3 strains were from B. Rombaut (Vrije Universiteit Brussels, Brussels, Belgium). Enterovirus 71 (BrCr) and coxsackieviruses A16 (G-10) and A21 (Coe) were obtained from the Rijksinstituut voor Volksgezondheid en Milieu (The Netherlands).
Compounds.
The synthesis of TTP-8307 (436 g/mol) will be reported elsewhere. The purity of the compound used in the present study was determined to be >96% by spectroscopic (nuclear magnetic resonance) and chromatographic (liquid chromatography-mass spectrometry) techniques. TBZE-029 and enviroxime were synthesized as reported elsewhere (10). Guanidine hydrochloride was from Sigma (Bornem, Belgium). All compounds were solubilized in dimethyl sulfoxide at 20 mM and stored at 4°C. For working solutions, the dimethyl sulfoxide stocks were diluted in minimal essential medium to the desired concentration.
In vitro RNA transcription and transfection.
Prior to in vitro RNA transcription, plasmid p53CB3/T7 was linearized with SalI (Promega, Leiden, The Netherlands). The digest was purified (gel and PCR purification kit; Promega), and 2.5 μg of DNA was used for in vitro RNA transcription (Ribomax large-scale RNA production system; Promega). The transcription reaction was carried out at 37°C for 4 h, after which the reaction mixture was DNase treated, and RNA was purified (RNA Cleanup System; Qiagen, Venlo, The Netherlands). Transfections were carried out in 25-cm2 culture flasks in Vero cells, grown to ∼75% confluence. Reaction mixtures, containing 2 ml of OptiMEM (Gibco), 2.5 μg of purified RNA, and 10 μl of DMRIE-C transfection reagent (Invitrogen) were incubated for 4 h at 37°C. Subsequently, the medium was replaced with fresh growth medium and incubated until the cultures exhibited an extensive cytopathic effect (CPE). At this point, the flasks were subjected to three rounds of freezing-thawing, and the collected supernatant was titrated for infectious virus content by endpoint titration.
Multicycle CPE reduction assays.
The antiviral activity of the selected compound was initially determined by using an MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenol)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt]-based CPE reduction assay. Briefly, cells grown to confluence in 96-well plates were infected with 100 50% cell culture infective doses (CCID50) of virus. After an adsorption period of 2 h at 37°C (35°C for rhinovirus), the virus was removed, and serial dilutions of the compounds were added. The cultures were further incubated at 37°C for 3 days, until complete CPE was observed in the infected and untreated virus control (VC). After removal of the medium, 90 μl of culture medium and 10 μl of MTS/PMS (Promega) were added to each well. After an incubation period of 2 h at 37°C, the optical density at 498 nm (OD498) of each well was read in a microplate reader. CPE values were calculated as follows: % CPE = 100 × ([ODCC - ODvirus+compound]/[ODCC - ODVC]). In these formulae, ODCC corresponds to the optical density of the uninfected and untreated cell cultures, ODVC represents the infected and untreated cell cultures, and ODvirus+compound are virus-infected cell cultures treated with a given concentration of compound. The 50% effective concentration (EC50) was defined as the concentration of compound that resulted in 50% protection against virus-induced CPE and was calculated by using logarithmic interpolation.
Viral plaque assays.
For determination of viral plaques, Vero cells, grown to confluence in six-well plates, were infected with CVB3 at 37°C with slight shaking at 55 rpm. After 2 h, the virus was removed, the cells were washed twice with phosphate-buffered saline (PBS), and the growth medium was replaced with agar (final concentration, 0.4%) in the presence or absence of compound. After 3 to 4 days, plaques were visualized. Briefly, cells were fixed with 2 ml of a solution containing 4% formaldehyde, after which the agar was removed. A 2% Giemsa solution was used to stain the cells.
Analysis of viral RNA accumulation with subgenomic replicon pCB53/T7-Luc.
Accumulation of viral (+)RNA was monitored by transfecting cells (in the presence or absence of 25 μM TTP-8307) with RNA derived from the SalI-linearized plasmid pCB53/T7-Luc, which contains a subgenomic CVB3 replicon, carrying a luciferase gene in place of the capsid coding P1 region. At the indicated times posttransfection, the cells were washed three times with PBS and lysed with 75 μl of lysis buffer. The luciferase activity was measured in a liquid scintillation counter with the luciferase assay system according to the recommendations of the manufacturer (Promega). The luciferase activity was expressed in (relative) light units.
Quantitative analysis of CVB3 RNA by real-time qRT-PCR assays.
Real-time quantitative reverse transcription-PCR (qRT-PCR) was performed with an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). Primers and probes were developed using Primer Express software (Applied Biosystems) as described elsewhere (9). The following primers and probe were used: a forward primer specific for nucleotides 2937 to 2957 (5′-ACG AAT CCC AGT GTG TTT TGG-3′), a reverse primer specific for nucleotides 3003 to 2982 (5′-TGC TCA AAA ACG GTA TGG ACA T-3′), and a TaqMan probe specific for nucleotides 2960 to 2977 (5′-FAM-CGA GGG AAA CGC CCC GCC-TAMRA-3′). Each reaction was performed in 25 μl of a PCR reagent mixture (One-Step qRT-PCR mix; Eurogentec, Seraing, Belgium) containing 900 nM concentrations of each primer and 200 nM concentrations of the specific TaqMan probe. The PCR consisted of an RT step (30 min at 48°C), a Taq activation step (10 min at 95°C), and 50 cycles of denaturation (15 s at 94°C) and annealing/extension (1 min at 60°C). The RNA copy number in each sample was determined by a standard curve generated from increasing copy numbers of a synthetic transcript corresponding to 67 nucleotides of the CVB3 genome.
Time of drug addition studies.
Vero cells, grown to confluence in 24-well culture plates, were infected with 104 CCID50 of coxsackievirus B3. After an adsorption period of 1 h at 37°C, virus was removed and replaced with 500 μl of growth medium. At 1-h intervals, 500 μl of medium containing a 2× compound solution was added (final concentration, 25 μM). At 8 h postinfection, the supernatant, as well as intracellular RNA of the infected cultures, was collected, and viral RNA was quantified by means of real-time qRT-PCR. The levels of viral RNA were compared to their untreated controls.
Analysis of viral polyprotein processing in vivo.
BGM cells, grown to confluence in 24-well plates, were infected with coxsackievirus B3 at a multiplicity of infection of 50. At 5 h postinfection, the medium was replaced with 300 μl of methionine-free medium. Thirty minutes later, the cultures were pulse-labeled in methionine-free medium containing 1 μl of Met35[S]/well in the absence or presence of TTP-8307 (25 μM final concentration) for 30 min. At 6 h postinfection cells were washed once with PBS and lysed in 75 μl of cold lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 0.05% sodium dodecyl sulfate (SDS). Translation products were analyzed on a 12.5% polyacrylamide gel containing SDS. The gels were fixed in 30% methanol-10% acetic acid, rinsed in dimethyl sulfoxide, fluorographed with 20% 2,5-diphenyloxazole in dimethyl sulfoxide, dried, and exposed to Kodak XAR film.
Generation of TTP-8307-resistant coxsackievirus.
Drug-resistant virus was generated by growing virus in the presence of gradually increasing concentrations of TTP-8307 on confluent Vero cultures in 48-well culture plates. After 4 to 5 days of culture, culture supernatant was collected from cultures that exhibited full CPE in the presence of the highest concentration of compound used. This virus was used for a successive round of infection, a procedure that was repeated until full CPE was noticed at concentrations of TTP-8307 (20 μM) that did not allow replication of wild-type virus. Subsequently, the resistant virus pool was subjected to plaque purification (in the presence of 20 μM compound), and individual clones were used for sequencing.
Site-directed mutagenesis.
Four mutant CVB3 clones were constructed, containing single amino acid replacements at positions 8, 45, 54, and 57 in protein 3A. The four clones were designated mutant 1 (3A[I8T]), mutant 2 (3A[V45A]), mutant 3 (3A[I54F]), and mutant 4 (3A[H57Y]). The corresponding synthetic oligonucleotides (and their complementary reverse oligonucleotides) were used for site-directed mutagenesis: (i) 5′-GGA CCA CCA GTA TAC AGA GAG ACC AAA ATT AGC GTT GCA CC-3′, (ii) 5′-GAA AAA GGA TGG TTG GCT CCT GAG ATC AAC TCC ACC C-3′, (ii) 5′-C TCC ACC CTC CAA TTT GAG AAA CAT GTC AGT CGG G-3′, and (iv) 5′-CC CTC CAA ATT GAG AAA TAT GTC AGT CGG GCT TTC-3′. The mutated sequences are underlined. Site-directed mutagenesis was performed with plasmid pCB53/T7 using the XL-Blue large site-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands), according to the manufacturer's instructions. After mutagenesis, the individual clones were verified by sequencing. Next, a 711-bp fragment containing the desired mutations was isolated using the enzymes BssHII and XbaI and reintroduced in an original, nonmutagenized clone of the same plasmid pCB53/T7. From these mutants, RNA transcripts and infectious viruses were generated as described earlier.
Sequencing.
PCR fragments that cover the entire CVB3 genome were generated and analyzed by using the cycle sequencing method (ABI Prism BigDye terminator cycle sequencing ready reaction kit). Both DNA strands were sequenced. Sequencing data were obtained with an ABI 373 automated sequence analyzer (Applied Biosystems), and sequences were analyzed with the Vector NTI software (Invitrogen).
RESULTS
TTP-8307 inhibits replication of several enteroviruses.
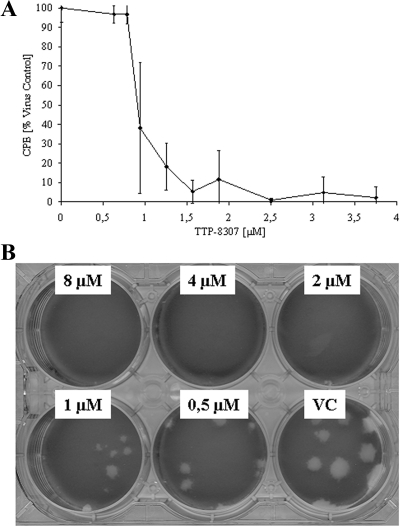
TTP-8307 was identified in a screening campaign as a selective inhibitor of CVB3 replication in Vero cells. The effect of TTP-8307 (Fig. 1A) was next evaluated against a selection of entero- and rhinoviruses in an MTS-based CPE reduction assay. TTP-8307 inhibits the replication of coxsackievirus B3 and the three poliovirus Sabin strains, as well as coxsackieviruses A16 and A21 (Table 1). TTP-8307 inhibits human rhinoviruses (HRVs) 2, 29, 39, 45, 63, and 85, but not other rhinovirus serotypes (HRV serotypes 9, 14, 15, 41, 42, 70, 72, 86, and 89) or enterovirus 71. Coxsackievirus B3 (CVB3), the prototype of the non-polio enteroviruses, was used to further study the particular characteristics of the antiviral activity of TTP-8307. TTP-8307 inhibited CVB3 replication in a dose-dependent manner (Fig. 2) when monitored either by (i) CVB3-induced CPE formation (Fig. 2A) or (ii) CVB3-induced plaque formation (Fig. 2B),
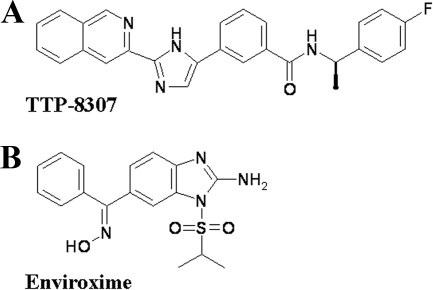
FIG. 1.
Structural formulae of TTP-8307 (A) and enviroxime (B).
TABLE 1.
Antiviral effect of TTP-8307 against selected entero- and rhinoviruses
| Virus | Mean EC50 (μM)a ± SD | |
|---|---|---|
| TTP-8307 | Enviroxime | |
| Human enterovirus A | ||
| CVA16 (G-10) | 0.85 ± 0.08 | 0.12 ± 0.01 |
| EV71 (BrCr) | >60 | 0.15 ± 0.06 |
| Human enterovirus B | ||
| CVB3 (Nancy) | 1.2 ± 0.1 | 1.9 ± 1.5 |
| Human enterovirus C | ||
| CVA21 (Coe) | 5.34 ± 0.93 | 0.48 ± 0.42 |
| Polioviruses | ||
| PV1 (Sabin) | 0.51 ± 0.05 | 0.19 ± 0,24 |
| PV2 (Sabin) | 0.58 ± 0.05 | 0.06 ± 0,01 |
| PV3 (Sabin) | 0.27 ± 0.23 | 0.04 ± 0,02 |
| Major group rhinoviruses | ||
| HRV9 | >50 | 0.038 ± 0.001 |
| HRV14 | >50 | 0.11 ± 0.04 |
| HRV15 | >50 | 0.042 ± 0.03 |
| HRV39 | 0.65 ± 0.51 | 0.028 ± 0.01 |
| HRV41 | >50 | 0.027 ± 0.003 |
| HRV42 | >50 | 0.11 ± 0.05 |
| HRV45 | 0.99 ± 0.04 | 0.25 ± 0.03 |
| HRV63 | 0.091 ± 0.054 | 0.027 ± 0.001 |
| HRV70 | >50 | 0.11 ± 0.02 |
| HRV72 | >50 | 0.72 ± 0.03 |
| HRV85 | 1.22 ± 0.21 | 0.033 ± 0.006 |
| HRV86 | >50 | 0.14 ± 0.03 |
| HRV89 | >50 | 0.10 ± 0.04 |
| Minor group rhinoviruses | ||
| HRV2 | 0.80 ± 0.12 | 0.026 ± 0.022 |
| HRV29 | 0.77 ± 0.38 | 0.029 ± 0.005 |
FIG. 2.
Dose-dependent inhibition of viral replication by TTP-8307. Vero cell cultures infected with CVB3 were treated with different concentrations of TTP-8307, and the effect on viral replication was monitored at day 3 postinfection. (A) Effect on virus-induced CPE formation (using an MTS-based cell protection assay). (B) Plaque reduction. The data represent averages ± the standard deviations (SD) from three independent experiments.
TTP-8307 acts at a stage that coincides with CVB3 viral RNA replication and polyprotein synthesis/processing.
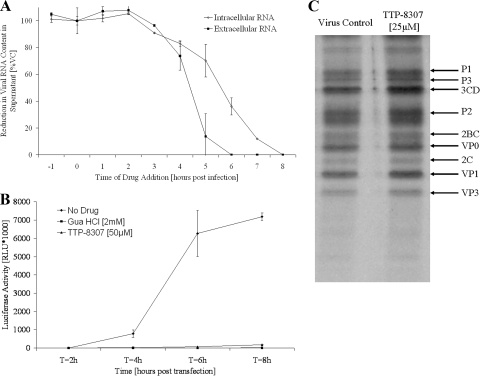
Time of drug addition studies were carried out to obtain a first indication about the stage in the viral replication cycle where TTP-8307 exerts its antiviral activity. Maximal inhibition of viral replication was maintained when the drug was added within the first 3 h postinfection (Fig. 3A). The addition of the drug at a time point later than 3 h postinfection, which coincides with the onset of viral RNA synthesis, resulted in a gradual decrease in antiviral activity. It can thus be concluded that TTP-8307 does not hamper early (attachment, entry, and uncoating) or late (assembly and release) events but rather interferes with intermediate processes, such as viral RNA replication, viral polyprotein synthesis, and/or processing.
FIG. 3.
TTP-8307 inhibits accumulation of CVB3 viral RNA without affecting polyprotein processing. (A) Time of drug addition. TTP-8307 (25 μM) was added to CVB3-infected cell cultures at 1-h intervals, starting at 1 h before infection. At 8 h postinfection, the time needed for a single replication cycle, intra- and extracellular viral RNA was collected and quantified by means of real-time qRT-PCR. Values were standardized to the 0-h time point. The data represent averages ± the SD from two independent experiments. (B) Analysis of viral RNA accumulation with subgenomic replicon pCB53/T7-Luc. Accumulation of viral (+)RNA was monitored after transfection of BGM cells (in the presence or absence of TTP-8307) with RNA derived from a chimeric subgenomic replicon (pCB53/T7-Luc). At the indicated times posttransfection, the luciferase activity was quantified and expressed in (relative) light units [(R)LU]. The data represent averages ± the SD from three independent experiments. (C) Effect of TTP-8307 on polyprotein processing. BGM cells grown to confluence in 24-well plates were infected with CVB3. At 5h30 min postinfection, cells were pulse-labeled for 30 min with Met35[S] after starvation in methionine-free medium for 30 min, either in the presence (25 μM) or absence of TTP-8307. Cells were lysed and translation products were analyzed by SDS-polyacrylamide gel electrophoresis. The data represent averages ± the SD from three independent experiments.
TTP-8307 inhibits accumulation of CVB3 viral RNA.
The accumulation of viral RNA in the absence or presence of TTP-8307 was monitored upon transfection of BGM cells with an infectious subgenomic replicon of CVB3 (in which the P1 region was replaced with a luciferase marker). Transfection in the presence of TTP-8307 (25 μM) led to a complete inhibition of viral RNA accumulation, whereas an increase in luciferase activity was measured in the absence of compound (Fig. 3B). The replication inhibitor guanidine hydrochloride (2 mM) was included as a reference compound and resulted in the inhibition of viral RNA accumulation as well.
TTP-8307 does not affect CVB3 polyprotein synthesis or processing.
Viral protein synthesis and polyprotein processing were monitored in a pulse-labeling experiment in the presence (25 μM) or absence of TTP-8307. From Fig. 3C it is evident that a similar pattern of viral proteins was observed in the presence or absence of TTP-8307. Thus, neither the rate of protein synthesis nor the processing of viral proteins was affected by the compound. This observation, together with the observed effect of TTP-8307 in the subgenomic replicon, points to the synthesis of viral RNA as the potential target of action of TTP-8307.
TTP-8307 resistant CVB3 clones share mutations in the nonstructural protein 3A.
To identify the molecular target of TTP-8307, drug-resistant CVB3 variants were selected. To this end, CVB3 was cultured successively in the presence of increasing concentrations of TTP-8307. After 10 passages, five independently cultured pools of CVB3 were obtained that replicated efficiently in the presence of TTP-8307 at concentrations that exceeded the EC50 more than 10-fold. The virus pools thus obtained were plaque purified in the presence of 20 μM TTP-8307 and one or several clones from each pool were picked up. As such, 10 clones were selected for genotyping (Table 2). Except for clone 1A, all clones carried mutations in two or more different proteins, including 2A, 2B, 2C, 3A, and/or 3D. A remarkable observation, however, was that all clones carried at least one mutation in protein 3A. Furthermore, each of the following four mutations in 3A recurred in different clones (either or not if they derived from the same pool): I8T, V45A, V54A, and H57Y. In contrast, none of the mutations observed in the other proteins occurred more than once in clones that were derived from independently cultured pools. Moreover, three of the four identified mutations in 3A were located in a region that was previously shown to accumulate mutations involved in resistance to the 3A inhibitor enviroxime (see Discussion). The contribution of the 3A mutations to the drug-resistant phenotype was therefore studied in more detail.
TABLE 2.
Mutations in TTP-8307-resistant CVB3 clones
| Passage line (pool) | Clone | Amino acid mutation(s) | ||
|---|---|---|---|---|
| 2A | 2B | 2C | 3A | 3D |
| 1 | A | I8T, V45A | ||
| B | N26D | H57Y | ||
| C | N26D | H57Y | ||
| 2 | A | E59Q | I112V | I54F |
| B | E64A | I8T | ||
| C | E120Q | S109G | I54F | D397E |
| 3 | A | R41K | V45A | |
| B | H57Y | V8I, G124R | ||
| 4 | A | L47Q, K51N | H57Y | |
| 5 | A | I92V | H57Y |
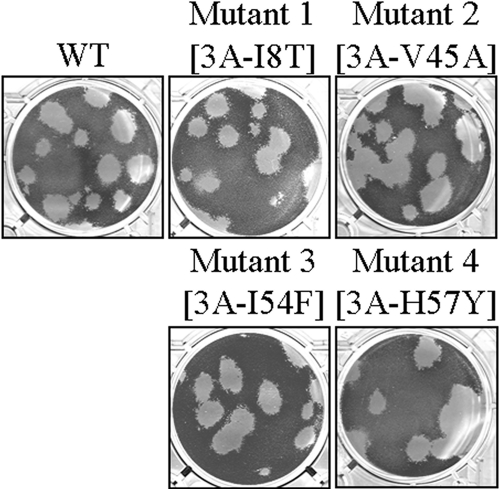
Recombinant CVB3 3A mutants have plaque phenotypes similar to those of wild-type virus.
A plasmid (pCB53/T7) encoding an infectious full-length CVB3 genome was used to generate four recombinant clones carrying the four identified mutations in protein 3A individually. These clones were designated mutant 1 (3A[I8T]), mutant 2 (3A[V45A]), mutant 3 (3A[I54F]), and mutant 4 (3A[H57Y]). Plaque assays with the mutant viruses revealed that infection with these mutant viruses do not result in altered plaque phenotypes compared to wild-type CVB3 (Fig. 4).
FIG. 4.
Plaque phenotypes of wild-type CVB3 and recombinant clones.
Recombinant CVB3 clones carrying mutations in 3A are resistant to inhibition by TTP-8307 and enviroxime.
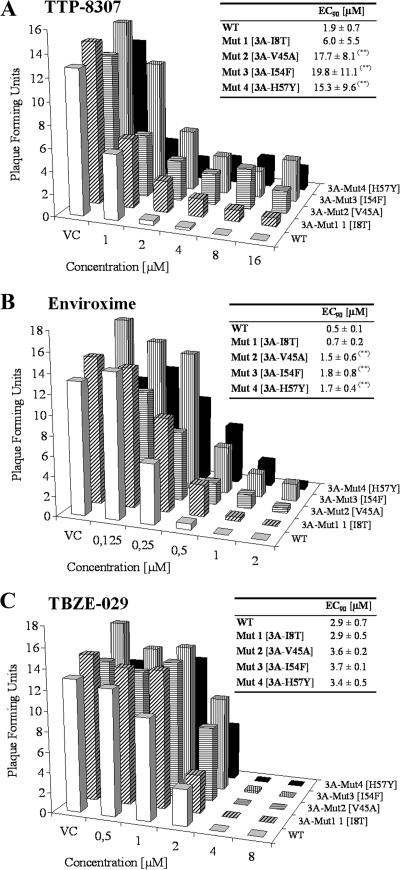
Next, the four recombinant mutant viruses were evaluated for their ability to form plaques in the presence of various concentrations of TTP-8307. Two reference molecules were included as a control: enviroxime (previously reported to inhibit enterovirus replication by targeting 3A) and the 2C inhibitor TBZE-029 (9). The graphs depicted in Fig. 5 represent PFU after infection of Vero cells with wild-type virus or the constructed 3A mutants at a given concentration for a given compound.
FIG. 5.
Viral replication of wild-type and recombinant viruses in the presence of TTP-8307, enviroxime and TBZE-029. Each graph represents the number of PFU produced by wild-type CVB3 or by any of the four constructed 3A mutants—3A[I8T], 3A[V45A], 3A[I54F], and 3A[H57Y]—at a given compound concentration. Calculated EC90 values represent averages ± the SD from five independent experiments. **, P < 0.05.
In the absence of drug, all clones (as well as the wild type) resulted in the formation of, on average, 10 × 104 to 15 × 104 plaques per ml. TTP-8307 at a concentration of 8 μM or higher completely prevented the formation of plaques in virus-infected cultures, whereas the recombinant mutant clones were still able to form plaques in the presence of TTP-8307 at these concentrations. In particular, mutants 2 and 3 (and to a lesser extent mutant 4) carrying amino acid mutations V45A and I54F (and H57Y) were still able to generate progeny virus at higher drug concentrations. Mutant 1 (I8T) remained relatively sensitive to inhibition by TTP-8307. A comparable pattern of (lack of) sensitivity of the variant mutants was observed for enviroxime. Enviroxime- and TTP-8307-resistant mutants can thus be considered cross-resistant. Finally, the sensitivity of the different recombinant viruses was assessed in the presence of TBZE-029, a compound that we recently reported to target the nonstructural protein 2C. As expected, TBZE-029 was equipotent in its inhibition of wild-type and mutant viruses. Taken together, these data indicate that in particular 3A mutations V45A, I54F, and H57Y confer resistance to TTP-8307.
DISCUSSION
We identified a novel compound, TTP-8307, that potently inhibits the replication of several enteroviruses, including coxsackievirus B3 and poliovirus by interfering with the synthesis of viral RNA. Contemplating the need for antivirals in the end stages of the worldwide polio eradication (5, 11), TTP-8307 might be considered as an interesting compound for lead optimization, given its potent inhibitory effect on the replication of the three PV Sabin strains.
To determine the viral target of TTP-8307, resistant CVB3 variants were selected. Each clone carried at least one of the following mutations in protein 3A: I8T, V45A, I54F, or H57Y. These mutations were reintroduced in an infectious CVB3 full-length clone and the antiviral sensitivity of the resulting viruses was studied. Mutants 2, 3, and 4 (carrying 3A mutations V45A, I54F, or H57Y) were shown to form plaques in the presence of concentrations of TTP-8307 that did not allow replication of wild-type virus, confirming that these mutations contribute to the observed resistance phenotype. The calculated EC90 values of TTP-8307 for inhibition of these mutant viruses were ca. 8- to 10-fold higher than for wild-type CVB3. Moreover, cross-resistance was observed with enviroxime. The specificity of this resistance profile was corroborated by the fact that a 2C-targeting compound (9, 10) inhibited the replication of the mutants as efficiently as that of the wild-type virus. Mutant I8T allowed for some very low replication in the presence of TTP-8307 and enviroxime. The viruses carrying the engineered mutations proved phenotypically (formation of plaques) comparable to the wild-type virus, suggesting no deleterious effects of the 3A mutations on viral replication. It should be noted, however, that the reconstructed mutants carrying single amino acid mutations in 3A did not exhibit the same high degree of resistance that we observed with the naturally selected clones. In fact, all naturally selected clones were selected and plaque purified in the presence of 20 μM TTP-8307 and, hence, had EC90 values of >20 μM. None of the reconstructed viruses carrying single amino acid mutations, however, reached EC90 values of ≥20 μM. It may therefore be assumed that other sequence variations (in proteins different than 3A) may contribute to a further increase in the level of resistance to TTP-8307.
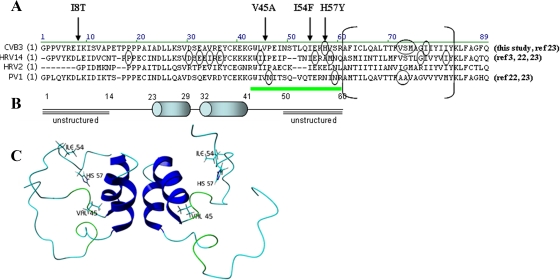
An amino acid sequence alignment of the 3A proteins of CVB3, PV1, HRV2, and HRV14 is depicted in Fig. 6. Globally, two major regions were reported to be important for resistance to enviroxime (3, 22, 23). The first region involves residues in the hydrophobic domain of 3A, located near its C terminus, whereas the second region is located between amino acids 40 and 60. This latter region, preceding the hydrophobic domain (underlined in green in Fig. 6), has been predicted to be unstructured, based on the nuclear magnetic resonance structure of the soluble domain of PV protein 3A (42). Remarkably, the 3A mutations that we identified in the present study as major determinants for resistance to TTP-8307 (V45A, I54F, and H57Y) were also located in this region (arrows in Fig. 6A; residues highlighted in Fig. 6C). Moreover, the very same H57Y mutation that was identified in TTP-8307-resistant CVB3 was also detected in enviroxime-resistant CVB3. In contrast to enviroxime, no mutations were detected in the hydrophobic domain of 3A in TTP-8307-resistant CVB3. This may be explained by the fact that TTP-8307 and enviroxime interact with the same region of 3A but that, given their different chemical structure, the precise molecular interactions with the various amino acids in this region may be (partially) different. Single amino acid mutations in the nonstructured region (amino acids 40 to 60) were shown to be sufficient to confer a certain degree of resistance to enviroxime (22, 23), corroborating our present observations. Florez de Sessions et al. observed that a chimeric CVB3 carrying an HRV2 internal ribosome entry site and that was adapted for growth in a neuroblastoma cell line carried the mutation V45A in 3A (16). Similar observations were described by Harris and Racaniello (19), who identified mutations in 3A at amino acid positions that correspond to CVB3 residues 42 and 44 in HRV39 that was adapted for growth in mouse cells. None of these mutations substantially affected viral plaque formation (16, 19), which is in line with our observations. The unstructured region preceding the hydrophobic domain may thus allow for adaptation of the virus to selective pressure of different kinds (e.g., antivirals or a changing host) without compromising viral growth.
FIG. 6.
(A) Sequence alignment of 3A proteins from CVB3, PV1, HRV2, and HRV14. The residues that are circled were earlier identified as amino acid mutations in enviroxime (or analog)-resistant variants (3, 22, 23). The arrows indicate mutations in TTP-8307 resistant CVB3, identified in the present study. The residues between brackets form a 22-residue hydrophobic region involved in membrane anchoring of 3A/3AB. The region underlined in green is suggested to be a “hot spot” for viral mutations when virus is selected in the presence of TTP-8307 or enviroxime. (B) Predicted solution structure of the N-terminal soluble domain of the PV 3A protein. The two cylinders represent two α-helices that form an α-helical hairpin. Adapted from Wessels et al. (46) with permission. (C) Location of the three predominant amino acid mutations involved in CVB3 resistance to TTP-8307 in a structural model of protein 3A.
A question that remains to be answered is what determines the spectrum of activity of TTP-8307. An alignment of the 3A amino acid sequence of TTP-8307-sensitive and -resistant viruses did not reveal any particular amino acids or regions in 3A that determined whether or not a virus would be inhibited by TTP-8307 (data not shown). For example, CVA16 and enterovirus 71 (TTP-8307 sensitive and resistant, respectively) share the N-terminal 3A amino acid sequence entirely, despite their difference in sensitivity. A possible explanation might be that 3A is a key protein in the replication inhibition, but that proteins other than 3A are involved in this inhibition as well.
Therefore, despite the fact that 3A was identified as a prime target for TTP-8307 as well as for enviroxime, the precise mechanism of action for inhibition of viral inhibition by these compounds remains to be elucidated. In contrast to 2CATPase, 3C(D)pro, and 3Dpol, protein 3A is not thought to be associated with any enzymatic activity and, hence, the inhibition of this protein by a compound cannot be assessed in a simple in vitro enzymatic assay. Rather than hampering a catalytic reaction, 3A-targeting drugs are likely to mediate hindrance of certain interactions of 3A with other (viral or cellular) proteins in the viral replication complex, an idea than can be supported given the multitude of interactions that have been ascribed to this protein (43, 50). For enviroxime, it was recognized that replication inhibition is probably not only occurring through targeting of 3A but may also depend on interactions with other proteins in the replication complex (3). More precisely, a HRV14 mutant was identified that carried, in addition to 3A mutations, mutations in 3Dpol. These additional mutations resulted in a higher degree of resistance. These findings are in line with our observations that several of the identified mutants carry amino acid changes in proteins other than 3A (Table 2). However, these mutations were identified in various proteins (2A, 2B, 2C, and 3D) and at different residue positions. Based on the hypothesis that TTP-8307 and enviroxime interfere with 3A through inhibition of certain interactions with other proteins, one could postulate that mutations in 3A are necessary for resistance, but that the level of resistance is increased by additional mutations in other proteins (that interact with 3A). Interestingly, recently CVB3 was selected that was resistant to amiloride; resistance was shown to map to the 3Dpol, but the resistant variant carried also a mutation at 3A residue I54 (similar to clones 4 and 6 identified in the present study) (21). The contribution of this 3A mutation to the resistant phenotype was however, not further determined.
In conclusion, we identified a novel enterovirus replication inhibitor that targets the nonstructural protein 3A. Mutations conferring the highest levels of resistance mapped to the nonstructured region preceding the hydrophobic domain, a region that was also reported to contain mutations in enviroxime-resistant viruses. The precise mechanism of viral inhibition by targeting 3A remains to be addressed in future studies.
Acknowledgments
This study was supported by the VIZIER integrated project (LSHG-CT-2004-511960) from the European Union 6th PCRDT. A.M.D.P. is a postdoctoral fellow of the FWO-Vlaanderen.
We thank Miette Stuyck for excellent technical assistance, A. Chimirri (University of Messina, Messina, Italy) for synthesizing TBZE-029, and G. Pürstinger (University of Innsbruck, Innsbruck, Austria) for the synthesis of enviroxime.
Footnotes
▿
Published ahead of print on 23 February 2009.
REFERENCES
- 1.Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271**:**23134-23137. [DOI] [PubMed] [Google Scholar]
- 2.Beard, C. W., and P. W. Mason. 2000. Genetic determinants of altered virulence of Taiwanese foot-and-mouth disease virus. J. Virol. 74**:**987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown-Augsburger, P., L. M. Vance, S. K. Malcolm, H. Hsiung, D. P. Smith, and B. A. Heinz. 1999. Evidence that enviroxime targets multiple components of the rhinovirus 14 replication complex. Arch. Virol. 144**:**1569-1585. [DOI] [PubMed] [Google Scholar]
- 4.Choe, S. S., D. A. Dodd, and K. Kirkegaard. 2005. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337**:**18-29. [DOI] [PubMed] [Google Scholar]
- 5.Couzin, J. 2006. Report concludes polio drugs are needed—after disease is eradicated. Science 311**:**1539. [DOI] [PubMed] [Google Scholar]
- 6.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97**:**13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong, D. C., and S. E. Reed. 1980. Inhibition of rhinovirus replication in in organ culture by a potential antiviral drug. J. Infect. Dis. 141**:**87-91. [DOI] [PubMed] [Google Scholar]
- 8.De Palma, A. M., W. Heggermont, K. Lanke, B. Coutard, M. Bergmann, A. M. Monforte, B. Canard, E. De Clercq, A. Chimirri, G. Puerstinger, J. Rohayem, F. van Kuppeveld, and J. Neyts. 2008. The thiazolobenzimidazole TBZE-029 inhibits enteroviral replication by targeting a short region immediately downstream motif C in the nonstructural protein 2C. J. Virol. 82**:**4720-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Palma, A. M., W. Heggermont, P. Leyssen, G. Purstinger, E. Wimmer, E. De Clercq, A. Rao, A. M. Monforte, A. Chimirri, and J. Neyts. 2007. Anti-enterovirus activity and structure-activity relationship of a series of 2,6-dihalophenyl-substituted 1H,3H-thiazolo[3,4-_a_]benzimidazoles. Biochem. Biophys. Res. Commun. 353**:**628-632. [DOI] [PubMed] [Google Scholar]
- 10.De Palma, A. M., G. Puerstinger, E. Wimmer, A. K. Patick, K. Andries, B. Rombaut, E. De Clercq, and J. Neyts. 2008. Comparative activity of a selected series of anti-picornavirus compounds against poliovirus replication in vitro. Emerg. Infect. Dis. 14**:**545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Palma, A. M., I. Vliegen, E. De Clercq, and J. Neyts. 2008. Selective inhibitors of picornavirus replication. Med. Res. Rev. 28**:**823-884. [DOI] [PubMed] [Google Scholar]
- 12.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75**:**8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71**:**9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14**:**894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74**:**6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florez de Sessions, P., E. Dobrikova, and M. Gromeier. 2007. Genetic adaptation to untranslated region-mediated enterovirus growth deficits by mutations in the nonstructural proteins 3AB and 3CD. J. Virol. 81**:**8396-8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita, K., S. S. Krishnakumar, D. Franco, A. V. Paul, E. London, and E. Wimmer. 2007. Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication. Biochemistry 46**:**5185-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65**:**2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris, J. R., and V. R. Racaniello. 2005. Amino acid changes in proteins 2B and 3A mediate rhinovirus type 39 growth in mouse cells. J. Virol. 79**:**5363-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269**:**27004-27014. [PubMed] [Google Scholar]
- 21.Harrison, D. N., E. V. Gazina, D. F. Purcell, D. A. Anderson, and S. Petrou. 2008. Amiloride derivatives inhibit coxsackievirus B3 RNA replication. J. Virol. 82**:**1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz, B. A., and L. M. Vance. 1995. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J. Virol. 69**:**4189-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinz, B. A., and L. M. Vance. 1996. Sequence determinants of 3A-mediated resistance to enviroxime in rhinoviruses and enteroviruses. J. Virol. 70**:**4854-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lama, J., and L. Carrasco. 1992. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J. Biol. Chem. 267**:**15932-15937. [PubMed] [Google Scholar]
- 25.Lama, J., and L. Carrasco. 1995. Mutations in the hydrophobic domain of poliovirus protein 3AB abrogate its permeabilizing activity. FEBS Lett. 367**:**5-11. [DOI] [PubMed] [Google Scholar]
- 26.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3aB as substrate for viral proteinases and as cofactor for RNA polymerase 3Dpol. J. Biol. Chem. 269**:**66-70. [PubMed] [Google Scholar]
- 27.Lama, J., M. A. Sanz, and L. Carrasco. 1998. Genetic analysis of poliovirus protein 3A: characterization of a non-cytopathic mutant virus defective in killing Vero cells. J. Gen. Virol. 79(Pt. 8)**:**1911-1921. [DOI] [PubMed] [Google Scholar]
- 28.Lama, J., M. A. Sanz, and P. L. Rodriguez. 1995. A role for 3AB protein in poliovirus genome replication. J. Biol. Chem. 270**:**14430-14438. [DOI] [PubMed] [Google Scholar]
- 29.Madan, V., A. Castello, and L. Carrasco. 2008. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell Microbiol. 10**:**437-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minor, P. D., P. Morgan-Capner, and P. Muir. 2000. Enteroviruses, p. 427-449. In A. J. Zuckerman, J. E. Banatvala, and J. R. Pattison (ed.), Principles and practice of clinical virology. John Wiley & Sons, Ltd., New York, NY.
- 31.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269**:**27015-27020. [PubMed] [Google Scholar]
- 32.Neznanov, N., A. Kondratova, K. M. Chumakov, B. Angres, B. Zhumabayeva, V. I. Agol, and A. V. Gudkov. 2001. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 75**:**10409-10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunez, J. I., E. Baranowski, N. Molina, C. M. Ruiz-Jarabo, C. Sanchez, E. Domingo, and F. Sobrino. 2001. A single amino acid substitution in nonstructural protein 3A can mediate adaptation of foot-and-mouth disease virus to the guinea pig. J. Virol. 75**:**3977-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pacheco, J. M., T. M. Henry, V. K. O'Donnell, J. B. Gregory, and P. W. Mason. 2003. Role of nonstructural proteins 3A and 3B in host range and pathogenicity of foot-and-mouth disease virus. J. Virol. 77**:**13017-13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul, A. V., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3Dpol: stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269**:**29173-29181. [PubMed] [Google Scholar]
- 36.Plotch, S. J., and O. Palant. 1995. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 69**:**7169-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards, O. C., and E. Ehrenfeld. 1998. Effects of poliovirus 3AB protein on 3D polymerase-catalyzed reaction. J. Biol. Chem. 273**:**12832-12840. [DOI] [PubMed] [Google Scholar]
- 38.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, and A. G. Mosser. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317**:**145-153. [DOI] [PubMed] [Google Scholar]
- 39.Rossmann, M. G., and J. E. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58**:**533-573. [DOI] [PubMed] [Google Scholar]
- 40.Sawyer, M. H. 2002. Enterovirus infections: diagnosis and treatment. Semin. Pediatr. Infect. Dis. 13**:**40-47. [DOI] [PubMed] [Google Scholar]
- 41.Semler, B. L., C. W. Anderson, R. Hanecak, L. F. Dorner, and E. Wimmer. 1982. A membrane-associated precursor to poliovirus VPg identified by immunoprecipitation with antibodies directed against a synthetic heptapeptide. Cell 28**:**405-412. [DOI] [PubMed] [Google Scholar]
- 42.Strauss, D. M., L. W. Glustrom, and D. S. Wuttke. 2003. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J. Mol. Biol. 330**:**225-234. [DOI] [PubMed] [Google Scholar]
- 43.Teterina, N. L., E. Levenson, M. S. Rinaudo, D. Egger, K. Bienz, A. E. Gorbalenya, and E. Ehrenfeld. 2006. Evidence for functional protein interactions required for poliovirus RNA replication. J. Virol. 80**:**5327-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wessels, E., D. Duijsings, K. H. Lanke, W. J. Melchers, C. L. Jackson, and F. J. van Kuppeveld. 2007. Molecular determinants of the interaction between coxsackievirus protein 3A and guanine nucleotide exchange factor GBF1. J. Virol. 81**:**5238-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wessels, E., D. Duijsings, R. A. Notebaart, W. J. Melchers, and F. J. van Kuppeveld. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 79**:**5163-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wessels, E., R. A. Notebaart, D. Duijsings, K. Lanke, B. Vergeer, W. J. Melchers, and F. J. van Kuppeveld. 2006. Structure-function analysis of the coxsackievirus protein 3A: identification of residues important for dimerization, viral rna replication, and transport inhibition. J. Biol. Chem. 281**:**28232-28243. [DOI] [PubMed] [Google Scholar]
- 47.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72**:**6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang, W., A. Cuconati, A. V. Paul, X. Cao, and E. Wimmer. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1**:**892-904. [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69**:**3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin, J., Y. Liu, E. Wimmer, and A. V. Paul. 2007. Complete protein linkage map between the P2 and P3 nonstructural proteins of poliovirus. J. Gen. Virol. 88**:**2259-2267. [DOI] [PubMed] [Google Scholar]