Dynamics of Malaria Drug Resistance Patterns in the Amazon Basin Region following Changes in Peruvian National Treatment Policy for Uncomplicated Malaria (original) (raw)
Abstract
Monitoring changes in the frequencies of drug-resistant and -sensitive genotypes can facilitate in vivo clinical trials to assess the efficacy of drugs before complete failure occurs. Peru changed its national treatment policy for uncomplicated malaria to artesunate (ART)-plus-mefloquine (MQ) combination therapy in the Amazon basin in 2001. We genotyped isolates collected in 1999 and isolates collected in 2006 to 2007 for mutations in the Plasmodium falciparum dihydrofolate reductase (Pf_dhfr_) and dihydropteroate synthase (Pf_dhps_) genes, multidrug resistance gene 1 (Pf_mdr-1_), the chloroquine (CQ) resistance transporter gene (Pf_crt_), and the Ca2+ ATPase gene (Pf_ATP6_); these have been shown to be involved in resistance to sulfadoxine-pyrimethamine (SP), MQ, CQ, and possibly ART, respectively. Microsatellite haplotypes around the Pf_dhfr_, Pf_dhps_, Pf_crt_, and Pf_mdr-1_ loci were also determined. There was a significant decline in the highly SP resistant Pf_dhfr_ and Pf_dhps_ genotypes from 1999 to 2006. In contrast, a CQ-resistant Pf_crt_ genotype increased in frequency during the same period. Among five different Pf_mdr-1_ allelic forms noted in 1999, two genotypes increased in frequency while one genotype decreased by 2006. We also noted previously undescribed polymorphisms in the Pf_ATP6_ gene as well as an increase in the frequency of a deletion mutant during this period. In addition, microsatellite analysis revealed that the resistant Pf_dhfr_, Pf_dhps_, and Pf_crt_ genotypes have each evolved from a single founder haplotype, while Pf_mdr-1_ genotypes have evolved from at least two independent haplotypes. Importantly, this study demonstrates that the Peruvian triple mutant Pf_dhps_ genotypes are very similar to those found in other parts of South America.
Several countries in South America have been obliged to change their malaria treatment policies (14, 15, 27), because drug resistance has spread rapidly across much of the continent. Chloroquine (CQ) resistance was first documented in 1960 in Venezuela (4), with widespread resistance across the Amazon by the mid-1980s (4). Sulfadoxine-pyrimethamine (SP) was introduced in the 1970s to counter CQ resistance, but in the early 1980s low levels of SP resistance were reported in in vivo efficacy trials, and failure rates had increased to >25% by the mid-1990s. As a result, CQ and SP were removed as first-line therapies for Plasmodium falciparum in many countries and were replaced by the more expensive artesunate (ART)-plus-mefloquine (MQ) combination therapy (ACT) (14). Until the early 1990s, CQ was the first-line therapy for uncomplicated P. falciparum malaria in Peru, and it remains the first-line therapy for Plasmodium vivax (15). Peru's national policy for the treatment of P. falciparum malaria changed from CQ to SP therapy for the Amazon basin region of Loreto in 1995 (13, 14) and for the North Coast region in June 1999 (13), but the usefulness of SP in the Peruvian Amazon was short-lived. ACT was implemented in 2001 and remains the primary treatment for uncomplicated P. falciparum infections in the Peruvian Amazon.
As malaria treatment policies have shifted to drug therapies with as yet unknown targets (i.e., artemisinin derivatives), it has become vital to monitor efficacy using passive surveillance, in vitro assays, and, when possible, molecular markers. Yet it is equally important that active surveillance be used to monitor how changes in public and open-market antimalarial usage influence circulating malaria parasite populations (12). In some instances, changes in drug policy have led to an increase in sensitive phenotypes, suggesting that mutant parasites have lower biological fitness than sensitive parasites in the absence of drug pressure (12, 34). Therefore, it is possible that antimalarials retired due to the emergence of drug resistance may eventually regain some usefulness. To this end, molecular surveillance data can provide highly useful information for policy makers in understanding the population dynamics of drug-resistant genotypes.
Genetic markers are being utilized to monitor the origins and spread of antimalarial drug resistance (1). Several loci have been identified for the monitoring of resistance to SP, CQ, atovaquone, MQ, and other important drugs that are still being utilized to treat uncomplicated malaria worldwide. Specific point mutations in P. falciparum crt (Pf_crt_) and in Pf_dhfr_ and Pf_dhps_ have been well characterized for CQ and SP resistance, respectively (7). Genotyping of specific polymorphisms before and during drug use is an excellent means of tracking the onset of resistance. Additionally, the characterization of microsatellite loci around these genes gives further traction to the molecular epidemiological approach in determining the origin and spread of the resistant phenotypes. Microsatellite loci have shown that both CQ-resistant and predominant SP-resistant genotypes originated independently in South America and Southeast Asia and that the Asian resistant genotypes spread to other parts of the world, including Africa (25, 33). Therefore, microsatellite markers are a useful tool for documenting the origins and tracking the spread of drug-resistant genotypes.
In this study, we have compared the frequencies of mutations in the Pf_dhfr_ and Pf_dhps_, Pf_mdr-1_, Pf_crt_, and Pf_ATP6_/SERCA (sarco/endoplasmic reticulum Ca2+ ATPase) genes from parasite isolates collected in the Amazon basin of Peru in 1999 (before ACT implementation) and in 2006 to 2007 (5 to 6 years after ACT implementation) in concert with an examination of the limited in vitro drug sensitivity data. Our findings demonstrate important shifts in the drug-resistant genotypes associated with resistance to SP, MQ, CQ, and possibly ART after the introduction of ACT. Additionally, we characterized microsatellite markers surrounding the genes in order to address the origins of drug-resistant genotypes in the Amazon basin.
MATERIALS AND METHODS
Study sites.
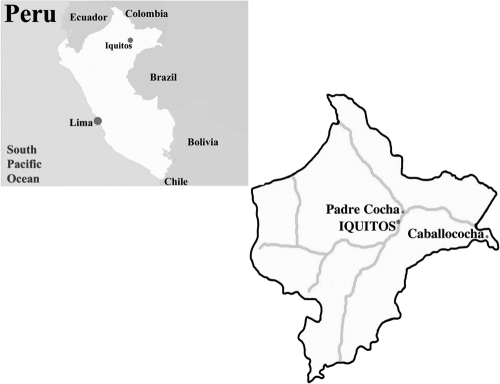
The Department of Loreto comprises almost one-fourth of the landmass of Peru and has an ecosystem characteristic of the Amazon lowlands (Fig. 1) (2). The rural population of Loreto, estimated at approximately 474,000 inhabitants, is clustered in towns and villages located in the Amazon tributary system. Iquitos is located 120 m above sea level (73°W, 3°S) at the juncture of the Ucayali and Napo Rivers, forming the Amazon River proper, and is accessible only by air or river. Iquitos has a tropical climate, with a mean temperature of 27.5°C, a mean annual rainfall of 4 m, and a mean annual precipitation of 2.7 m. The climate in this area is typical of the Amazon basin, with a rainy season from November to May and a dry season from June through October. The peak transmission rate for P. falciparum occurs during the rainy season (2).
FIG. 1.
Map depicting the locations of the various study sites where isolates were collected during the course of this study.
The city of Iquitos is the largest urban center in Loreto, with a population of approximately 345,000. The population is predominantly mixed Spanish and American Indian (mestizo). Household economies are sustained by agriculture, fishing, or tourism. The 2006-to-2007 study samples came from Iquitos, while the 1999 study samples were from Padre Cocha and Caballococha. The village of Padre Cocha is located on the Nanay River, approximately 5 km (10 min by boat) from Iquitos, with a population of 1,400 and relatively low levels of migration in and out of the village (26) (Fig. 1). The town of Caballococha is located in the northeastern Peruvian Amazon region, approximately 30 km from the Brazilian and Colombian borders, with a population of 3,300 (13) (Fig. 1).
Collection of malaria isolates.
The 1999 isolates were collected during an in vivo efficacy trial for SP (approval no. MCMR-RCQ-HR 70-1n from the Walter Reed Army Institute of Research) that contributed to the removal of this combination therapy from Peruvian Ministry of Health clinics (13, 26; D. J. Bacon et al., unpublished data). The 2006 and 2007 malaria isolates were collected as part of an ongoing febrile surveillance protocol (approval no. NMRCD.2000.0006 from the Naval Medical Research Center Detachment, Lima, Peru) from patients with symptoms typical of malaria who visited clinics located in the jungle city of Iquitos. Once they had been diagnosed with malaria by microscopy, blood was spotted onto filter paper for characterization of single-nucleotide polymorphisms (SNPs) and microsatellites. Venous blood was collected into 3-ml EDTA tubes for laboratory studies from febrile patients when P. falciparum malaria was diagnosed by microscopic examination. A total 600 μl of blood was taken from each tube under sterile conditions in a laminar-flow hood for drug susceptibility testing. One milliliter was used for the cryopreservation of parasites. Of the remainder, some was spotted onto Whatman 3M chromatography paper (Whatman, Inc., Sanford, ME) for PCR; some was used to prepare thick and thin smears for parasite confirmation and quantification; and the rest was frozen at −80°C.
Malaria control isolates.
Asexual P. falciparum cultures of reference clones D6/Sierra Leone (CQ sensitive and MQ resistant), W2/Indochina (CQ resistant and MQ sensitive; kindly provided by Dennis Kyle of WRAIR, Division of Experimental Therapeutics, Silver Spring, MD), and 3D7 (CQ and MQ sensitive; ATCC/MR4 clone originally collected from a traveler at the Amsterdam airport) were used as controls. Additional control strains, including 7G8/Brazil and Dd2/Indochina, were provided by MR4.
In vitro drug susceptibility testing.
Chloroquine diphosphate (molecular weight [MW], 515.9) (Sterling-Winthrop Research Institute, Rensselaer, NY), mefloquine hydrochloride (MW, 414.8) (Ash Stevens Inc., Detroit, MI), and quinine hydrochloride (MW, 782.9) (source unknown) were used for the in vitro test and were kindly provided by WRAIR and the WRAIR Chemical Repository. Plates were prepared and stored as previously described (3). Briefly, a total of 600 μl of fresh blood collected from patients with P. falciparum malaria was diluted with complete RPMI 1640 medium (Invitrogen), mixed, dispensed at 100 μl/well into predosed 96-well Nunc tissue culture plates, incubated at 37°C in a candle jar for 72 h, and then frozen at −80°C until the assay was performed using SYBR green I as previously described (3).
Genotyping of drug-resistant markers.
Genomic DNA was extracted either from 200-μl aliquots of in vitro-grown reference strains or from fresh whole blood by using a whole-blood DNA isolation kit (Qiagen, Chatsworth, CA). At study sites where whole blood was not collected, samples were spotted onto Isocode sticks or Whatman no. 1 filter paper and eluted using DNA/RNase-free water. PCR was performed on a GeneAmp 9700 system (Applied Biosystems, Foster City, CA) using the specific methods provided below for each gene of interest. Products were purified by using a QIAquick PCR purification kit (Qiagen) and were visualized prior to DNA sequencing. Five microliters of P. falciparum genomic DNA was used for each PCR with gene-specific primers (21). The DNA of purified PCR products was sequenced using the BigDye Terminator kit, version 3.1, and an ABI Prism Avant genetic analyzer, model 3100. All primers used for the PCR and sequencing of the five genes are listed in Table 1.
TABLE 1.
Primers used for PCR and DNA sequencing
| Gene | Primer orientation,a name | Primer sequence | Reference |
|---|---|---|---|
| Pf_mdr-1_ | |||
| SNP 86 | F, 727up | 5′-GTGTTTGGTGTAATATTAAAG-3′ | 17 |
| R, 1080Dn | 5′-CAAACGTGCATTTTTTATTAATG-3′ | ||
| SNP 1034 | F, mdr1 | 5′-GCTATTGATTATAAAAATAAAGGAC-3′ | 20 |
| R, mdr2 | 5′-CCA AAT TTG ATA TTT TCA TAT ATG GAC-3′ | ||
| Pf_dhfr_ | F, AMP1 | 5′-TTTATATTTTCTCCTTTTTA-3′ | 22 |
| R, AMP2 | 5′-CATTTTATTATTCGTTTTCT-3′ | ||
| Pf_dhps_ | F, M3717 | 5′-CCATTCCTCATGTGTATACAACAC-3′ | 31 |
| R, #186 | 5′-GTTTAATCACATGTTTGCACTTTC-3′ | ||
| 165-R | 5′-TTTTCATTTTGTTGTTCATCATG-3′ | ||
| Pf_crt_ | F, 23327up | 5′-CATTGTCTTCCACATATATGACATAAA-3′ | 17 |
| R, 24076dn | 5′-TTGGTAGGTGGAATAGATTCTCTT-3′ | ||
| ATP6 | F, ATP6-11 | 5′-TTTTCTTGGTTCTTTGCTCTTCC-3′ | 9 |
| R, ATP6-12 | 5′-TAGGCAAGCACCTTATCTTTATC-3′ | ||
| F, ATP6-21 | 5′-AATATGAACAGTGATCCTCAGAC-3′ | ||
| R, ATP6-22 | 5′-AATCCACCAGAACATGACGTAAT-3′ | ||
| F, ATP6-31 | 5′-TACCTAGTGCTGTTGCTGGTAA-3′ | ||
| R, ATP6-32 | 5′-TTGAAGCTTTAACGGATGATGGA-3′ | ||
| CaF6b | 5′-AGGTGCACCTGAGAATATAATAA-3′ | ||
| F, ATP6-41 | 5′-TAAAGCTAATTCGGTACTGTCTC-3′ | ||
| R, ATP6-42 | 5′-ATGCTCATACATACGATGTTGAG-3′ |
Pf_dhfr_ and Pf_dhps_.
For Pf_dhfr_, the A16V, C50R, N51I, C59R, S108T/N, and I164L mutations and the presence of the Bolivian repeat (BR) insertion were determined (4, 11, 29). For Pf_dhps_, the S436A, A437G, K540E, A581G, and A613T/S mutations were determined (31).
Pf_crt_.
The Pf_crt_ sequence was amplified by using a nested-PCR strategy with direct DNA sequencing analysis (17) in order to identify polymorphisms at amino acids 72 to 76 and 97 (6).
Pf_mdr-1_.
Two PCRs were used to determine polymorphisms at five locations in Pf_mdr-1_. The previously described primers 727up and 1080dn (17) were used to amplify a 350-bp product spanning codons 84 to 186 (17). A second reaction using primers mdr1 and mdr2 was used to amplify a 747-bp product in order to determine mutations at positions 1034, 1042, and 1246 (20).
Plasmodium falciparum Ca2+ ATPase gene (Pf_ATP6_).
The P. falciparum Ca2+ ATPase gene was analyzed in order to identify five previously characterized mutations (F263L, L264D/K, E431K, A623E, and S769N) (9). Three microliters of genomic DNA was used in each of four different PCRs containing 0.25 mM (final concentration) deoxynucleoside triphosphate mixture, 1× PCR buffer, 4 mM MgCl2, 0.5 μM each primer, and 2.5 U of Amplitaq Gold polymerase (Applied Biosystems, Foster City, CA). Amplification conditions were as follows: 94°C for 9 min; 38 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 3 min or at 60°C for 2 min, 30 s (for the first fragments and for the last two fragments, respectively), and extension at 72°C for 2 min; and a final extension at 72°C for 10 min. PCR products were purified and DNA sequenced as described above.
Pf_mdr-1_ copy number.
The Pf_mdr-1_ copy number was assessed using a LightCycler 480 real-time PCR instrument (Roche Applied Science). The β-tubulin gene was used as a copy number control for relative quantification assays. The primer and TaqMan probe sequences were the same as those of Price et al. (23), and fluorescent tags were changed to 6-carboxyfluorescein and Cy5 for Pf_mdr-1_ and β-tubulin, respectively. Reference strain 3D7 was used as a single-copy control for relative quantification. Individual 20-μl reaction mixtures consisting of 10 μl of LightCycler 480 Probes Master (Roche Diagnostics, Indianapolis, IN), 600 nM each primer, 300 nM probe, and 5 μl of purified genomic DNA were used under the following conditions: 5 min of initial denaturation at 95°C, followed by 45 cycles of 95°C for 30 s, 53°C for 10 s, and 72°C for 5 s. The copy number was determined using the relative quantification module in the LightCycler 480 software.
Genetic analysis.
DNA sequences were annotated using Sequencher, version 4.7 (Gene Codes Corporation). Nucleotide and amino acid sequences were aligned and compared to reference strains by using Clustal X (28).
Microsatellite analysis.
Microsatellite analysis was conducted for all of the samples from 1999 and 2006 to 2007 that were genotyped. All samples were assayed for five microsatellite loci that span approximately 11 kb on chromosome 4 around Pf_dhfr_, nine loci that span 17 kb on chromosome 8 around Pf_dhps_, four loci that span 11 kb on chromosome 7 around Pf_crt_, and six loci that span 8 kb on chromosome 5 around Pf_mdr1_. The primers used to amplify microsatellite loci have been described previously (16, 18, 19, 33). Single-reaction thermal cycling conditions and nested-PCR thermal cycling conditions have been detailed previously (18, 24). PCR Master Mix from Promega (Madison, WI) with a total reaction volume of 15 μl was used for PCRs. PCR products were assayed for size on Applied Biosystems 3100 and 3130XL capillary sequencers. The data were scored using GeneMapper software, versions 3.5 and 3.7 (Applied Biosystems, Foster City, CA). Microsatellite data were grouped into haplotypes (variation in ≤2 loci compared to reference group A1) and haplogroups (variation in >2 loci compared to the reference group). For example, a designation of A1 (reference haplotype) means that this could have been a founder lineage, and other haplogroups, such as B1 and C1, differ from this founder lineage at >2 loci. When the microsatellite locus genotype was different by only <2 loci, it was considered a minor variant and labeled as a haplotype variant (A1, A2, and A3, etc).
Statistical analysis.
The genotypic frequencies of samples collected in 1999 and in 2006 to 2007 were compared. The frequency procedure (proc freq) in SAS, version 9.1.3, was used for all comparisons (SAS Institute Inc., NC). The exact version of Pearson's chi-square test was used for all comparisons in order to provide conformity in the interpretation of results. Unlike Fisher's exact test, this statistical approach allows for tables greater than 2 by 2, which was appropriate for our sample set, since we had more than two genotypes for some genes. In addition, Pearson's chi-square test is still robust when confronted with small sample sizes, which was appropriate for our samples, because a few genotypes were seen rarely.
We used an α of 0.05 as our threshold of statistical significance for comparisons that included all genotype counts at both time points. However, when there were more than two genotypes at each time point, the Pearson chi-square test did not define the individual impact of any particular genotype on the overall P value reported. Therefore, we conducted multiple comparisons for genes for which there were more than two genotypes. In such comparisons, a genotype of interest was compared to the combined sample counts of the remaining genotypes in order to examine the influence of each genotype on the overall reported P value. For these subtests, we reduced our threshold α by dividing it by the number of multiple comparisons being conducted. Thus, we set the significance at an α of 0.0167 for the comparisons of dhfr and dhps genotypes (three comparisons), at an α of 0.01 for Pf_mdr-1_ (five comparisons), and at an α of 0.00625 for Pf_ATP6_ (eight comparisons).
RESULTS
Isolates and molecular marker determination.
A total of 105 isolates of P. falciparum collected in 1999 and 62 isolates collected in 2006 to 2007 were subjected to molecular marker analysis in order to identify polymorphisms in five genes (Pf_crt_, Pf_mdr-1_, Pf_dhfr_, Pf_dhps_, and Pf_ATP6/SERCA_) shown previously to confer drug resistance. We characterized microsatellite loci around the four genes Pf_crt_, Pf_mdr-1_, Pf_dhfr_, and Pf_dhps_ in order to investigate the relationships among genotypes in the population and also to explore haplotype distribution over time. One sample appeared to have a polyclonal infection, as shown by multiple microsatellite haplotypes at multiple loci for Pf_dhfr_ and Pf_dhps_. This sample was not included in the data presented here or in the analysis portion of the study; thus, we present data for 104 isolates from 1999 and 62 isolates from 2006 and 2007.
Molecular markers for Pf_dhfr_ and Pf_dhps_.
Only two Pf_dhfr_ genotypes were found in the sample set from 1999: the 108N single mutant and the BR 51I 108N 164L quadruple mutant. The frequency of the Pf_dhfr_ BR 51I 108N 164L genotype among the 1999 isolates was determined to be 52% (54/104), while 48% (50/104) of the isolates had only the single 108N mutation (Table 2). We found that the frequency of the same quadruple mutant in 2006 had declined significantly, to 16% (P < 0.0000006), while the frequency of the 108N mutant had increased significantly, to 79% (P < 0.00008). A previously undetected quadruple mutant genotype in Pf_dhfr_ (BR 50R 51I 108N), which to date has been seen only outside of Peru, was found in 5% of the isolates from 2006 to 2007. This genotype was found only in conjunction with the Pf_dhps_ 437G 540E 581G triple mutant. For Pf_dhps_, 30% (31/104) of the isolates collected in 1999 had the 437G 540E 581G triple mutation; 22% (23/104) had the 437G 581G double mutation; and 48% (50/104) were wild type (Table 3). As with Pf_dhfr_, the presence of the triple and double mutants dropped to 6% (4/62) (P < 0.0003) and 11% (7/62) (P = 0.09), respectively, and the presence of the wild-type genotype increased to 83% (51/62) (P < 0.0001) in 2006 to 2007.
TABLE 2.
Pf_dhfr_ genotypes and microsatellite markers from isolates collected in 1999 and 2006 to 2007
| Yr of collection (no. of isolates) | No. of samples froma: | Pf_dhfr_ genotypeb | Microsatellite haplotype at the following locusc: | Haplogroup | Frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | PC | Iquitos | BR | C50R | N51I | S108N | I164L | −5.30 | −3.87 | −0.3 | 0.52 | 5.87 | ||
| 1999 (104) | 25 | 28 | Yes | C | I | N | L | 223 | 216 | 101 | 97 | 122 | DHFR-A1 | 0.51 |
| 1 | 0 | Yes | C | I | N | L | 223 | 220 | 101 | 97 | 122 | DHFR-A2 | 0.01 | |
| 0 | 31 | No | C | N | N | I | 223 | 195 | 125 | 97 | 108 | DHFR-B1 | 0.30 | |
| 14 | 5 | No | C | N | N | I | 202 | 209 | 97 | 103 | 108 | DHFR-C1 | 0.18 | |
| 2006-2007 (62) | 8 | Yes | C | I | N | L | 223 | 216 | 101 | 97 | 122 | DHFR-A1 | 0.13 | |
| 1 | Yes | C | I | N | L | 223 | 216 | 101 | 97 | 120 | DHFR-A3 | 0.016 | ||
| 1 | Yes | C | I | N | L | 225 | 216 | 101 | 97 | 122 | DHFR-A4 | 0.016 | ||
| 3 | Yes | R | I | N | I | 202 | 209 | 97 | 99 | 120 | DHFR-D1 | 0.05 | ||
| 44 | No | C | N | N | I | 223 | 195 | 125 | 97 | 108 | DHFR-B1 | 0.71 | ||
| 1 | No | C | N | N | I | 223 | 195 | 113 | 97 | 108 | DHFR-B2 | 0.016 | ||
| 4 | No | C | N | N | I | 202 | 209 | 97 | 103 | 108 | DHFR-C1 | 0.065 |
TABLE 3.
Pf_dhps_ genotypes and microsatellite markers from isolates collected in 1999 and 2006 to 2007
| Yr of collection (no. of isolates) | No. of samples froma: | Pf_dhps_ genotypeb | Microsatellite haplotype at the following locusc: | Haplogroup | Frequency | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | PC | Iquitos | S436A | A437G | K540E | A581G | A613S/T | −7.4 | −2.47 | −1.64 | 0.80 | 0.006 | 0.144 | 1.59 | 6.19 | 9.79 | ||
| 1999 (104) | 7 | 24 | S | G | E | G | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 180 | 232 | DHPS-A1 | 0.3 |
| 0 | 1 | S | G | K | G | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 180 | 232 | DHPS-A1 | 0.01 | |
| 19 | 3 | S | G | K | G | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 178 | 232 | DHPS-A2 | 0.21 | |
| 0 | 31 | S | A | K | A | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 180 | 232 | DHPS-A1 | 0.3 | |
| 14 | 5 | S | A | K | A | A | 285 | 247 | 140 | 130 | 134 | 190 | 189 | 180 | 220 | DHPS-B1 | 0.18 | |
| 2006-2007 | 1 | S | G | E | G | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 178 | 232 | DHPS-A2 | 0.016 | |
| (62) | 3 | S | G | E | G | A | 310 | 266 | 140 | 124 | 134 | 175 | 197 | 183 | 232 | DHPS-A3 | 0.05 | |
| 7 | S | G | K | G | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 178 | 232 | DHPS-A2 | 0.11 | ||
| 46 | S | A | K | A | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 180 | 232 | DHPS-A1 | 0.74 | ||
| 1 | S | A | K | A | A | 310 | 266 | 140 | 124 | 134 | 171 | 197 | 232 | DHPS-Ad | 0.016 | |||
| 2 | S | A | K | A | A | 310 | 247 | 140 | 130 | 134 | 190 | 189 | 180 | 222 | DHPS-B2 | 0.032 | ||
| 2 | S | A | K | A | A | 292 | 270 | 140 | 137 | 134 | 166 | 181 | 171 | 220 | DHPS-C1 | 0.032 |
Considering both genes together, the presence of the septuplet Pf_dhfr_ BR 51I 108N 164L and Pf_dhps_ 437G 540E 581G and the presence of the sextuplet Pf_dhfr_ BR 51I 108N 164L and Pf_dhps_ 437G 581G dropped from 30% (31/104) and 22% (23/104), respectively, in 1999 to 6% (7/62) and 11% (7/62), respectively, in 2006. A new sextuplet (Pf_dhfr_ BR 50R 108N 164L and Pf_dhps_ 437G 581G) emerged in isolates collected in 2006 to 2007 (5% [3/62]).
Microsatellite haplotypes around Pf_dhfr_ and Pf_dhps_.
The Pf_dhfr_ BR 51I 108N 164L quadruple mutant belonged to a single haplogroup, Pf_dhfr_A, with minor variants defined as Pf_dhfr_-A1 to -A4. The Pf_dhfr_-A1 haplotype was the predominant haplotype for the quadruple mutant genotypes at both of the study sites from 1999 (Table 2). Between 1999 and 2006 to 2007, at least two new minor frequency variants, Pf_dhfr_-A3 and -A4, emerged for the quadruple mutant genotype. Interestingly, the newly found Pf_dhfr_ mutant genotype BR 50R 51I 108N was similar to the Pf_dhfr_-C1 haplotype but differed at two loci and was assigned a new haplogroup, D1.
The 108N single mutant belonged to two distinct haplogroups: Pf_dhfr_-B and Pf_dhfr_-C. Haplogroup Pf_dhfr_-C1 was found at both sites in 1999 and remained in the population at a low frequency in 2006 and 2007. Pf_dhfr_-B1 was found in Padre Cocha (1999) and in the majority of the Iquitos 108N mutant samples from 2006 to 2007; Padre Cocha and Iquitos are in close proximity. It should be noted, however, that Pf_dhfr_-B1 was not found in Caballococha, which is 311 km east of Padre Cocha and Iquitos. A minor variant, Pf_dhfr_-B2, has emerged since 1999.
All isolates from 1999 with the triple mutant Pf_dhps_ genotype 437G 540E 581G showed a single haplotype (Pf_dhps_-A1), and there was no difference between the two study sites (Table 3). By 2006 to 2007, this haplotype was no longer found for the triple mutant genotype, and only the minor variant haplotypes Pf_dhps_-A2 and Pf_dhps_-A3 were found. The double mutant (437G 581G) genotypes at both time points also belonged to haplogroup A.
Wild-type genotypes existed in four distinct haplotypes (Pf_dhps_-A1, -B1, -C1, and -B2). Interestingly, in 1999 a wild-type genotype shared the haplotype Pf_dhps_-A1 with the triple mutant genotype. The isolates with wild-type genotypes collected in 1999 from Padre Cocha had this A1 haplotype, but a majority of wild-type isolates from Caballococha had mostly B1, suggesting that wild-type genotypes had some geographic differentiation.
Molecular markers and in vitro tests for Pf_crt_.
No wild-type Pf_crt_ genotypes were present at either time point. Two common genotypes were seen in both year groups (Table 4). In 1999, the CVMNT genotype (underlined letters indicate mutations) was found in 46% (48/104) of the isolates and the SVMNT genotype was found in 54% (56/104). SVMNT (68%) was more common in Caballococha than CVMNT (32%), but these two genotypes were present at approximately equal frequencies in Padre Cocha. Among isolates collected in 2006 to 2007, the CVMNT genotype was found in only 34% (21/62) while the SVMNT genotype increased to 66% (41/62); the shifts in these genotypes were not statistically significant.
TABLE 4.
Pf_crt_ genotypes and microsatellite markers from isolates collected in 1999 and 2006 to 2007
| Yr of collection (no. of isolates) | No. of samples froma: | Pf_crt_ genotypeb | Microsatellite haplotype at the following locusc: | Haplogroup | Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | PC | Iquitos | C72S | 73 | M74I | N75E | K76T | H97Q | −5 | −4.3 | 1 | 6 | |
| 1999 (104) | 26 | C | V | M | N | T | H | 186 | 233 | 160 | 305 | CRT-A1 | 0.25 |
| 2 | C | V | M | N | T | H | 183 | 233 | 160 | 305 | CRT-A2 | 0.02 | |
| 1 | C | V | M | N | T | H | 152 | 233 | 160 | 305 | CRT-A3 | 0.01 | |
| 1 | C | V | M | N | T | H | 186 | 231 | 155 | 289 | CRT-B1 | 0.01 | |
| 12 | 5 | C | V | M | N | T | H | 186 | 231 | 155 | 305 | CRT-B2 | 0.16 |
| 1 | C | V | M | N | T | H | 152 | 231 | 155 | 305 | CRT-B3 | 0.01 | |
| 24 | 26 | S | V | M | N | T | H | 183 | 233 | 160 | 305 | CRT-A2 | 0.48 |
| 2 | S | V | M | N | T | H | 152 | 233 | 160 | 305 | CRT-A3 | 0.02 | |
| 1 | S | V | M | N | T | H | 183 | 231 | 160 | 305 | CRT-A4 | 0.01 | |
| 1 | S | V | M | N | T | H | 152 | 233 | 160 | 294 | CRT-A5 | 0.01 | |
| 1 | S | V | M | N | T | H | 135 | 233 | 160 | 305 | CRT-A6 | 0.01 | |
| 1 | S | V | M | N | T | H | 186 | 231 | 155 | 305 | CRT-B2 | 0.01 | |
| 2006-2007 (62) | 2 | C | V | M | N | T | H | 186 | 233 | 160 | 305 | CRT-A1 | 0.03 |
| 1 | C | V | M | N | T | H | 183 | 233 | 160 | 305 | CRT-A2 | 0.016 | |
| 1 | C | V | M | N | T | H | 186 | 201 | 155 | 305 | CRT-A7 | 0.016 | |
| 1 | C | V | M | N | T | H | 186 | 233 | 155 | 305 | CRT-A8 | 0.016 | |
| 16 | C | V | M | N | T | H | 186 | 231 | 155 | 305 | CRT-B2 | 0.26 | |
| 40 | S | V | M | N | T | H | 183 | 233 | 160 | 305 | CRT-A2 | 0.65 | |
| 1 | S | V | M | N | T | H | 186 | 231 | 155 | 305 | CRT-B2 | 0.016 |
CQ and quinine IC50s were determined for the fresh isolates collected in 2006 to 2007 containing the CVMNT or SVMNT genotype. The mean IC50s of CQ were 75 ± 44 nM for the CVMNT group and 89 ± 35 nM for the SVMNT group. The IC50s of quinine for the CVMNT and SVMNT genotypes were 66 ± 26 nM and 70 ± 25 nM, respectively.
Microsatellite haplotypes around Pf_crt_.
The microsatellite haplotypes were found to be similar and closely related for both the CVMNT and SVMNT genotypes in 1999 and 2006 to 2007 (Table 4). The predominant Pf_crt_-A2 SVMNT haplotype was seen in both Padre Cocha and Caballococha in 1999 as well as in Iquitos in 2006 to 2007. However, by 2006 to 2007 there was notably less variation in SVMNT haplotypes. In addition, the SVMNT haplotype A2 is closely related to the Pf_crt_-A1 haplotype, which was seen in the majority of CVMNT genotypes from Padre Cocha in 1999. The CVMNT genotypes from Caballococha were represented by haplogroup B (Table 4). By 2006 to 2007, the majority of CVMNT genotypes had the B2 haplotype.
Molecular markers, copy number characterization, and in vitro tests for Pf_mdr-1_.
Five Pf_mdr-1_ genotypes were found in 1999 and in 2006 to 2007 (Table 5). In 1999, several genotypes were distributed in different proportions between the two study locations. For instance, allelic variants NGFSDD and NGFCDD were found exclusively in Padre Cocha, while genotype NDFSDD was detected only in one sample from Caballococha. This skewed distribution could be attributed to a small sample size. The most common genotypes in 1999, regardless of sampling site, were NDFCDD (46/104; 44%) and NDFCDY (44/104; 42%). In 2006 to 2007, the most common genotypes were NGFSDD (18/62; 29%) and NDFCDD (32/62; 52%) (Table 5). Interestingly, the frequencies of NGFSDD and NDFCDD increased from 1999 to 2006 to 2007 (from 15% to 29% [P < 0.0065] and from 44% to 52% [with no significant difference], respectively), while the frequency of NDFCDY dropped significantly (from 42% to 14% [P < 0.0002]).
TABLE 5.
Pf_mdr-1_ genotypes and microsatellite markers from isolates collected in 1999 and 2006 to 2007
| Yr of collection (no. of isolates) | No. of samples froma: | Pf_mdr-1_ genotypeb | Microsatellite haplotype at the following locusc: | Haplogroup | Frequency | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | PC | Iquitos | N86Y | D142G | Y184F | S1034C | N1042D | D1246Y | −4.2 | −3.4 | −1.2 | id | 0.56 | 3.8 | |
| 1999 (104) | 1 | N | D | F | S | D | D | 204 | 127 | 197 | 206 | 192 | 169 | MDR-A1 | 0.01 |
| 4 | N | G | F | S | D | D | 196 | 133 | 203 | 206 | 178 | 189 | MDR-B1 | 0.038 | |
| 4 | N | G | F | S | D | D | 196 | 135 | 203 | 206 | 178 | 189 | MDR-B2 | 0.038 | |
| 4 | N | G | F | S | D | D | 196 | 127 | 203 | 206 | 178 | 189 | MDR-B3 | 0.038 | |
| 26 | N | D | F | C | D | D | 204 | 127 | 197 | 206 | 192 | 169 | MDR-A1 | 0.25 | |
| 1 | N | D | F | C | D | D | 204 | 127 | 200 | 206 | 192 | 169 | MDR-A2 | 0.01 | |
| 2 | N | D | F | C | D | D | 204 | 133 | 197 | 206 | 192 | 169 | MDR-A3 | 0.02 | |
| 1 | N | D | F | C | D | D | 204 | 133 | 200 | 206 | 192 | 169 | MDR-A4 | 0.01 | |
| 1 | N | D | F | C | D | D | 204 | 119 | 197 | 206 | 192 | 169 | MDR-A5 | 0.01 | |
| 1 | N | D | F | C | D | D | 204 | 121 | 197 | 206 | 192 | 169 | MDR-A6 | 0.01 | |
| 4 | N | D | F | C | D | D | 196 | 133 | 203 | 206 | 178 | 189 | MDR-B1 | 0.038 | |
| 1 | N | D | F | C | D | D | 196 | 135 | 203 | 206 | 178 | 189 | MDR-B2 | 0.01 | |
| 7 | N | D | F | C | D | D | 196 | 127 | 203 | 206 | 178 | 189 | MDR-B3 | 0.067 | |
| 1 | N | D | F | C | D | D | 196 | 133 | 197 | 206 | 178 | 189 | MDR-B4 | 0.01 | |
| 1 | N | D | F | C | D | D | 196 | 127 | 203 | 206 | 178 | 169 | MDR-B5 | 0.01 | |
| 19 | 14 | N | D | F | C | D | Y | 204 | 127 | 197 | 206 | 192 | 169 | MDR-A1 | 0.32 |
| 4 | 3 | N | D | F | C | D | Y | 204 | 133 | 197 | 206 | 192 | 169 | MDR-A3 | 0.067 |
| 2 | N | D | F | C | D | Y | 204 | 127 | 197 | 206 | 192 | 189 | MDR-A7 | 0.02 | |
| 1 | N | D | F | C | D | Y | 196 | 133 | 203 | 206 | 178 | 189 | MDR-B1 | 0.01 | |
| 1 | N | D | F | C | D | Y | 196 | 127 | 203 | 206 | 178 | 169 | MDR-B5 | 0.01 | |
| 1 | N | G | F | C | D | D | 196 | 127 | 203 | 206 | 178 | 189 | MDR-B3 | 0.01 | |
| 2006-2007 (62) | 1 | N | D | F | S | D | D | 196 | 133 | 203 | 206 | 178 | 189 | MDR-B1 | 0.016 |
| 10 | N | G | F | S | D | D | 196 | 133 | 203 | 206 | 178 | 189 | MDR-B1 | 0.16 | |
| 1 | N | G | F | S | D | D | 196 | 135 | 203 | 206 | 178 | 189 | MDR-B2 | 0.016 | |
| 1 | N | G | F | S | D | D | 196 | 133 | 203 | 206 | 111 | 189 | MDR-B6 | 0.016 | |
| 1 | N | G | F | S | D | D | 204 | 133 | 203 | 206 | 178 | 189 | MDR-B7 | 0.016 | |
| 1 | N | G | F | S | D | D | 196 | 133 | 203 | 206 | 194 | 189 | MDR-B8 | 0.016 | |
| 1 | N | G | F | S | D | D | 204 | 133 | 203 | 206 | 194 | 189 | MDR-B9 | 0.016 | |
| 3 | N | G | F | S | D | D | 196 | 133 | 205 | 206 | 178 | 189 | MDR-B10 | 0.05 | |
| 25 | N | D | F | C | D | D | 204 | 127 | 197 | 206 | 192 | 169 | MDR-A1 | 0.40 | |
| 2 | N | D | F | C | D | D | 204 | 127 | 200 | 206 | 192 | 169 | MDR-A2 | 0.03 | |
| 1 | N | D | F | C | D | D | 204 | 127 | 197 | 206 | 194 | 169 | MDR-A8 | 0.016 | |
| 3 | N | D | F | C | D | D | 196 | 133 | 203 | 206 | 178 | 189 | MDR-B1 | 0.05 | |
| 1 | N | D | F | C | D | D | 196 | 135 | 203 | 206 | 178 | 189 | MDR-B2 | 0.016 | |
| 6 | N | D | F | C | D | Y | 204 | 127 | 197 | 206 | 192 | 169 | MDR-A1 | 0.1 | |
| 1 | N | D | F | C | D | Y | 204 | 127 | 200 | 206 | 192 | 169 | MDR-A2 | 0.016 | |
| 1 | N | D | F | C | D | Y | 204 | 127 | 197 | 206 | 194 | 169 | MDR-A8 | 0.016 | |
| 1 | N | D | F | C | D | Y | 196 | 127 | 197 | 206 | 192 | 169 | MDR-A9 | 0.016 | |
| 1 | N | G | F | C | D | D | 196 | 133 | 203 | 206 | 178 | 189 | MDR-B1 | 0.016 | |
| 1 | N | G | F | C | D | D | 196 | 127 | 205 | 206 | 178 | 169 | MDR-C1 | 0.016 |
The copy number of Pf_mdr-1_ was determined for all isolates. Regardless of when isolates were collected, only a single copy of Pf_mdr-1_ was detected. Additionally, MQ IC50s were determined for the fresh isolates collected in 2006 to 2007. MQ IC50s for fresh isolates from 2006 to 2007 with the NDFCDY, NDFCDD, and NGFSDD genotypes were determined to be 39 ± 11 nM, 28 ± 17 nM, and 21 ± 10 nM, respectively.
Microsatellite haplotypes around Pf_mdr-1_.
There are three main microsatellite haplogroups for Pf_mdr-1_: A, B, and C. These haplogroups differ from each other at the majority of the loci. Haplogroups A and B are seen for the majority of the samples, while haplogroup C is seen only for one NGFCDD isolate from 2006 to 2007. We find that haplogroup A is predominant for the NDFCDD triple mutant and NDFCDY quadruple mutant genotypes from both sampling periods. It should be noted that there are low-frequency representatives of the Pf_mdr-1_-B haplogroup for these two genotypes in 1999 and for the triple mutant in 2006. All of the NDFCDD samples from Caballococha belonged to the Pf_mdr-1_-B haplogroup, whereas most of the NDFCDD parasites from Padre Cocha belonged to the Pf_mdr1_-A haplogroup. The NGFSDD genotypes at both time points belonged to the B haplogroup. The single samples with the NDFSDD genotype at each time point fell into two different haplogroups: A in 1999 and B in 2006 to 2007. Interestingly, each NGFCDD sample (n = 3) had a different haplotype.
Molecular markers of Pf_ATP6_/SERCA.
DNA from three exons of the Pf_ATP6_ gene was sequenced for the 104 isolates collected in 1999 and 57 of the 62 isolates collected in 2006 to 2007 (Table 6; see also Table S1 in the supplemental material). Five of the isolates were not included in the analysis, because they failed to produce the PCR fragment containing the L402V SNP. For 1999, two nonsynonymous mutations (L402V and S466N), one synonymous mutation (C1031C), and one complete codon deletion (G884) were found, none of which had been described previously (9). Among isolates collected in 2006 to 2007, we found five mutations (L402V, S466N, A630S, C1031C, and V1168I) and one codon deletion (G884). All of the mutations seen in 1999 were also seen in 2006, and two of the mutations found in 2006 to 2007 (630S and 1168I) were not seen in 1999. The S769N mutation, which was shown previously to be linked to increases in in vitro levels of resistance to artemether (9), was not seen in any samples from either year. There was a significant increase in the frequency of the G884 deletion (P < 0.005) from 1999 to 2006 and 2007. The possible role of the deletion at G884 in drug susceptibility is not currently known. When these mutants were grouped into allelic types, at least eight genotypes were found, as shown in Table 6. In samples collected in 2006 to 2007, at least two new genotypes were found: genotype 3 (402V 630E 1168I) at a frequency of 7.02% and genotype 7 (G884 1030C) at 3.51%. The genotype with a single deletion at G884 significantly increased in frequency, from 39.42% in 1999 to 68.45% in 2006 to 2007 (P < 0.0005). Other allelic types showed a trend toward a decrease in frequency.
TABLE 6.
Genotypes of Pf_ATP_ 6 found in the Peruvian Amazon basin in 1999 and 2006 to 2007
| Genotypea | Frequency (%) in: | 2006-2007 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 263 | 264 | 402 | 431 | 466 | 623 | 630 | 769 | 884 | 1031 | 1168 | 1999 | |
| L | F | L | E | S | A | A | S | G | C | V | 1.92 | 3.51 |
| L | F | V | E | S | A | A | S | G | C | V | 20.19 | 1.75 |
| L | F | V | E | S | A | S | S | G | C | I | 0 | 7.02 |
| L | F | L | E | N | A | A | S | —b | C | V | 18.27 | 5.26 |
| L | F | L | E | N | A | A | S | — | Cc | V | 0.96 | 0 |
| L | F | L | E | S | A | A | S | — | C | V | 39.42 | 68.45 |
| L | F | L | E | S | A | A | S | — | Cc | V | 0 | 3.51 |
| L | F | L | E | S | A | A | S | G | Cc | V | 19.23 | 10.53 |
DISCUSSION
This is one of the first studies to comprehensively determine the molecular changes associated with five different genes implicated in CQ, SP, MQ, and ART resistance in South America before and after a change in treatment policy. This study highlights several important observations, as follows. (i) The removal of SP in the Peruvian Amazon basin after a policy change has led to a decline in the frequency of the highly pyrimethamine resistant quadruple mutant Pf_dhfr_ genotype (BR 51I 108N 164L) and the sulfadoxine-resistant triple mutant Pf_dhps_ genotype (437G 540E 581G), confirming a recent report (34). (ii) A small number of isolates with a new Pf_dhfr_ quadruple mutant genotype (BR 50R 51I 108N), which had not been reported in Peru before, had migrated to the Peruvian Amazon by 2006 to 2007. (iii) It is demonstrated that the Pf_dhps_ triple mutant genotype circulating in South America may have come from a single common founder. (iv) The highly resistant SVMNT genotype in P. falciparum has increased in frequency but does not appear to have reached fixation in the Peruvian Amazon despite continuous selective pressure due to widespread use of CQ for the treatment of P. vivax in this region. (v) Shifts in Pf_mdr_-1 genotypes after the introduction of ACT are demonstrated for the first time. (vi) Microsatellite data demonstrate at least two different founder lineages for the evolution of the NDFCDD and NDFCDY mutant genotypes and a single lineage for the evolution of the NGFSDD and NGFCDD genotypes. (vii) Microsatellite data demonstrate some interesting patterns of geographic isolation of haplotypes. (viii) Molecular data show some important trends in the evolution of the SERCA gene, implicated in ART resistance, since the implementation of ACT in the Peruvian Amazon.
The results presented here clearly illustrate significant reductions in the frequencies of both the Pf_dhfr_ and Pf_dhps_ mutant genotypes, a finding consistent with a recent study that showed a reduction in the frequency of these resistant genotypes after the removal of SP (34). Thus, we have shown clear evidence for the decline in frequency of SP-resistant genotypes following a policy change in Peru—as has been demonstrated for CQ-resistant parasites in Malawi and China (32). Our results also show an increase in the frequency of the Pf_dhfr_ 108N mutants in combination with wild-type Pf_dhps_ genotypes, suggesting that the highly resistant genotypes do have a fitness cost in the absence of drug pressure and are not maintained in the population. In addition, the microsatellite results show that the existing wild-type Pf_dhps_ and 108N single mutant Pf_dhfr_ lineages persist and increase in frequency over time, rather than increasing due to migration from another source. These findings demonstrate the significance of molecular surveillance studies to monitor drug-resistant parasites even after a change in the drug policy. In the absence of such surveillance, it is difficult to know whether a policy change really alters the profile of drug-resistant parasites. It has been shown that resistant genotypes continue to persist at high frequencies in some populations even after a policy change (10, 16). The reason for the continued existence of resistant parasites at a high frequency after a policy change has been discussed in detail previously (16, 34).
Two major genotypes for Pf_dhfr_ (BR 51I 108N 164L and 50R 51I 108N) and one major genotype for Pf_dhps_ (437G 540E 581G) were associated with pyrimethamine and sulfadoxine resistance, respectively, in South America (4, 11). A previous study by Cortese et al. (4) using data from only two microsatellite loci around both Pf_dhfr_ and Pf_dhps_ and a limited number of field parasite isolates suggested that (i) the two Pf_dhfr_ mutant genotypes may have originated in the southern Bolivian/Brazilian area and may have independent origins, and (ii) the Pf_dhps_ mutants have a common origin in the Amazon. Independent origins for the two Pf_dhfr_ mutant genotypes have been verified recently in population-based studies from Peru and Venezuela (16, 34); however, additional population-level data to support the single-origin hypothesis for the Pf_dhps_ triple mutant genotype was needed. Our data from Peru clearly show that this genotype is very similar to the Venezuelan Pf_dhps_ genotype (16), suggesting a common origin for this Pf_dhps_ genotype on the continent. Additional population-level data from other sites in South America are still needed to confirm this hypothesis.
Interestingly, the non-Peruvian Pf_dhfr_ genotype BR 50R 51I 108N was found for the first time in Peru (3/62 samples in 2006 to 2007). Given that this genotype was not detected in the samples from 1999, we speculate that either this genotype was present at a very low frequency and thus was not detected in 1999 or it was introduced more recently due to population migration. It remains to be seen whether this genotype will be maintained or increase in frequency in the population.
Even though CQ is no longer prescribed for uncomplicated P. falciparum malaria in Peru, it continues to be the standard of care for P. vivax, which causes approximately 80% of the cases of malaria in this region of the Amazon. As a result, one would predict that continued use of this drug in this population may lead to fixation of the CQ-resistant SVMNT genotype in Peru, as has been observed in Venezuela (S. M. Griffing et al., unpublished data). Indeed, this fixation may eventually occur, but at this point we documented only a slight increase (not statistically significant) in the frequency of the SVMNT genotype in Peru from 1999 to 2006 and 2007.
The microsatellite profile of the SVMNT genotype has remained unchanged over time. Our findings support previous work suggesting that the SVMNT genotype present in Peru is related to the same genotype in Brazil (30, 33). It appears as though the CVMNT genotypes may have at least two founding lineages in the Peruvian Amazon. One parsimonious hypothesis we can generate from the microsatellite data is that a single or possibly multiple CVMNT genotypes are ancestors of the SVMNT genotypes. This is consistent with the findings of Wootton et al. (33), who utilized isolates from Brazil, Peru, Ecuador, and Colombia. We observed one or two haplotypes at high frequencies with a few rare variants for the CVMNT and SVMNT genotypes in 2006. These haplotype frequency distributions are characteristic of strong selection of the CVMNT and SVMNT genotypes in this population.
Pf_mdr-1_ has been implicated in resistance to CQ, MQ, and ART. However, the molecular changes that confer resistance associated with this gene are not well understood, especially for South America. Further, it is not known whether Pf_mdr-1_ mutant genotypes have originated from a single lineage or from multiple lineages. The data from this study show that there is a shift in the Pf_mdr-1_ mutant genotypes after the introduction of ART plus MQ. This is evident in a significant increase in the frequency of the NGFSDD genotype and a significant decline in the frequency of the NDFCDY genotype from 1999 to 2006 and 2007. We do not know whether these shifts are due to increased drug pressure from ART and/or MQ or to another factor such as migration or genetic drift. The limited amount of in vitro data that we have does not support the hypothesis that an increase in the frequency of the NGFSDD genotype is due to increased resistance to MQ. It remains to be determined why this shift in the molecular profile of Pf_mdr-1_ is occurring.
It should be noted that a previous study was conducted that assessed the molecular changes in Pf_mdr-1_ in 60 isolates collected in Padre Cocha in 1999 (8). This study reported a single genotype in Pf_mdr1_ (NFCDD) based on restriction fragment length polymorphism analysis. We were able to identify other genotypes by DNA sequencing. This highlights the limitation associated with the use of PCR and restriction fragment length polymorphism analysis to determine polymorphism.
The microsatellite data around Pf_mdr-1_ clearly establish that at least two founder lineages have contributed to the different mutant genotypes present in this population. The NDFCDY quadruple mutant and NDFCDD triple mutant genotypes may have evolved from the NDFSDD double mutant lineages via the Pf_mdr-1_-A and Pf_mdr-1_-B haplogroups independently. We speculate that a single mutation of position 1034 from S to C could have led the double NDFSDD genotype to contribute to the evolution of the NDFCDD triple mutant genotype and that an additional mutation at position 1246 (D to Y) may have given birth to the NDFCDY quadruple mutant genotype. These two progressive mutations may have occurred independently at least twice, as evidenced by the presence of both in the Pf_mdr-1_-A and Pf_mdr-1_-B haplogroups for the NDFCDD and NDFCDY genotypes. On the other hand, the NGFSDD triple mutant genotype may have evolved from the NDFSDD double mutant genotype and may have eventually yielded the NGFCDD quadruple mutant genotype, since the triple and quadruple mutant genotypes belong only to the Pf_mdr-1_-B haplogroup. It is also possible that a mutation from D to G at position 142 in the NDFCDD triple mutant genotype could have led to the NGFCDD quadruple mutant genotype, and the microsatellite haplotypes in Table 5 are consistent with this possibility as well.
For each gene, some mutant genotypes and haplotypes were found at only one of the two sites in 1999. For example, the Pf_dhfr_ 108N mutants from Caballococha had the C1 haplotype, while B1 was predominant for Padre Cocha. Similarly, the isolates with the wild-type Pf_dhps_ genotype in Caballococha belonged predominantly to the B1 haplotype, while those in Padre Cocha belonged to A1. The Pf_crt_ CVMNT genotypes with haplogroup A were predominant in Padre Cocha and those with haplogroup B in Caballococha. The Pf_mdr-1_ NDFCDY genotype with haplogroup A was found only in Padre Cocha, while in Caballococha the same genotype was found only with haplogroup B. A similar situation was also seen for the NDFCDD genotypes. We also saw geographic differentiation of Pf_mdr-1_ genotypes. If we discount genotypes found in <2 samples, then the NGFSDD genotype was found only in Padre Cocha. This result suggests that particular genotypes and lineages were not introduced as of 1999 or have not been maintained in Caballococha. The data support the general isolation of Padre Cocha P. falciparum populations from those in Caballococha. The overall similarity in the haplotypes present for most of the genotypes in Padre Cocha and Iquitos is expected, given their close geographic proximity.
In a previous study from French Guyana, the S769N mutation in the SERCA gene was implicated in in vitro resistance to ART (9); however, this mutation was not found in Peru. Three nonsynonymous mutations in the SERCA gene (L402V, S466N, and V1168I) and one synonymous mutation (C1031C) newly found in this study have not been reported previously (5, 9). Among the SERCA genotypes, a notable change was the substantial increase in the frequencies of G884 deletion mutants and the L402V A630S V1168I genotype by 2006 to 2007. We do not know whether this reflects a change due to drug pressure from ACT use or due to additional unknown factors. Additional studies will be required to understand the potential functional significance of these mutations, if any.
In conclusion, this study has provided comprehensive data illustrating changes in the allele frequencies of five major genes involved in resistance to different antimalarials in the Peruvian Amazon. These data will contribute to the understanding of the evolution of drug-resistant P. falciparum parasites in this region.
Supplementary Material
[Supplemental material]
Acknowledgments
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of the Navy, the Department of Defense, the Department of Health and Human Services, or the U.S. government.
This work was supported in part by funds provided by the Department of Defense-Global Emerging Infectious System (DoD-GEIS) under unit 847705.82000.25GB,B0016. The work was also supported in part by the Antimicrobial Drug Resistance Working Group, the Centers for Disease Control and Prevention, and the Atlanta Research and Education Foundation, Atlanta VA Medical Center. One of the authors is a military service member (D.J.B.). This work was prepared as part of his official duties. S. M. Griffing is supported under a National Science Foundation Graduate Research Fellowship. A. A. Escalante is supported by grant R01GM084320 from the National Institutes of Health.
The study protocol was approved by the Naval Medical Research Center Institutional Review Board in compliance with all applicable Federal regulations governing the protection of human subjects.
We acknowledge Shannon K. McClintock for support in the statistical analysis of the results.
Footnotes
▿
Published ahead of print on 2 March 2009.
REFERENCES
- 1.Anderson, T. J. C., and C. Roper. 2005. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 94**:**269-280. [DOI] [PubMed] [Google Scholar]
- 2.Aramburú Guarda, J., C. Ramal Asayag, and R. Witzig. 1999. Malaria reemergence in the Peruvian Amazon region. Emerg. Infect. Dis. 5**:**209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacon, D. J., C. Latour, C. Lucas, O. Colina, P. Ringwald, and S. Picot. 2007. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob. Agents Chemother. 51**:**1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortese, J. F., A. Caraballo, C. E. Contreras, and C. V. Plowe. 2002. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 186**:**999-1006. [DOI] [PubMed] [Google Scholar]
- 5.Dahlström, S., M. I. Veiga, P. Ferreira, A. Martensson, A. Kaneko, B. Andersson, A. Bjorkman, and J. P. Gil. 2008. Diversity of the sarco/endoplasmic reticulum Ca2+-ATPase orthologue of Plasmodium falciparum (PfATP6). Infect. Genet. Evol. 8**:**340-345. [DOI] [PubMed] [Google Scholar]
- 6.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. Ursos, A. B. Sidhu, B. Naude, K. W. Deitsch, X. Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6**:**861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57**:**117-145. [DOI] [PubMed] [Google Scholar]
- 8.Huaman, M. C., N. Roncal, S. Nakazawa, T. T. Long, L. Gerena, C. Garcia, L. Solari, A. J. Magill, and H. Kanbara. 2004. Polymorphism of the Plasmodium falciparum multidrug resistance and chloroquine resistance transporter genes and in vitro susceptibility to aminoquinolines in isolates from the Peruvian Amazon. Am. J. Trop. Med. Hyg. 70**:**461-466. [PubMed] [Google Scholar]
- 9.Jambou, R., E. Legrand, M. Niang, N. Khim, P. Lim, B. Volney, M. T. Ekala, C. Bouchier, P. Esterre, T. Fandeur, and O. Mercereau-Puijalon. 2005. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet 366**:**1960-1963. [DOI] [PubMed] [Google Scholar]
- 10.Khim, N., C. Bouchier, M. T. Ekala, S. Incardona, P. Lim, E. Legrand, R. Jambou, S. Doung, O. M. Puijalon, and T. Fandeur. 2005. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob. Agents Chemother. 49**:**3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kublin, J. G., R. S. Witzig, A. H. Shankar, J. Q. Zurita, R. H. Gilman, J. A. Guarda, J. F. Cortese, and C. V. Plowe. 1998. Molecular assays for surveillance of antifolate-resistant malaria. Lancet 351**:**1629-1630. [DOI] [PubMed] [Google Scholar]
- 12.Laufer, M. K., P. C. Thesing, N. D. Eddington, R. Masonga, F. K. Dzinjalamala, S. L. Takala, T. E. Taylor, and C. V. Plowe. 2006. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 355**:**1959-1966. [DOI] [PubMed] [Google Scholar]
- 13.Magill, A. J., J. Zegarra, C. Garcia, W. Marquiño, and T. K. Ruebush II. 2004. Efficacy of sulfadoxine-pyrimethamine and mefloquine for the treatment of uncomplicated Plasmodium falciparum malaria in the Amazon basin of Peru. Rev. Soc Bras Med. Trop. 37**:**279-281. [DOI] [PubMed] [Google Scholar]
- 14.Marquiño, W., M. Huilca, C. Calampa, E. Falconi, C. Cabezas, R. Naupay, T. K. Ruebush II, J. R. MacArthur, L. M. Barat, F. E. Oblitas, M. Arrunategui, G. Garavito, M. L. Chafloque, B. Pardave, S. Gutierrez, N. Arrospide, and C. Carrillo. 2003. Efficacy of mefloquine and a mefloquine-artesunate combination therapy for the treatment of uncomplicated Plasmodium falciparum malaria in the Amazon Basin of Peru. Am. J. Trop. Med. Hyg. 68**:**608-612. [DOI] [PubMed] [Google Scholar]
- 15.Marquiño, W., J. R. MacArthur, L. M. Barat, F. E. Oblitas, M. Arrunategui, G. Garavito, M. L. Chafloque, B. Pardave, S. Gutierrez, N. Arrospide, C. Carrillo, C. Cabezas, and T. K. Ruebush II. 2003. Efficacy of chloroquine, sulfadoxine-pyrimethamine, and mefloquine for the treatment of uncomplicated Plasmodium falciparum malaria on the north coast of Peru. Am. J. Trop. Med. Hyg. 68**:**120-123. [PubMed] [Google Scholar]
- 16.McCollum, A. M., K. Mueller, L. Villegas, V. Udhayakumar, and A. A. Escalante. 2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51**:**2085-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehlotra, R. K., H. Fujioka, P. D. Roepe, O. Janneh, L. M. Ursos, V. Jacobs-Lorena, D. T. McNamara, M. J. Bockarie, J. W. Kazura, D. E. Kyle, D. A. Fidock, and P. A. Zimmerman. 2001. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with Pf_crt_ polymorphism in Papua New Guinea and South America. Proc. Natl. Acad. Sci. USA 98**:**12689-12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair, S., D. Nash, D. Sudimack, A. Jaidee, M. Barends, A. C. Uhlemann, S. Krishna, F. Nosten, and T. J. Anderson. 2007. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol. Biol. Evol. 24**:**562-573. [DOI] [PubMed] [Google Scholar]
- 19.Nash, D., S. Nair, M. Mayxay, P. N. Newton, J. P. Guthmann, F. Nosten, and T. J. Anderson. 2005. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc. Biol. Sci. 272**:**1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai, D. R., G. Hijar, Y. Montoya, W. Marouino, T. K. Ruebush II, C. Wongsrichanalai, and K. C. Kain. 2003. Lack of prediction of mefloquine and mefloquine-artesunate treatment outcome by mutations in the Plasmodium falciparum multidrug resistance 1 (pfmdr1) gene for P. falciparum malaria in Peru. Am. J. Trop. Med. Hyg. 68**:**107-110. [PubMed] [Google Scholar]
- 21.Plowe, C. V., J. F. Cortese, A. Djimde, O. C. Nwanyanwu, W. M. Watkins, P. A. Winstanley, J. G. Estrada-Franco, R. E. Mollinedo, J. C. Avila, J. L. Cespedes, D. Carter, and O. K. Doumbo. 1997. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 176**:**1590-1596. [DOI] [PubMed] [Google Scholar]
- 22.Plowe, C. V., A. Djimde, M. Bouare, O. Doumbo, and T. E. Wellems. 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 52**:**565-568. [DOI] [PubMed] [Google Scholar]
- 23.Price, R. N., A.-C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364**:**438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roper, C., R. Pearce, B. Bredenkamp, J. Gumede, C. Drakeley, F. Mosha, D. Chandramohan, and B. Sharp. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361**:**1174-1181. [DOI] [PubMed] [Google Scholar]
- 25.Roper, C., R. Pearce, S. Nair, B. Sharp, F. Nosten, and T. Anderson. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305**:**1124. [DOI] [PubMed] [Google Scholar]
- 26.Roper, M. H., R. S. Torres, C. G. Goicochea, E. M. Andersen, J. S. Guarda, C. Calampa, A. W. Hightower, and A. J. Magill. 2000. The epidemiology of malaria in an epidemic area of the Peruvian Amazon. Am. J. Trop. Med. Hyg. 62**:**247-256. [DOI] [PubMed] [Google Scholar]
- 27.Ruebush, T. K., II, W. Marquiño, J. Zegarra, R. Villaroel, J. C. Avila, C. Díaz, and E. Beltrán. 2003. Practical aspects of in vivo antimalarial drug efficacy testing in the Americas. Am. J. Trop. Med. Hyg. 68**:**391-397. [PubMed] [Google Scholar]
- 27a.Tanabe, K., N. Sakihama, T. Hattori, L. Ranford-Cartwright, I. Goldman, A. A. Escalante, and A. A. Lal. 2004. Genetic distance in housekeeping genes between Plasmodium falciparum and Plasmodium reichenowi and within P. falciparum. J. Mol. Evol. 59**:**687-694. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25**:**4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasconcelos, K. F., C. V. Plowe, C. J. Fontes, D. Kyle, D. F. Wirth, L. H. Pereira da Silva, and M. G. Zalis. 2000. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase of isolates from the Amazon region of Brazil. Mem. Inst. Oswaldo Cruz 95**:**721-728. [DOI] [PubMed] [Google Scholar]
- 30.Vieira, P. P., M. U. Ferreira, M. G. Alecrim, W. D. Alecrim, L. H. da Silva, M. M. Sihuincha, D. A. Joy, J. Mu, X. Z. Su, and M. G. Zalis. 2004. Pf_crt_ polymorphism and the spread of chloroquine resistance in Plasmodium falciparum populations across the Amazon Basin. J. Infect. Dis. 190**:**417-424. [DOI] [PubMed] [Google Scholar]
- 31.Wang, P., C. S. Lee, R. Bayoumi, A. Djimde, O. Doumbo, G. Swedberg, L. D. Dao, H. Mshinda, M. Tanner, W. M. Watkins, P. F. Sims, and J. E. Hyde. 1997. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol. Biochem. Parasitol. 89**:**161-177. [DOI] [PubMed] [Google Scholar]
- 32.Wang, X., J. Mu, G. Li, P. Chen, X. Guo, L. Fu, L. Chen, X. Su, and T. E. Wellems. 2005. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am. J. Trop. Med. Hyg. 72**:**410-414. [PubMed] [Google Scholar]
- 33.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X. Z. Su. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418**:**320-323. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, Z., S. M. Griffing, A. M. de Oliveira, A. M. McCollum, W. M. Quezada, N. Arrospide, A. A. Escalante, and V. Udhayakumar. 2008. Decline in sulfadoxine-pyrimethamine-resistant alleles after change in drug policy in the Amazon region of Peru. Antimicrob. Agents Chemother. 52**:**739-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]