Ubiquitin-mediated proteolysis of HuR by heat shock (original) (raw)
Abstract
The RNA-binding protein HuR regulates the stability and translation of numerous mRNAs encoding stress-response and proliferative proteins. Although its post-transcriptional influence has been linked primarily to its cytoplasmic translocation, here we report that moderate heat shock (HS) potently reduces HuR levels, thereby altering the expression of HuR target mRNAs. HS did not change HuR mRNA levels or de novo translation, but instead reduced HuR protein stability. Supporting the involvement of the ubiquitin–proteasome system in this process were results showing that (1) HuR was ubiquitinated in vitro and in intact cells, (2) proteasome inhibition increased HuR abundance after HS, and (3) the HuR kinase checkpoint kinase 2 protected against the loss of HuR by HS. Within a central, HS-labile ∼110-amino-acid region, K182 was found to be essential for HuR ubiquitination and proteolysis as mutant HuR(K182R) was left virtually unubiquitinated and was refractory to HS-triggered degradation. Our findings reveal that HS transiently lowers HuR by proteolysis linked to K182 ubiquitination and that HuR reduction enhances cell survival following HS.
Keywords: post-transcriptional gene regulation, proteasome, protein stability, ribonucleoprotein complex, ubiquitination
Introduction
In response to environmental changes, mammalian cells maintain homeostasis by altering the collections of expressed proteins. Protein expression patterns can be modified transcriptionally, but are also tightly controlled through post-transcriptional mechanisms (Moore, 2005). Following cell damage, changes in mRNA turnover and translation are particularly influential towards altering the subsets of expressed cellular proteins (Keene, 2007). These events are governed by factors that associate with the mRNA, most prominently RNA-binding proteins (RBPs) and microRNAs (miRNAs). Although the effect of stress on miRNA function is still poorly understood, RBPs are solidly established regulators of mRNA stability and translation following cellular damage. Acting upon specific RNA sequences (typically present in the 5′- or 3′-untranslated regions (UTRs)), RBPs can modulate mRNA turnover and translation after stress. Some stress-regulated RBPs are specialized in one post-transcriptional function; for instance, RBPs tristetraprolin (TTP), KSRP, and BRF1 promote target mRNA decay (reviewed in Abdelmohsen et al, 2008). However, many RBPs influence both the stability and translation of target mRNAs. For example, AUF1 primarily promotes mRNA degradation, but can also stabilize and promote the translation of target mRNAs (Xu et al, 2001; Liao et al, 2007); TIA-1 primarily inhibits target mRNA translation, but was also shown to inhibit mRNA decay (Kedersha and Anderson, 2002; Yamasaki et al, 2007); and NF90 can modulate both the stability and the translation of target mRNAs (Kuwano et al, 2008).
The RBP HuR can also promote mRNA stability and influence translation (Brennan and Steitz, 2001; Gorospe, 2003). The ubiquitous member of the Hu family (which also comprises the primarily neuronal RBPs HuB, HuC, and HuD), HuR, has three RNA recognition motifs (RRMs) through which it binds to target mRNAs, many of them bearing U-rich sequences (Chen et al, 2002; López de Silanes et al, 2003). In stress-treated and untreated cells HuR was shown to stabilize many target mRNAs, including those that encode stress-response and proliferative proteins such as p21, c-fos, cyclins A2, B1, D1, inducible nitric oxide synthase (iNOS), vascular endothelial growth factor (VEGF), sirtuin 1 (SIRT1), tumor necrosis factor (TNF)-α, bcl-2, mcl-1, cyclooxygenase (COX)-2, γ-glutamylcysteine synthetase h (γ-GCSh), MAP kinase phosphatase-1 (MKP-1), interleukin-3, and urokinase plasminogen activator (uPA) and its receptor (uPAR) (Fan and Steitz, 1998; Peng et al, 1998; Wang et al, 2000a, 2000b; Ming et al, 2001; Chen et al, 2002; Tran et al, 2003; Lal et al, 2004; Abdelmohsen et al, 2007a, 2007b and 2008; Kuwano et al, 2008). In addition, stress conditions also promoted the translation of HuR target mRNAs encoding prothymosin α, p53, hypoxia-inducible factor-1α (HIF-1α), cationic amino acid transporter 1 (CAT-1), and MKP-1 (Mazan-Mamczarz et al, 2003; Lal et al, 2005; Bhattacharyya et al, 2006; Kuwano et al, 2008; Galbán et al, 2008).
In response to stress-causing agents, the post-transcriptional effect of HuR on its target mRNAs has been tightly correlated to HuR's translocation to the cytoplasm. This process implicates several transport machinery components [CRM1, transportins 1 and 2, and importin-1α (Gallouzi and Steitz, 2001; Guttinger et al, 2004; Rebane et al, 2004)], and is influenced by HuR phosphorylation (Doller et al, 2007; Kim et al, 2008). HuR's post-transcriptional influence has also been linked to its modification, including its phosphorylation at S88, S100, and T118 by the checkpoint kinase 2 (CHK2), which modulated HuR's association with target mRNAs (Abdelmohsen et al, 2007a), and methylation at R217 by CARM1 (Li et al, 2002). In addition, a few instances of altered whole-cell HuR levels have been reported. For example, HuR was elevated in early-passage, proliferating fibroblasts, but was expressed at very low levels in senescent, terminally growth-arrested fibroblasts (Wang et al, 2001). Similarly, HuR abundance increased during muscle differentiation (Figueroa et al, 2003; van der Giessen et al, 2003), and was found to be markedly higher in cancer specimens compared with healthy control tissues (López de Silanes et al, 2003). However, the mechanisms that influence the steady-state levels of HuR have not been explored. The transcriptional regulation of HuR expression remains virtually unknown; on the post-transcriptional level, HuR binds the HuR mRNA (Pullmann et al, 2007), but little else is known regarding the regulation of HuR expression post-transcriptionally.
A recent survey of stress agents revealed that mild heat shock (HS), but not other stresses, transiently and potently decreased HuR abundance. Whole-cell HuR levels were not previously found to fluctuate in response to an acute stimulus. In earlier studies, HS (45°C) was shown to trigger the cytoplasmic accumulation of HuR into stress granules (SGs), disrupted HuR's association with mRNAs on polysomes, and increased HuR's interaction with polyadenylated mRNA in the nucleus (Gallouzi et al, 2000; Kedersha and Anderson 2002), but HS-induced alterations in total HuR abundance were not formally reported. Here, we present evidence that HS lowers whole-cell HuR protein levels by promoting the degradation of HuR through HuR ubiquitination. We map this post-translational modification to HuR residue K182 and find that cells expressing the mutant HuR(K182R) show impaired HuR ubiquitination and enhanced cell death following HS. This is the first report to implicate the ubiquitin (Ub)–proteasome pathway in the regulation of HuR abundance, HuR function, and HuR-enhanced cell survival.
Results
HS markedly reduces HuR levels
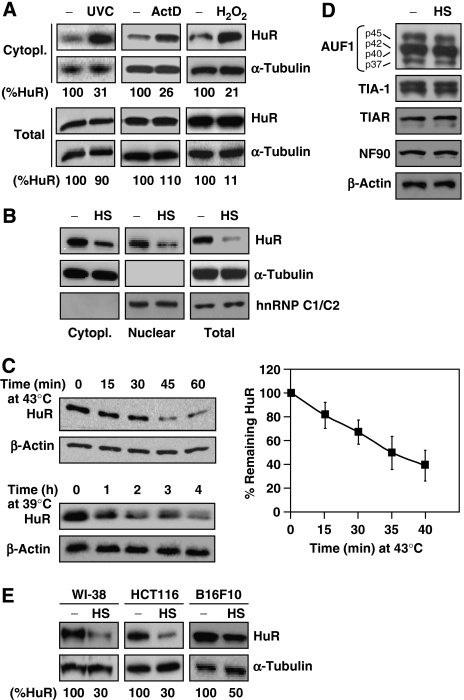
Exposure of HeLa (human cervical carcinoma) cells to stresses such as actinomycin D (ActD), hydrogen peroxide (H2O2), or short-wavelength ultraviolet light (UVC) increased the levels of cytoplasmic (Cytopl.) HuR, but had no measurable influence on whole-cell (Total) HuR levels (Figure 1A). These findings agree with previous reports showing that HuR translocates to the cytoplasm in response to various stimuli; as reported earlier, HuR is predominantly nuclear and therefore these treatments did not affect nuclear HuR levels significantly (Keene, 1999; Wang et al, 2000b; and data not shown). By contrast, exposure to moderate HS (43°C for 1 h) potently lowered total HuR levels, with the nuclear and cytoplasmic HuR levels showing comparable reductions (Figure 1B). Despite slight variation among experiments, this decrease was dependent on the length and magnitude of HS, as HuR was progressively reduced and the effect was more pronounced at 43°C than at milder temperatures (39°C) (Figure 1C). HuR was specifically reduced by HS, as other RBPs remained unchanged following the same intervention (Figure 1D). HS also decreased HuR in other cell types, indicating that the effect was not limited to HeLa cells (Figure 1E).
Figure 1.
Decline in HuR levels following heat shock. (A) HeLa cells were treated with UVC (20 J/m2, collected 4 h later), with actinomycin D (ActD, 2 μg/ml for 1 h), or with H2O2 (1 mM, 3 h). The levels of HuR and loading control α-Tubulin in whole-cell (Total, 2.5 μg) and cytoplasmic (Cytopl., 5 μg) lysates were assessed by western blot analysis. (B) Cells were treated with heat shock (HS, 43°C) for 1 h, whereupon Cytopl. (5 μg), Nuclear (2.5 μg), and Total (2.5 μg) lysates were prepared and the levels of HuR and the cytoplasmic (α-Tubulin) and nuclear (hnRNP C1/C2) markers were tested by western blot analysis. (C) Kinetics of HuR reduction after HS at 43°C (top) and 39°C (bottom); graph, means±s.d. (_n_>3) of HuR signals after HS at 43°C. (D) Effect of HS (43°C, 1 h) on the levels of the RNA-binding proteins shown (whole-cell lysates); four AUF1 isoforms are indicated; β-actin was included as loading control. (E) Effect of HS (43°C, 1 h) on the levels of HuR and loading control α-Tubulin in human and mouse cell lines. % HuR, densitometric analysis of signals from HuR and loading control proteins.
Consequences of lowering HuR levels on mRNA expression programs
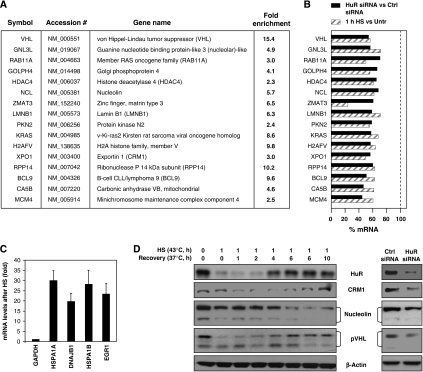
In order to assess the importance of the HS-mediated reduction in HuR levels, we compared the collections of HuR-associated mRNAs and mRNAs whose levels were reduced by HS. HuR-associated mRNAs were identified by ribonucleoprotein immunoprecipitation followed by microarray analysis (RNP IP, Supplementary Table I). In parallel, the transcripts present in total cellular RNA showing altered levels after HS were also studied using microarrays (Supplementary Tables II and III). Joint analysis of these data revealed 57 mRNAs that both were associated with HuR and showed reduced levels following HS. Of these transcripts, 29 were studied further using specific primer pairs. This mRNA subset was validated by comparing the concentration in HuR IP samples with that in IgG IP samples (‘Fold Enrichment' column) (Figure 2A) and by comparing their abundance in total RNA (‘% mRNA' column) in HS-treated cells relative to untreated cells (hatched bars) and in cells with HuR silenced by transfection with HuR-directed siRNA relative to cells transfected with control siRNAs (solid bars) (Figure 2B). As shown, 16 target mRNAs were reduced both after HS and after HuR silencing; analysis of the 13 other mRNAs is shown in Supplementary Figure S1.
Figure 2.
Analysis of HuR target mRNAs after HS. Microarray analyses were performed to identify HuR-associated mRNAs (Supplementary Table I) and mRNAs showing altered levels after HS (1 h, 43°C) (Supplementary Tables II and III). (A, B) The mRNAs listed (shown to be associated with HuR and to decrease after HS by array analysis) were validated using RT-qPCR and specific primer pairs; the individual enrichment of each mRNA in anti-HuR IPs compared with IgG IPs is indicated [Fold Enrichment, (A)], and the influence of HS (relative to no treatment) and the influence of HuR silencing by siRNA transfection (relative to control Ctrl siRNA transfection) were quantified [% mRNA, (B)]. (C) The levels of control HS-inducible mRNAs were tested by RT-qPCR to monitor the effectiveness of the HS conditions. (D) Left, Western blot analysis of the levels of proteins encoded by HuR target mRNAs showing reduction after HS (CRM1, Nucleolin, pVHL) and loading marker β-actin. Protein levels were monitored after 1 h at 43°C and several time points after HS as at 37°C (Recovery). Right, protein levels by 48 h after transfecting cells with control (Ctrl) or HuR-directed siRNAs.
The effect of HS was verified for several known HS-inducible mRNAs (Figure 2C). To validate further the data in Figure 2B, we also analyzed the levels of proteins encoded by HuR-associated mRNAs whose levels declined after HS. As shown, the concentrations of the proteins exportin1/CRM1, nucleolin (an RBP), and pVHL (a tumor suppressor), all encoded by HuR-associated mRNAs, decreased after HS (and in some cases later increased), with different kinetics after a period of incubation at 37°C (‘Recovery', Figure 2D left). A similar downregulation was seen after HuR silencing (Figure 2D right). The levels of β-actin were also tested; although β-actin mRNA is a target of HuR (Dormoy-Raclet et al, 2007), it showed no reduction under the conditions employed here (48 h after HuR silencing or 1 h HS at 43°C). Together, HuR downregulation after HS correlated with a transient and potent decrease in the levels of HuR target mRNAs and the proteins they encode, that was recapitulated by silencing HuR. Thus, we set out to elucidate the molecular details underlying the transient lowering of HuR by HS.
HS specifically reduces HuR protein stability
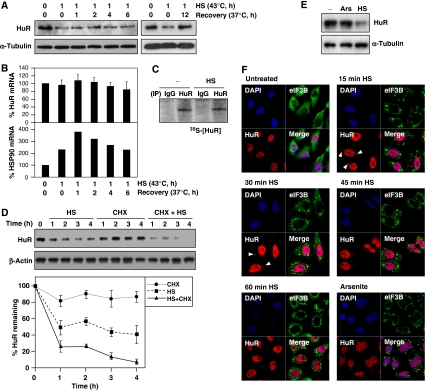
Following 1 h HS, HuR levels remained low for several hours even though cells were allowed to recover at 37°C; full restoration of HuR abundance was typically seen by 12 h of recovery (Figure 3A). Throughout this time, the levels of a control HS-regulated transcript (HSP90 mRNA, Figure 3B, bottom) fluctuated as expected, but HuR mRNA levels remained unchanged (Figure 3B, top). These findings suggested that HS did not influence the transcription or stability of the HuR mRNA. Furthermore, the de novo synthesis of HuR also appeared unchanged; this parameter was measured by incubating HeLa cells briefly (15 min) with 35S-labeled methionine and cysteine, then performing IP with anti-HuR antibody (or IgG in control IP reactions). As shown in Figure 3C, the signal intensity of the newly translated HuR was comparable between the HS (1 h, 43°C) and untreated (−) groups. Similarly, HS for 1 h did not affect the translation of a housekeeping protein (not shown). We then tested whether HS influenced the stability of HuR by monitoring the rate of HuR loss after incubating cells with the inhibitor of protein synthesis cycloheximide (CHX, Figure 3D). As shown, HuR levels remained unchanged in untreated cells (CHX only group), indicating that HuR is not labile at normal temperature. By contrast, HuR levels declined rapidly in the HS group, and even more rapidly in the HS + CHX group, indicating that HS accelerated HuR decay.
Figure 3.
HS transiently localizes HuR in SGs and reduces HuR protein stability. (A) HeLa cells were subjected to HS (1 h) or no treatment, whereupon they were collected or returned to 37°C for the times shown (Recovery); the levels of HuR and α-Tubulin in whole-cell lysates were tested by western blot analysis. (B) The levels of HuR mRNA or the control HS-inducible HSP90 mRNA were measured by RT-qPCR at the times shown in cells that were treated with HS with or without recovery as explained in panel (A). (C) Influence of HS on the de novo HuR translation (35S-[HuR]). (D) The levels of HuR were measured in cells treated with HS (HS), incubated with 10 μg/ml cycloheximide (CHX), or exposed to HS in the presence of CHX (CHX + HS). The levels of HuR and loading control β-actin were measured by western blot analysis. (E) Western blot analysis of HuR expression levels in whole-cell lysates prepared from cells that were treated with sodium arsenite (Ars, 400 μM, 30 min, as positive control) or HS (43°C, 1 h). (F) Cells were treated as in panel (E), and the presence of stress granules (SGs, arrowheads) was assessed by immunofluorescence at the times shown after HS or arsenite treatments. Nuclei were visualized using DAPI, and SGs with the specific marker eIF3B; the overlap of the two signals (Merge) is shown.
As HS was reported to induce HuR accumulation into SGs, we hypothesized that downregulation of HuR by HS might be linked to SG formation, and thus tested the possible role of SGs in this process. Cells treated with the SG-triggering agent arsenite (Ars), which does not lower HuR levels (Figure 3E), were included as positive control (Figure 3F; Kedersha and Anderson, 2002). HuR localized in SGs by 15 and 30 min (arrowheads), but disappeared by 60 min (Figure 3F). CHX treatment blocked the formation of SGs (Supplementary Figure S2), but HuR was still reduced in this group (Figure 3D), suggesting that HuR degradation was independent of its localization in SGs. Together, these findings reveal that HS did not affect HuR biosynthesis and instead influenced HuR protein stability and subcytoplasmic localization.
HuR kinase CHK2 protects against HuR decay after HS
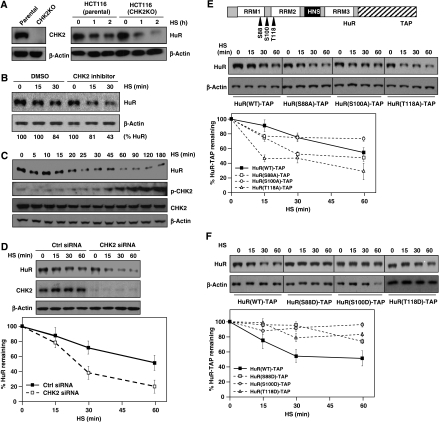
While screening cell types to study the reduction in HuR levels by HS (Figure 1E), we observed that HuR was more efficiently reduced in HCT116 cells carrying somatic deletions of the HuR kinase CHK2 (Jallepalli et al, 2003; Abdelmohsen et al, 2007a) than in the parental HCT116 counterparts (Figure 4A and B). Supporting the notion that CHK2 might ameliorate HuR degradation, HeLa cells treated with a CHK2 inhibitor showed a similarly accelerated loss of HuR after HS (Figure 4B). Although CHK2 was phosphorylated (and hence activated) by HS, HuR degradation preceded the increase in CHK2 phosphorylation above basal levels (Figure 4C). In keeping with a role for CHK2 in preventing HuR degradation during HS, the decline in HuR by HS was accelerated when CHK2 was silenced by siRNA (Figure 4D).
Figure 4.
CHK2 reduces HuR loss by HS. (A) Western blot analysis of the levels of CHK2, HuR, and loading control in untreated (left) or HS-treated (43°C, right) colon carcinoma HCT116 cells expressing CHK2 (Parental) or CHK2-null through somatic knockout (CHK2KO). (B) Western blot analysis of HuR and β-actin expression levels in HeLa cells pre-treated with a CHK2 inhibitor (10 μM, 1 h), then exposed to HS for the times indicated; signal intensities were quantified by densitometry. (C) Western blot analysis of the levels of HuR, phosphorylated (p-)CHK2, total CHK2, and β-actin in HeLa cells treated with HS for the times indicated. (D) At 48 h after transfection of CHK2-directed or control (Ctrl) siRNAs, HeLa cells were treated with HS for the times shown and the levels of HuR, CHK2, and loading control β-actin were assessed by western blot analysis. In panels (D–F), the western blotting signals were quantified by densitometry and the means ±s.d. from three independent experiments are shown. (E) Top, schematic of the sites of CHK2 phosphorylation on HuR. By 48 h after transfection of plasmids to express HuR–TAP fusion proteins [WT or bearing mutations in putative CHK2 phosphorylation sites (S88A, S100A, T118A)], the levels of HuR–TAP fusion proteins were assessed by western blotting. (F) By 48 h after transfection of plasmids to express HuR–TAP fusion proteins [WT or bearing phosphomimic mutations (S88D, S100D, T118D)], HuR–TAP fusion protein levels were assessed as explained in (E).
CHK2 phosphorylates HuR at three residues (S88, S100, T118; Figure 4E), and through these modifications CHK2 was proposed to modulate HuR binding to target mRNAs (Abdelmohsen et al, 2007a, 2007b). Preliminary evidence suggested that HuR is phosphorylated by HS in a CHK2-dependent manner (Supplementary Figure S3), but in-depth analysis of this process awaits the availability of specific antibodies recognizing phosphorylated S88, S100, and T118. We hypothesized that if CHK2 slows down the loss of HuR after HS, HuR mutants refractory to CHK2 phosphorylation could be more labile. Indeed, comparison of non-phosphorylatable and wild-type HuR–TAP fusion proteins revealed that HuR(S88A)–TAP and HuR(T118A)–TAP decreased more rapidly after HS (Figure 4E). To further test the notion that HuR phosphorylation at CHK2 target sites reduced HuR decay by HS, we generated plasmids to express HuR–TAP fusion proteins mimicking phosphorylated residues S88, S100, and T118. As shown in Figure 4F, fusion proteins HuR(S88D)–TAP, HuR(S100D)–TAP, and HuR(T118D)–TAP were markedly protected against HS-triggered loss. These observations further strengthen the view that phosphorylation of HuR by CHK2 slows down HuR degradation by HS.
Ubiquitination linked to HuR stability after HS
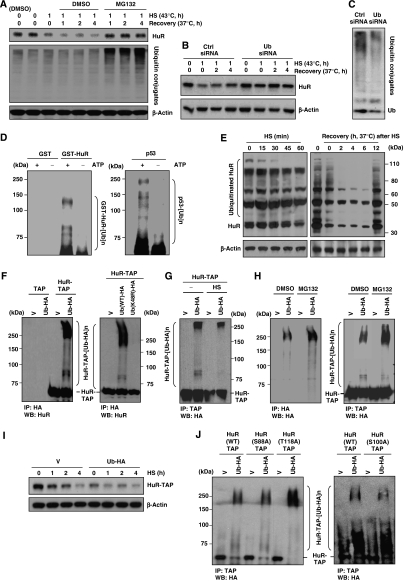
To identify the mechanisms whereby HS accelerates the degradation of HuR, a number of inhibitors of proteases [e.g., inhibitors of calpains, caspases (Z-VAD-FMK), and lysosomal proteases (chloroquine and ammonium chloride)] were tested and found not to block the degradation of HuR by HS (not shown). Only inhibition of the proteasome using MG132 (20 μM) enhanced HuR levels, increasing HuR signals relative to those in the control (DMSO) group (Figure 5A). It should be noted that pretreatment with MG132 before HS as well as joint MG132 and HS treatments were severely toxic for HeLa cells (unlike for WI-38 cells (Bonelli et al, 2004)), and therefore the effects of these treatments could not be studied (not shown). Similarly, when the Ub–proteasome degradation system was suppressed by silencing Ub (Ub siRNA group), we observed a stabilization of HuR after HS (Figure 5B). As anticipated, this intervention not only reduced the levels of Ub (an 8.5-kDa protein), but also lowered the subset of ubiquitinated proteins in HeLa cells (Ub conjugates, Figure 5C). Proteasome activity remained elevated during HS and during recovery at 37°C (Supplementary Figure S4). Collectively, this evidence suggested that HuR degradation by HS was linked to HuR ubiquitination and prompted us to test directly whether HuR was ubiquitinated.
Figure 5.
Analysis of HuR ubiquitination in vitro and in vivo. (A) The levels of HuR, ubiquitinated proteins, and loading control β-actin were studied by western blot analysis in cells that were treated (temperatures and times) as shown, in the presence of MG132 (20 μM) or vehicle control (DMSO). (B) At 48 h after transfecting either control (Ctrl) siRNA or a ubiquitin-targeting (Ub) siRNA, HuR and loading control β-actin were assessed in cells incubated as shown. (C) Western blot analysis of the levels of ubiquitin and total ubiquitin-conjugated proteins 48 h after transfection with Ctrl or Ub siRNAs. (D) Left, in vitro polyubiquitination of HuR was measured using a control protein (GST) and a GST-HuR fusion protein in the absence or presence of ATP; Right, in vitro polyubiquitination of purified p53; kDa, sizes of molecular weight markers. (E) Western blot analysis (modified as detailed in the Supplementary data) of endogenous ubiquitinated HuR after treatment of HeLa cells with HS (left) and during recovery from HS (right). (F) Left, HeLa cells were cotransfected with a plasmid expressing an HA-tagged ubiquitin (Ub-HA) or the corresponding control vector (V), together with a plasmid expressing either HuR–TAP or the vector control (TAP); polyubiquitinated HuR–TAP was assessed 48 h later by HA IP, followed by HuR western blot (WB) analysis. Right, cells were processed as shown on the left of panel (F), but a mutant variant of ubiquitin that cannot oligomerize [Ub(K48R)-HA] was also tested; polyubiquitinated HuR–TAP was assessed 48 h later by HA IP, followed by HuR WB analysis. (G) Cells cotransfected with HuR–TAP along with either Ub-HA-expressing or V plasmids were left without further treatment or were treated with HS (43°C for 15 min), whereupon the levels of ubiquitinated HuR–TAP were detected by TAP IP (using Rabbit IgG) and WB analysis using an anti-HA antibody. (H) Left, in cells transfected with Ub-HA-expressing plasmid, Ub-HA levels were tested in the absence (DMSO) or presence of MG132 (20 μM for 4 h) by IP using an anti-HA antibody, followed by HA detection by WB. Right, cells co-transfected with pHuR–TAP and Ub-HA were used to detect the levels of ubiquitinated HuR–TAP in the absence (DMSO) or presence of MG132 (20 μM for 4 h) after TAP IP (using rabbit IgG), and then to detect Ub using an anti-HA antibody. (I) HuR abundance in cells expressing normal ubiquitin levels or overexpressing ubiquitin as HA-Ub; protein levels were studied at the times shown following HS. (J) The ubiquitination of HuR–TAP fusion proteins bearing point mutations S88A and S118A (left) or S100A (right) was tested by cotransfection of plasmids as explained for panel (F), except that the HuR mutants tested were those described in Figure 4E. Protein signals were visualized by TAP IP, followed by WB analysis using an anti-HA antibody.
In vitro evidence of HuR ubiquitination was obtained using a cell-free assay (Materials and methods). As shown in this system, Ub residues were readily incorporated onto GST-HuR (and not onto GST), but this reaction only occurred in the presence of ATP, in agreement with the energy dependence of the ubiquitination process (Figure 5D); ubiquitination of p53 was included as positive control (Figure 5D).
In vivo evidence that endogenous HuR was ubiquitinated was first sought using standard western blotting conditions, without success. Using a modified western blotting protocol (which included lysis under strong inhibition of anti-deubiquitinating enzymes, use of greater amounts of lysate, longer transferring times, and concentrated primary antibody; details in Supplementary data), we could detect endogenous ubiquitinated HuR (Figure 5E) in cells that were untreated, HS-treated, and recovering from HS. Silencing of HuR revealed that these bands indeed corresponded to polyubiquitinated HuR (Supplementary Figure S5). As the routine use of this cumbersome procedure posed major limitations to the molecular characterization of HuR ubiquitination, additional experiments were conducted using cells cotransfected with plasmids to express HA-tagged Ub (Ub-HA) and tandem affinity purification-tagged HuR (HuR–TAP); the corresponding control vector-transfected groups are V and TAP, respectively. As shown, ubiquitinated HuR–TAP was readily observed in the presence of Ub-HA, but not in the presence of HA alone, whereas no Ub-HA was incorporated into TAP alone (Figure 5F, left). Similarly, no Ub-HA was incorporated when using the mutant Ub(K48R) that could not form polyubiquitin chains (Figure 5F, right). HS treatment was found to decrease ubiquitinated endogenous HuR and HuR–TAP (Figure 5E and G); this was observed at all times studied, including at the earliest times when the analysis was possible (by 15 min of HS, not shown). Further characterization of this effect in intact cells included treatment with MG132 (4 h), which enhanced the levels of total ubiquitinated (Ub-HA-modified) proteins (Figure 5H, left) ubiquitinated HuR–TAP specifically (Figure 5H, right), and ubiquitinated endogenous HuR (Supplementary Figures S5 and S6). Moreover, experiments that complemented the increase in HuR seen after silencing Ub (Figure 5B and C) revealed that Ub overexpression actually reduced the levels of HuR–TAP (Figure 5I). Given that non-phosphorylatable HuR–TAP mutants had distinct stabilities (Figure 4E), we studied whether their ubiquitination levels differed. As shown in Figure 5J, the ubiquitination of HuR(S88A)–TAP was similar to that of HuR(WT)–TAP, in agreement with the similar stabilities of both proteins (Figure 4E). By contrast, the least stable mutant, HuR(T118A)–TAP, was more extensively ubiquitinated (left), whereas the more stable mutant, HuR(S100A)–TAP, was less ubiquitinated (right). Together, these results show that HuR is ubiquitinated in vitro and in vivo and link HuR ubiquitination to its degradation in response to HS.
Ubiquitination at Lys-182 promotes HuR decay and enhances cell survival after HS
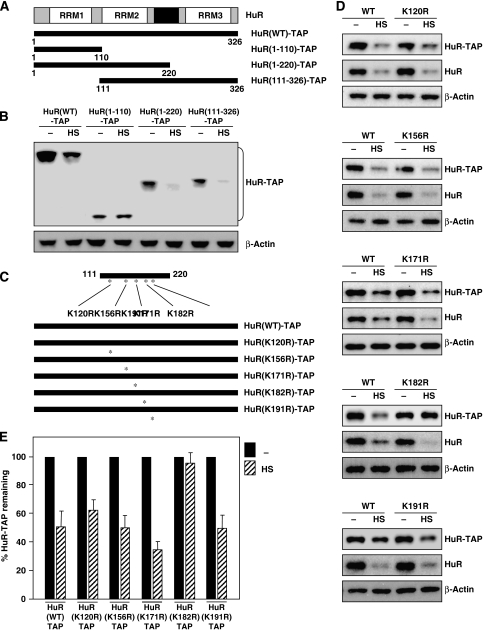
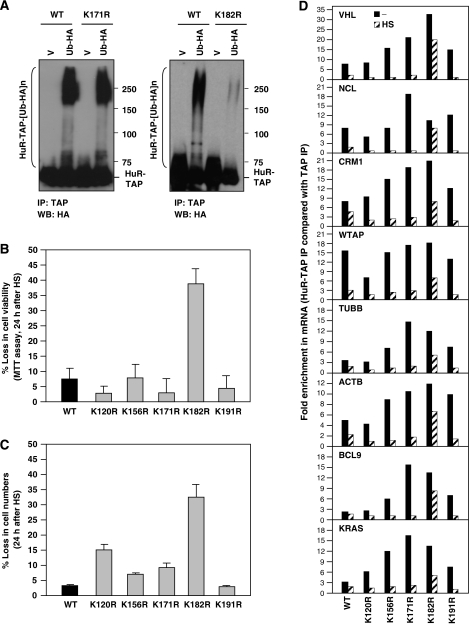
In order to map the residue(s) implicated in Ub-mediated HuR proteolysis, we first narrowed down the region of HuR involved in HS-mediated degradation. Depicted in Figure 6A are three TAP-linked segments of HuR that were tested initially. After transfection of the corresponding expression vectors, we assessed the stability of the resulting fusion proteins in response to HS. A segment spanning HuR amino acids 1–110 was stable after HS, but addition of amino acids 111–220 rendered it labile in response to HS (Figure 6B), indicating that the 111–220 region of HuR conferred instability after HS. This middle region contains five lysines, the amino acids where Ub moieties are ligated. After systematically mutating each lysine to an arginine (Figure 6C), we tested the stability of the full-length HuR–TAP carrying each of the resulting point mutations (K120R, K156R, K171R, K182R or K191R). As shown, in each transfection group, both the HuR(WT)–TAP and the endogenous HuR were labile after HS (Figure 6D). In addition, each of the HuR–TAP point mutants was also labile, with the exception of mutant K182R, which was refractory to degradation by HS (Figure 6D). The extent of degradation of each HuR–TAP variant following HS was quantified (Figure 6E). Ub-HA co-expression in HeLa cells transfected with each mutant verified that all HuR–TAP mutants were efficiently ubiquitinated [as shown for HuR(K171R)–TAP], but not HuR(K182R)–TAP, which was found virtually without ubiquitination (Figure 7A).
Figure 6.
HuR ubiquitination and degradation by HS requires Lys-182. (A) Schematic of truncated HuR segments (generated from HuR–TAP). (B) The effect of HS on the levels of the HuR segments shown in (A) were tested by measuring HuR–TAP signals on western blots using an anti-IgG antibody to recognize the TAP segments of the fusion proteins. (C) Schematic of the point mutations [Lys → Arg (K → R)] introduced into the five lysine residues within the labile region of HuR–TAP. (D) The relative stability of each HuR–TAP (K → R) mutant was tested by western blot analysis; the levels of endogenous HuR, HuR–TAP (WT relative to K120R, K156R, K171R, K182R or K191R), and loading control β-actin were tested by monitoring HuR signals in untreated (−) compared with HS-treated (HS) cells. (E) Quantification of the western blotting data shown in (D); data represent the means±s.d. from three independent experiments.
Figure 7.
In vivo ubiquitination of HuR–TAP and mutant TAP and influence on cell survival after HS. (A) Cells were transfected with HuR–TAP (WT), HuR–TAP (K171R) (left), or HuR–TAP (K182R) (right) along with V or Ub-HA-expressing plasmids. The levels of ubiquitinated HuR–TAP were detected by IP (using Rabbit IgG) followed by WB analysis using anti-HA antibody. (B, C) Cells were transfected with plasmids to overexpress the HuR–TAP fusion proteins shown [WT (black) and K → R mutants (gray)]; 48 h later, cells were subjected to HS (43°C, 1 h), and after an additional 24 h the surviving fraction was quantified using the MTT assay (B) or by direct cell counts (C). The loss in cell viability and numbers was represented as a percentage of untreated cells in each transfection group (means±s.d. from three independent experiments). (D) In cells transfected as described in panels (B, C), the association of each HuR–TAP chimeric protein with HuR target transcripts was tested by RNP IP. The fold enrichment represents mRNA association to each HuR–TAP relative to TAP; the average of two similar experiments is shown.
Importantly, while the HS employed throughout this study (43°C) was not cytotoxic, expression of K182R significantly reduced cell survival, causing a significant loss in cell viability (∼38% by the MTT assay, 32% by direct cell counts) by 24 h following HS in this population; in contrast, only <8% (by both assays) of loss in cell viability was seen in the WT HuR population (Figure 7B and C). The other transfection groups (K120R, K156R, K171R, K191R) showed significantly less toxicity than the K182R group, with viabilities closer to that seen for WT (Figure 7B and C). In keeping with its relatively higher stability, the K182R mutant remained associated to numerous HuR target mRNAs tested (Figure 7D and data not shown). Collectively, these results indicate that the transient HS-mediated degradation of HuR is prevented by CHK2, involves ubiquitination of K182, and confers a survival advantage against HS cytotoxicity.
Discussion
Despite extensive evidence that stress-regulated HuR function is linked to its nucleocytoplasmic shuttling in the absence of net changes in HuR levels, here we document the rapid and potent degradation of HuR in response to an acute damaging stimulus. HS (43°C) triggered a fast and robust reduction in HuR stability and the effect was reversed if cells were returned to 37°C. Given HuR's central role in the post-transcriptional regulation of many critical proteins (e.g., cell cycle control factors, stress-response proteins, and cytokines), we investigated in depth the mechanism of HuR downregulation by HS. Our results show that HuR is a ubiquitination substrate in vitro and in vivo, that CHK2 ameliorates HuR loss by HS, that residue K182 is particularly important for HuR ubiquitination and HS-mediated decay, and that the HS-triggered transient downregulation of HuR protects against cell death.
Post-translational modification of HuR by HS
Earlier reports showed that HuR associated with cytoplasmic poly(A)+ RNA in unstressed cells, displaying a prominent presence in the polysomal component (Gallouzi et al, 2000). HS altered the subcellular localization and function of HuR, mobilizing it to SGs, enhancing its association with poly(A)+ RNA in the nucleus, and promoting the nuclear export of HuR linked to the cytoplasmic localization of hsp70 mRNA (Gallouzi et al, 2000, Gallouzi and Steitz, 2001). Our analysis did not directly test the association of HuR with poly(A)+ RNA, but did show that HuR colocalized in SGs shortly after commencing HS (Figure 3F). Additionally, we did not observe specific decreases in cytoplasmic or nuclear HuR, but an overall reduction in both compartments. Although there appeared to be a reduction in whole-cell HuR levels after HS in the studies by Gallouzi and coworkers, they did not formally report overt decreases in HuR levels following HS. It is likely that these differences collectively arise from variations between the two experimental systems, including the fact that we employed milder HS without serum (43°C, 1 h) whereas Gallouzi and coworkers used 45°C with serum.
Our results also agree with those of an earlier study in which inhibition of the proteasome elevated HuR levels in human diploid fibroblasts, although the mechanism responsible for this effect was not examined (Bonelli et al, 2004). We initially tested other post-translational modifications of HuR, including sumoylation, but did not find that HS influenced the levels of sumoylated HuR (Supplementary Figure S7). Instead, we found that HuR was ubiquitinated and was degraded through a process dependent on Ub and the proteasome. The Ub–proteasome system degrades many cellular proteins through the action of three enzymes: E1 activates Ub, triggering its transfer onto the Ub carrier enzyme E2, which in turn is transferred onto a substrate protein by an E3 Ub protein ligase and Ub becomes covalently linked. The repeated addition of Ub moieties results in the formation of a polyubiquitinated substrate protein, which is recognized by a large proteolytic complex, the 26S proteasome (Hershkom and Ciechanover, 1998). Here, we were able to detect endogenous as well ectopically expressed tagged HuR migrating with slower electrophoretic mobilities (Figures 5 and 7, Supplementary Figure S6), but it is unclear why HS did not appear to elevate the levels of polyubiquitinated moieties. It is possible that HS does not increase HuR ubiquitination per se, but instead signals the decay of already-ubiquitinated HuR protein.
Consequences of ubiquitination of RBPs AUF1 and HuR
Among the RBPs influencing the stability/translation of specific mRNA subsets, only AUF1 was previously shown to be ubiquitinated (Laroia et al, 1999). Laroia and colleagues showed that AUF1-associated AU-rich mRNAs were degraded after eukaryotic initiation factor (eIF)4G dissociated from AUF1, and that AUF1 was ubiquitinated and degraded (along with associated mRNAs, as proposed by the authors) via the proteasome. Interestingly, in this model system, hsp70 (which was elevated in response to HS) sequestered AUF1 in the nuclear–perinuclear region and blocked the decay of AUF1 and AUF1-bound mRNAs (Laroia et al, 1999). These investigators further showed that overexpression of a deubiquitinating protein (DUB) or inhibition of the proteasome prevented the decay of labile mRNAs (Laroia et al, 2002). Together with these two reports, our findings suggest a functional interplay between HuR and AUF1 upon the levels of many predictedly shared target mRNAs: following HS, the AUF1-bound hsp70 would be unable to trigger the degradation of AUF1-bound mRNAs, while the loss of HuR through Ub-mediated proteolysis would preclude the mRNA-stabilizing influence of HuR. Thus, the balance between decay and stability for an mRNA that is the target of HuR and AUF1 could depend directly on its relative affinity for each RBP. It is as yet unclear whether the proteasomal degradation of HuR also degrades HuR-bound mRNAs, although this possibility seems unlikely, given the wealth of evidence supporting HuR's mRNA-stabilizing influence (Brennan and Steitz, 2001). Nonetheless, as HS affects each protein differently (prevents AUF1 degradation and promotes HuR degradation), perhaps HuR proteolysis does not itself degrade target mRNAs, but the very decrease of HuR facilitates the binding of AUF1 to shared target mRNAs for which both RBPs compete. Such mRNAs could initially be protected from degradation while AUF1 is nuclear/perinuclear and bound to hsp70, but may later be degraded through an AUF1-dependent process.
It is also worth noting that the lysine residue implicated in HuR ubiquitination (K182) lies within the RRM2 (spanning amino acid residues 106–186). The post-translational modification of HuR at this site raises the possibility that ubiquitination at K182 interferes with HuR's association with a target mRNA and may also help to ‘release' it, in conjunction with triggering HuR degradation. Moreover, the findings that RRM2 mutants S88A and T118A are significantly more heat-labile than HuR(WT) and that all phosphomimic mutants (S88D, S100D, T118D) are markedly more heat-resistant than WT (Figure 4E and F) further support the notion that CHK2 phosphorylation of HuR protects against HS-triggered HuR decay. In this regard, the ubiquitination-deficient mutant K182R showed enhanced binding to target mRNAs after HS (Figure 7D). Although this mutant is logically expected to continue forming RNP complexes after HS (as it is refractory to HS-triggered decay), the influence of post-translational modifications at S88, S100, T118, and K182 upon binding to target mRNAs deserves further experimental scrutiny.
RBPs mediating post-transcriptional gene expression by HS
Heat stress induces the misfolding and denaturation of cellular proteins, causing cytotoxicity and triggering the HS response (De Maio, 1999; Voellmy, 2006). A prominent component of the HS response is the activation of a family of HS factors (HSFs), which bind to HS elements (HSEs) and direct the transcription of HS proteins (HSPs) and other stress and immune response proteins (recently reviewed by Wheeler and Wong, 2007). In addition, hyperthermia has long been recognized to regulate gene expression through post-transcriptional mechanisms (Yost et al, 1990). Global studies of the influence of HS on mRNA stability have revealed that large subsets of mRNAs are stabilized or destabilized in response to HS in both mammalian and yeast cells (Fan et al, 2002; Grigull et al, 2004). Global analyses have also shown that HS broadly represses the translation of subsets of mRNAs (Lindquist, 1981), and that hyperthermia can act upon the RNA itself, modifying its secondary structure and thereby altering its translational status (Narberhaus et al, 2006). At the same time, both the stability and translation of HSPs are favored in response to HS (Panniers, 1994; Kaarniranta et al, 2000).
The post-transcriptional changes in gene expression following HS are controlled through the action of various RBPs interacting with HS-regulated mRNAs. For example, HS inhibits the translation of capped mRNAs by dephosphorylating both eIF4E and the 4E-inactivating protein BP-1, thereby reducing eIF4E activity and availability (Feigenblum and Schneider, 1996). RBPs associating with specific regions of the 5′ and 3′UTRs, such as AUF1, TIAR, TIA-1, and TTP, have also been linked to HS-regulated gene expression (Laroia et al, 1999; Gallouzi et al, 2000; Kedersha and Anderson, 2002). However, HS did not cause dramatic changes in the levels of TIAR, TIA-1, or TTP, instead triggering changes in their subcellular distribution and their aggregation in SGs (Kedersha and Anderson, 2002). In this regard, the lower HuR abundance after HS is a novel mechanism whereby an RBP can regulate subsets of target mRNAs. The changes in both RBP localization and RBP levels are in agreement with the post-transcriptional operon model (Keene and Tenenbaum, 2002). According to this model, collections of functionally related genes can be coordinately regulated through the presence of shared RNA elements within the transcripts, which render them targets of a given RBP. Thus, by reducing HuR expression levels, HS can effectively reduce the stability and/or alter the translation of HuR target mRNAs. Given the protective influence of a timely reduction of HuR following HS (Figure 7B and C), the corresponding decreases in proliferative proteins encoded by HuR target mRNAs (e.g., c-Fos, prothymosin-α, and Cyclins A2, B1, and D1) are likely to be advantageous, allowing cells to channel its resources and energy towards the repair of heat-damaged components. As we gain a deeper understanding of HuR's influence on cell survival after HS, we propose that the transient decrease in HuR abundance is a key adaptive mechanism whereby mammalian cells maintain homeostasis in response to thermal stress.
Materials and methods
Cell culture, treatments, and transfections
Human cervical carcinoma HeLa cells and mouse melanoma B16F10 cells were cultured in Dulbecco's modified essential medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS) and antibiotics; WI-38 cells were further supplemented with non-essential amino acids (Gibco). HCT116 colon cancer cells (WT, CHK2KO; Jallepalli et al, 2003) were cultured in McCoy's 5A medium supplemented with 10% FBS and antibiotics. For HS treatments, cells were washed with PBS, given preheated medium (DMEM without FBS), and further incubated for 1 h at 43°C unless indicated otherwise. Control cells were also washed with PBS and given pre-warmed medium (DMEM without FBS) for 1 h at 37°C. All small interfering (si)RNAs (Qiagen, listed in the Supplementary data) were used at a final concentration of 10 nM. The plasmids used are described in the Supplementary data. CHX and the CHK2 inhibitor were from Calbiochem; sodium arsenite was from Sigma. Cell survival was measured by the MTT assay (Sigma) and by direct cell counts using a hemocytometer.
mRNA and RNP analysis
Endogenous mRNA–protein complexes were precipitated as described previously (Abdelmohsen et al, 2007a). Total RNA as well as RNA in RNP IP reactions was reverse transcribed (RT) using random hexamers, oligo(dT) primer, and Superscript II RT (Invitrogen). The levels of specific mRNAs were measured by real-time quantitative (q)PCR using the QuantiTect SYBR green PCR kit (Qiagen) and analyzed on an ABI Prism 7000 detection system; transcript-specific primers are listed as Supplementary data.
Protein analysis
Western blot analysis was performed using standard procedures, as described (Abdelmohsen et al, 2007a and Supplementary data). Nascent (de novo) translation of HuR was studied as described (Galbán et al, 2008). Briefly, following incubation for 1 h at 43°C (HS) or 37°C (control), HeLa cells were incubated with 1 mCi L-[35S]methionine and L-[35S]cysteine (EasyTag ™EXPRESS, NEN/Perkin Elmer) per 60-mm plate for 15 min. Cells were then lysed in RIPA buffer and the IP reactions carried out in 1 ml TNN buffer (50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, and 0.5% NP-40) for 16 h at 4°C using IgG (BD Pharmingen) or anti-HuR antibodies. Following extensive washes in TNN buffer, the IP samples were resolved by SDS-PAGE, transferred onto PVDF filters, and visualized with a PhosphorImager (Molecular Dynamics). The analysis of ubiquitinated HuR is detailed in the Supplementary data.
Supplementary Material
Supplementary Results
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Figures
Supplementary Table Legends
Acknowledgments
We thank MJ Pazin, V Dixit, WH Wood 3rd, B Frank, S Subaran, and FE Indig for their assistance with these studies. HCT116 cells were kindly provided by F Bunz. This research was supported entirely by the NIA-IRP Program Z01-AG000511-10 and by set-aside funds of the NIA-IRP, NIH.
References
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M (2008) Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 389: 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Lal A, Kim HH, Gorospe M (2007b) Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 6: 1288–1292 [DOI] [PubMed] [Google Scholar]
- Abdelmohsen K, Pullmann R Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M (2007a) Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 25: 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Bonelli MA, Alfieri RR, Desenzani S, Petronini PG, Borghetti AF (2004) Proteasome inhibition increases HuR level, restores heat-inducible HSP72 expression and thermotolerance in WI-38 senescent human fibroblasts. Exp Gerontol 39: 423–432 [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA (2001) HuR and mRNA stability. Cell Mol Life Sci 58: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Xu N, Shyu AB (2002) Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol 22: 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio A (1999) Heat shock proteins: facts, thoughts, and dreams. Shock 11: 1–12 [DOI] [PubMed] [Google Scholar]
- Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W (2007) Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol Biol Cell 18: 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormoy-Raclet V, Ménard I, Clair E, Kurban G, Mazroui R, Di Marco S, von Roretz C, Pause A, Gallouzi IE (2007) The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich-element-dependent manner. Mol Cell Biol 27: 5365–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yang X, Wang W, Wood III WH, Becker KG, Gorospe M (2002) Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci USA 99: 10611–10616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA (1998) Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17: 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenblum D, Schneider RJ (1996) Cap-binding protein (eukaryotic initiation factor 4E) and 4E-inactivating protein BP-1 independently regulate cap-dependent translation. Mol Cell Biol 16: 5450–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Muñoz-Canoves P, Gorospe M, Muñoz A (2003) Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol 23: 4991–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán S, Kuwano Y, Pullmann R Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Lewis SM, Holcik M, Gorospe M (2008) RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol 28: 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA (2000) HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA 97: 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA (2001) Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895–1901 [DOI] [PubMed] [Google Scholar]
- Gorospe M (2003) HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle 2: 412–414 [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR (2004) Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Muhlhausser P, Koller-Eichhorn R, Brennecke J, Kutay U (2004) Transportin2 functions as importin and mediates nuclear import of HuR. Proc Natl Acad Sci USA 101: 2918–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkom A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Lengauer C, Vogelstein B, Bunz F (2003) The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J Biol Chem 278: 20475–20479 [DOI] [PubMed] [Google Scholar]
- Kaarniranta K, Holmberg CI, Helminen HJ, Eriksson JE, Sistonen L, Lammi MJ (2000) Protein synthesis is required for stabilization of hsp70 mRNA upon exposure to both hydrostatic pressurization and elevated temperature. FEBS Lett 475: 283–286 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30: 963–969 [DOI] [PubMed] [Google Scholar]
- Keene JD (1999) Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA 96: 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD (2007) RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA (2002) Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell 9: 1161–1167 [DOI] [PubMed] [Google Scholar]
- Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr, Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, Gorospe M (2008) Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev 22: 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R Jr, Martindale JL, Yang X, Gorospe M (2008) MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol 28: 4562–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M (2005) Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J 24: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 23: 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ (1999) Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284: 499–502 [DOI] [PubMed] [Google Scholar]
- Laroia G, Sarkar B, Schneider RJ (2002) Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc Natl Acad Sci USA 99: 1842–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA (2002) Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem 277: 44623–44630 [DOI] [PubMed] [Google Scholar]
- Liao B, Hu Y, Brewer G (2007) Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol 14: 511–518 [DOI] [PubMed] [Google Scholar]
- Lindquist S (1981) Regulation of protein synthesis during heat shock. Nature 293: 311–314 [DOI] [PubMed] [Google Scholar]
- López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M (2003) Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22: 7146–7154 [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galbán S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M (2003) RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA 100: 8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming XF, Stoecklin G, Lu M, Looser R, Moroni C (2001) Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol 21: 5778–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ (2005) From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518 [DOI] [PubMed] [Google Scholar]
- Narberhaus F, Waldminghaus T, Chowdhury S (2006) RNA thermometers. FEMS Microbiol Rev 30: 3–16 [DOI] [PubMed] [Google Scholar]
- Panniers R (1994) Translational control during heat shock. Biochimie 76: 737–747 [DOI] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu A-B (1998) RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J 17: 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullmann R Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M (2007) Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol 27: 6265–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebane A, Aab A, Steitz JA (2004) Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 10: 590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Maurer F, Nagamine Y (2003) Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol Cell Biol 23: 7177–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giessen K, Di-Marco S, Clair E, Gallouzi IE (2003) RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem 278: 47119–47128 [DOI] [PubMed] [Google Scholar]
- Voellmy R (2006) Feedback regulation of the heat shock response. Handbook Exp Pharmacol 172: 43–68 [DOI] [PubMed] [Google Scholar]
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M (2000a) HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 19: 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook NJ, Gorospe M (2000b) HuR Regulates p21 mRNA stabilization by ultraviolet light. Mol Cell Biol 20: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M (2001) Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 21: 5889–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DS, Wong HR (2007) Heat shock response and acute lung injury. Free Radic Biol Med 42: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen C, Shyu A-B (2001) Versatile role for hnRNPD isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol 21: 6960–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Stoecklin G, Kedersha N, Simarro M, Anderson P (2007) T-cell intracellular antigen-1 (TIA-1)-induced translational silencing promotes the decay of selected mRNAs. J Biol Chem 282: 30070–30077 [DOI] [PubMed] [Google Scholar]
- Yost HJ, Petersen RB, Lindquist S (1990) RNA metabolism: strategies for regulation in the heat shock response. Trends Genet 6: 223–227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Results
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Figures
Supplementary Table Legends