INTERFEROME: the database of interferon regulated genes (original) (raw)
Abstract
INTERFEROME is an open access database of types I, II and III Interferon regulated genes (http://www.interferome.org) collected from analysing expression data sets of cells treated with IFNs. This database of interferon regulated genes integrates information from high-throughput experiments with annotation, ontology, orthologue sequences from 37 species, tissue expression patterns and gene regulatory information to enable a detailed investigation of the molecular mechanisms underlying IFN biology. INTERFEROME fulfils a need in infection, immunity, development and cancer research by providing computational tools to assist in identifying interferon signatures in gene lists generated by high-throughput expression technologies, and their potential molecular and biological consequences.
INTRODUCTION
The interferons (IFNs) are a family of cytokines identified more than 50 years ago as potent antiviral proteins. In addition to antiviral, antibacterial and anti-parasitic host-defense functions they are now also recognized as crucial regulators of cell proliferation, differentiation, survival and death as well as activators of specialized cell functions particularly in the immune system (1,2) and play important roles in infectious and inflammatory diseases, autoimmunity and cancer (3). Intriguingly, their biological properties have led to their use as therapeutics in certain viral infections such as chronic hepatitis C (HCV), hepatitis B (HCB) virus infections and in combination with antiretroviral therapy in AIDS, in haematological cancers and solid tumuors and in multiple sclerosis (4); but they are targeted for blockade in autoimmune diseases such as systemic lupus erythematosus (5). Therefore, understanding the molecular pathways and genes regulated by IFNs will result in better understanding of disease mechanisms and effective clinical use of both IFNs and related cytokines that utilize similar signal transduction pathways.
There are three types of IFNs, namely types I (IFN α, β, δ, ε, ζ, κ, ν, τ, ω), II (IFNγ) and III (IFNλ) (3). Each IFN type has sequence similarity, signals via specific cell surface receptor complexes (IFNAR, IFNGR, IFNLR) and is produced by particular cells in response to specific stimuli (6,7). Despite these differences, all types of IFNs mediate their effects by activating multiple signaling pathways, some of which are common, including the well-characterized JAK-STAT pathway (8) as well as other pathways (9). The JAK-STAT signaling events lead to the activation of transcription factor complexes such as IFN stimulated gene factor 3 (ISGF3) and homo- or hetero-dimerized signal transducers and activators of transcription (STAT) molecules, and their subsequent binding to interferon response elements such as interferon stimulated response element (ISRE), gamma activated sequence (GAS) and STAT-binding sites in promoters of IRGs, resulting in the transcriptional activation of many IRGs (10). The advent of expression profiling technology has enabled us to ascertain a broad picture of gene expression changes resulting from the action on target cells of cytokines such as IFNs. Some IRGs are well characterized in relation to specific functions (e.g. the antiviral Mx proteins, 2, 5-OAS, PKR and RnaseL) (11,12). However, many IRGs have no known function or functions that have not been previously associated with IFNs. The challenge is now to firstly determine the scope of the genes regulated by IFNs, and to understand why some, and not others, are activated or repressed in specific circumstances and relate this to biological outcomes.
In this exercise, we have catalogued IRGs from all published reports where cells were treated with an IFN of any type, and incorporated these into a searchable database (http://www.interferome.org). The utility of the database is to enable the reliable identification of interferon regulated gene signatures from high-throughput data sets (i.e. microarray, proteomic data, etc.). It will also assist in identifying regulatory elements and enable comparison of normal tissue expression of IRGs in human and mouse. This will have implications for determining hitherto unknown role of IFNs or IRGs in specific circumstances whether regulating homoeostasis or disease pathogenesis or understanding the basis of patient responses to IFN or related therapies.
OVERVIEW
The INTERFEROME database enables the identification of interferon regulated genes and integrates IRG-specific sequence, annotation and regulatory information. IRGs in INTERFEROME were collected manually by analysing in-house IFN microarray data and published microarray/proteomic data sets. In addition, extensive manual literature mining and searching of regulatory databases, such as Transcription Regulatory Region Database (TRRD) (13), was performed to identify previously characterized IRGs.
Sequence and annotation data for IRGs were mined from public databases such as Ensembl v.49 (14), and NCBI Entrez Gene (15) using an automated data mining and integration software pipeline developed in our laboratory. The main component of this pipeline is the data downloader program, which is written in the Perl programming language and interacts with the application programming interfaces (APIs) provided by external databases. It utilizes the list of manually curated IRGs to extract corresponding sequence and annotation information from public databases and ensures that the sequence and annotation data in INTERFEROME is current. The sequence data collected includes; genes (59 308), amino acids (79 215), putative promoters (79 206), 3′- and 5′-UTR sequences. Annotation includes multiple gene identifiers and gene alias information, gene ontologies and orthologue information. Orthologues of human and mouse IRGs were collected from 37 species and will enable detailed phylogenetic analysis of these proteins. Basal tissue expression data for human and mouse IRG were collected from the Novartis Foundation GNF portal (16). Quality control of sequence data included BLAST2GO (17) analysis to ensure amino acid sequences corresponded to relevant IRGs. These diverse data sets were then processed to generate regulatory information such as transcription factor binding sites (TFBS), protein domains and motifs, gene ontologies and normalized tissue expression.
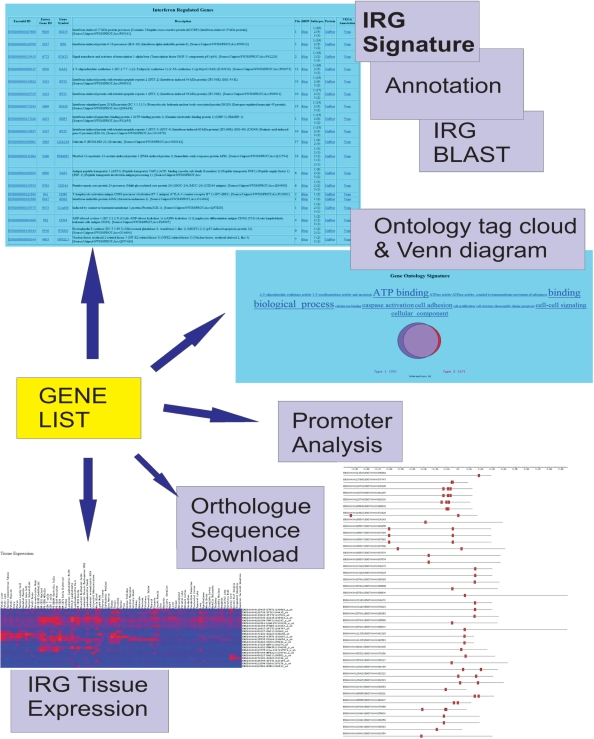
An example of possible search results is shown in the screenshots depicted in Figure 1. In addition to identifying IRG signatures in gene lists we also provide the ability to identify putative TFBS in proximal promoter regions of IRGs and the integration of expression information provides the ability to search for normal tissue expression of IRGs in 79 human and 61 mouse tissues. We have also generated BLAST databases of IRG sequences, integrated with NCBI BLAST software, enabling users to perform BLAST analysis (15). Addition of data sets and incorporation of new functionality will be announced in the INTERFEROME blog (http://www.interferome.org/blog). As new data sets are incorporated, INTERFEROME will grow significantly and enable more sophisticated analysis of IRGs.
Figure 1.
The functionality of the INTERFEROME database is summarized in Figure 1, which displays some of the screen captures resulting from searching the database with a gene list. Its main functionality is the IRG signature page which identifies IRGs that were present in the gene list and displays associated annotation. In addition INTERFEROME's expression and promoter analysis capability is highlighted. An example search is demonstrated in Supplementary Figures.
DESIGN AND IMPLEMENTATION
Data were collected from in-house IFN-treated microarrays and more than 28 publications (18–45) identified through literature searches where high-throughput analysis (microarray or proteomic) was performed on cells/tissues treated only with IFNs. The statistical analysis of the data sets in the relevant publications adhered to currently accepted methods and standards. Gene lists were analysed and genes that demonstrated 1.5-fold or more differential expression were identified as IRGs. This included up- and downregulated genes from 23 human data sets, five mouse datasets and single chimp, cow and sheep data sets. There were 28 type I IFN data sets, 11 type II and three type III data sets. The subtypes used include type I IFNs such as (IFNA1a, IFNA1, IFNA2, IFNA2a, IFNA2b, IFN conA, IFNB, IFNT and IFNE1), type II (IFNG) and type III (IFNL). Overall 1996 human and 1925 mouse IRGs were identified.
INTERFEROME uses an Apache web server, in a Linux environment and a collection of Perl CGI and PHP scripts providing the user interface coupled to a MySQL relational database management system. The PHP scripts include a tag cloud generator, a Venn visualizer and all the search form pages. Two CGI Perl programs which generate TFBS graphics [utilizing BioPerl graphics modules (46)] and hierarchical clustered heat maps [using Cluster 3.0 and matrix2png (47)] of tissue expression data provide the other data visualization methods.
USING INTERFEROME
Searching interferon regulated gene signatures
The main functionality of this database is to enable the identification interferon regulated genes in a gene list generated from a high-throughput microarray or proteomic experiment. This ability to detect an interferon signature is invaluable in identifying interferon regulated pathways involved in innate immune, inflammatory or anti-tumour responses and will enable the extraction of more meaningful information about the biology associated with a set of differentially regulated genes. A mouse or human gene list of up to 100 genes can be submitted to INTERFEROME, and provides the option of identifying either all IRGs or those that are induced by a specific interferon type (types I, II or III). The submitted gene list must be Ensembl IDs as the use of gene names and gene symbols may result in ambiguities (48). A link is provided to external gene identifier conversion resources to enable the conversion of gene identifiers (49). When the list is submitted to the INTERFEROME server, a new page with identified IRGs is generated (Supplementary Figure S1A and B). This page provides information, such as Ensembl ID, Entrez Gene ID and Gene Symbol, all of which are clickable links that directs the user to the relevant page in external database resources such as the Ensembl genome browser, NCBI Gene page or HGNC gene nomenclature page. In addition, a short description of each gene and its chromosomal location together with additional links to external bioinformatic resources such as iHOP (50), UniProt (51) and the manually curated VEGA database (52) are provided to enable the collection of additional gene and protein-specific information. A list of IFN types (I, II or III) that induce the gene is shown together with the number of data sets (in brackets) in which a particular gene was differentially regulated. This enables the differentiation of core IRGs (those that are induced by most IFNs in most situations) from those that are induced in specific situations. This page also displays a tag cloud of all the ontologies associated with the IRG list where ontologies (obtained from Ensembl and GO) with higher frequencies are shown in a larger font. Clicking on an ontology term redirects the user to the gene ontology organization web page (53) for that particular term. Finally, a Venn diagram visualization of type I versus type II is also generated to show the proportional induction by different IFNs. A collection of help pages offer assistance in using INTERFEROME and an example search is available in Supplementary Data.
Searching normal tissue expression of IRGs
The human and mouse expression data sets were downloaded from the GNF portal, mapped to IRGs and expression of each gene was normalized across tissue by log2 transformation with a mean of 0 and an SD of 1. When a list of either human or mouse IRGs is submitted, a results page with a heat map depicting the tissue expression of the genes in 79 human or 61 mouse tissues is displayed (Supplementary Figure S2). Red and blue denote up and down regulation of gene expression, respectively, and the intensity of the colour is proportional to the intensity of expression. This heat-map enables the identification of IRGs with tissue-specific expression. It should be noted that the heat map reflects the tissues where these IRGs are basally expressed, not after their induction by IFNs.
IRG promoter analysis
INTERFEROME stores more than 274 000 TFBS and enables the visualization of selected TFBS such as ISRE, STAT, IRF, ICSBP, TATA and NFKB known to be important in the regulation of IRGs. TFBSs are displayed in the 1000 bp immediately upstream of the transcription start site (TSS) and includes the 5′-UTR. The form also provides the user with the option of displaying selected TFBSs within all the known transcripts of a particular gene (Supplementary Figure S3A and B). Previous analysis of IRGs suggests that core promoters for most IRGs are located within the first 1000 bp upstream of the TSS (54). However, ∼64% of IRGs collected in INTERFEROME have ISRE elements within the 10 kb upstream of the TSS. Distal promoter elements that are beyond the first 1000 bp may be important for transcriptional regulation of IRGs, thus care should be taken in interpreting preliminary promoter analysis results and predictions should be validated by experimental techniques such as chromatin immuno-precipitation (ChIP). We utilized Transfac Professional version 12.1 to preprocess sequences and store binding sites and their positions within INTERFEROME. The Transfac database is a collection of position weight matrices for transcription factor binding sites (55). These matrices are generated from experimentally derived TFBS (using sequences that are known to be bound with high affinity by particular transcription factors) derived either by in vitro selection of tightly bound oligonucleotides (SELEX) or by ChIP or bandshift experiments that identify specific transcription factor–DNA interactions. These matrices summarize how many sequences have a given nucleotide at a given position in the transcription factor binding site. A position weight matrix (PWM) can be used to define a position-specific scoring matrix (PSSM), whose values are proportional to binding free energies (56). The search form enables the selection of core and matrix match cut-offs between 0 and 1. When these match values are closer to one, only higher stringency TFBS are displayed. The ability to specify the cut-off values enables the user to determine the optimal values necessary to minimize the identification of false-positive and false-negative TFBS.
Orthologue sequence download
A search form enabling the submission of a human or mouse IRG and retrieving its orthologous amino acid sequences stored in INTERFEROME is provided. Orthologue sequences are derived from 37 species from Ensembl version 49. When a human or mouse IRG is used to search the database a table with a list of orthologues is displayed and a link to download the sequences in the FASTA format is provided (Supplementary Figure S4).
Search IRG BLAST databases
We have generated IRG protein BLAST databases for IRG sequences from human, mouse and 37 vertebrate species, enabling protein or nucleotide BLAST (BLASTP or BLASTX) analysis to be performed on our sequence data sets. This functionality will enable other sequences/novel genes to be searched against IRGs and allow the identification of common domains or novel IRGs. The BLAST search forms provide the ability to specify settings and display graphical results similar to NCBI BLAST (15).
FUTURE DIRECTIONS
As expression, genomic and other sequence information is updated in external databases, we will reflect those changes in INTERFEROME. In addition as more interferon-treated expression data sets become available, we hope to add these to INTERFEROME and expand the IRG data sets. Each additional data set will increase the reliability and specificity of the IRG gene set. We propose to eventually provide normalized expression information on the IRG data sets and enable IFN subtype, dose, time and tissue or cell-type-specific IRG analysis. We will also provide more comprehensive IRG-specific regulatory and interaction information, statistical and computational methods to analyse _cis_-regulatory modules, pathway information, sophisticated data visualization methods and disease data sets to identify disease-specific IRG signatures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Health and Medical Research Council, Australia and CRC for chronic inflammatory disease. Funding for open access charge: National Health and Medical Research Council of Australia.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
The Bio21 UROP program.
REFERENCES
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Hertzog PJ, O’Neill LA, Hamilton JA. The interferon in TLR signaling: more than just antiviral. Trends Immunol. 2003;24:534–539. doi: 10.1016/j.it.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annu. Rev. Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 4.Gutterman JU. Cytokine therapeutics: lessons from interferon alpha. Proc. Natl Acad. Sci. USA. 1994;91:1198–1205. doi: 10.1073/pnas.91.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt KN, Ouyang W. Targeting interferon-alpha: a promising approach for systemic lupus erythematosus therapy. Lupus. 2004;13:348–352. doi: 10.1191/0961203304lu1025oa. [DOI] [PubMed] [Google Scholar]
- 6.de Weerd NA, Samarajiwa SA, Hertzog PJ. Type I interferon receptors: biochemistry and biological functions. J. Biol. Chem. 2007;282:20053–20057. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 7.Noppert SJ, Fitzgerald KA, Hertzog PJ. The role of type I interferons in TLR responses. Immunol. Cell Biol. 2007;85:446–457. doi: 10.1038/sj.icb.7100099. [DOI] [PubMed] [Google Scholar]
- 8.Brierley MM, Fish EN. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J. Interferon Cytokine Res. 2002;22:835–845. doi: 10.1089/107999002760274845. [DOI] [PubMed] [Google Scholar]
- 9.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 10.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 11.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr. Top. Microbiol. Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- 13.Kolchanov NA, Ignatieva EV, Ananko EA, Podkolodnaya OA, Stepanenko IL, Merkulova TI, Pozdnyakov MA, Podkolodny NL, Naumochkin AN, Romashchenko AG. Transcription regulatory regions database (TRRD): its status in 2002. Nucleic Acids Res. 2002;30:312–317. doi: 10.1093/nar/30.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinstock-Guttman B, Badgett D, Patrick K, Hartrich L, Santos R, Hall D, Baier M, Feichter J, Ramanathan M. Genomic effects of IFN-beta in multiple sclerosis patients. J. Immunol. 2003;171:2694–2702. doi: 10.4049/jimmunol.171.5.2694. [DOI] [PubMed] [Google Scholar]
- 19.Satoh J, Nanri Y, Tabunoki H, Yamamura T. Microarray analysis identifies a set of CXCR3 and CCR2 ligand chemokines as early IFNbeta-responsive genes in peripheral blood lymphocytes in vitro: an implication for IFNbeta-related adverse effects in multiple sclerosis. BMC Neurol. 2006;6:18. doi: 10.1186/1471-2377-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 21.Leaman DW, Chawla-Sarkar M, Jacobs B, Vyas K, Sun Y, Ozdemir A, Yi T, Williams BR, Borden EC. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J. Interferon Cytokine Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 22.Coelho LF, Magno de Freitas Almeida G, Mennechet FJ, Blangy A, Uze G. Interferon-alpha and -beta differentially regulate osteoclastogenesis: role of differential induction of chemokine CXCL11 expression. Proc. Natl Acad. Sci. USA. 2005;102:11917–11922. doi: 10.1073/pnas.0502188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S, Snyder M. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brierley MM, Marchington KL, Jurisica I, Fish EN. Identification of GAS-dependent interferon-sensitive target genes whose transcription is STAT2-dependent but ISGF3-independent. FEBS J. 2006;273:1569–1581. doi: 10.1111/j.1742-4658.2006.05176.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Choi Y, Bazer FW, Spencer TE. Identification of genes in the ovine endometrium regulated by interferon tau independent of signal transducer and activator of transcription 1. Endocrinology. 2003;144:5203–5214. doi: 10.1210/en.2003-0665. [DOI] [PubMed] [Google Scholar]
- 26.Holko M, Williams BR. Functional annotation of IFN-alpha-stimulated gene expression profiles from sensitive and resistant renal cell carcinoma cell lines. J. Interferon Cytokine Res. 2006;26:534–547. doi: 10.1089/jir.2006.26.534. [DOI] [PubMed] [Google Scholar]
- 27.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Indraccolo S, Pfeffer U, Minuzzo S, Esposito G, Roni V, Mandruzzato S, Ferrari N, Anfosso L, Dell’Eva R, Noonan DM, et al. Identification of genes selectively regulated by IFNs in endothelial cells. J. Immunol. 2007;178:1122–1135. doi: 10.4049/jimmunol.178.2.1122. [DOI] [PubMed] [Google Scholar]
- 29.Hilkens CM, Schlaak JF, Kerr IM. Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J. Immunol. 2003;171:5255–5263. doi: 10.4049/jimmunol.171.10.5255. [DOI] [PubMed] [Google Scholar]
- 30.Peng FW, Duan ZJ, Zheng LS, Xie ZP, Gao HC, Zhang H, Li WP, Hou YD. Purification of recombinant human interferon-epsilon and oligonucleotide microarray analysis of interferon-epsilon-regulated genes. Protein Expr. Purif. 2007;53:356–362. doi: 10.1016/j.pep.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Yan W, Lee H, Yi EC, Reiss D, Shannon P, Kwieciszewski BK, Coito C, Li XJ, Keller A, Eng J, et al. System-based proteomic analysis of the interferon response in human liver cells. Genome Biol. 2004;5:R54. doi: 10.1186/gb-2004-5-8-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh MK, Scott TF, LaFramboise WA, Hu FZ, Post JC, Ehrlich GD. Gene expression changes in peripheral blood mononuclear cells from multiple sclerosis patients undergoing beta-interferon therapy. J. Neurol. Sci. 2007;258:52–59. doi: 10.1016/j.jns.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Antoniou E, Liu Z, Hearne LB, Roberts RM. A microarray analysis for genes regulated by interferon-tau in ovine luminal epithelial cells. Reproduction. 2007;134:123–135. doi: 10.1530/REP-07-0387. [DOI] [PubMed] [Google Scholar]
- 34.Sanda C, Weitzel P, Tsukahara T, Schaley J, Edenberg HJ, Stephens MA, McClintick JN, Blatt LM, Li L, Brodsky L, et al. Differential gene induction by type I and type II interferons and their combination. J. Interferon Cytokine Res. 2006;26:462–472. doi: 10.1089/jir.2006.26.462. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, Liu H, Barahmand-pour F, Sivakumar P, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 37.Rani MR, Shrock J, Appachi S, Rudick RA, Williams BR, Ransohoff RM. Novel interferon-beta-induced gene expression in peripheral blood cells. J. Leukoc. Biol. 2007;82:1353–1360. doi: 10.1189/jlb.0507273. [DOI] [PubMed] [Google Scholar]
- 38.Lanford RE, Guerra B, Lee H, Chavez D, Brasky KM, Bigger CB. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- 39.Ramana CV, Gil MP, Han Y, Ransohoff RM, Schreiber RD, Stark GR. Stat1-independent regulation of gene expression in response to IFN-gamma. Proc. Natl Acad. Sci. USA. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramana CV, Gil MP, Schreiber RD, Stark GR. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 41.Gil MP, Bohn E, O’Guin AK, Ramana CV, Levine B, Stark GR, Virgin HW, Schreiber RD. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl Acad. Sci. USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoshida MS, Gorjao R, Lima C, Daher S, Curi R, Bevilacqua E. Regulation of gene expression in mouse trophoblast cells by interferon-gamma. Placenta. 2007;28:1059–1072. doi: 10.1016/j.placenta.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Taylor MW, Grosse WM, Schaley JE, Sanda C, Wu X, Chien SC, Smith F, Wu TG, Stephens M, Ferris MW, et al. Global effect of PEG-IFN-alpha and ribavirin on gene expression in PBMC in vitro. J. Interferon Cytokine Res. 2004;24:107–118. doi: 10.1089/107999004322813354. [DOI] [PubMed] [Google Scholar]
- 44.Schlaak JF, Hilkens CM, Costa-Pereira AP, Strobl B, Aberger F, Frischauf AM, Kerr IM. Cell-type and donor-specific transcriptional responses to interferon-alpha. Use of customized gene arrays. J. Biol. Chem. 2002;277:49428–49437. doi: 10.1074/jbc.M205571200. [DOI] [PubMed] [Google Scholar]
- 45.Zimmerer JM, Lesinski GB, Radmacher MD, Ruppert A, Carson W.E., III STAT1-dependent and STAT1-independent gene expression in murine immune cells following stimulation with interferon-alpha. Cancer Immunol. Immunother. 2007;56:1845–1852. doi: 10.1007/s00262-007-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 48.Petsko GA. What's in a name? Genome Biol. 2002;3 COMMENT1005. [Google Scholar]
- 49.Alibes A, Yankilevich P, Canada A, Diaz-Uriarte R. IDconverter and IDClight: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat. Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 51.The UniProt consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukahara T, Kim S, Taylor MW. REFINEMENT: a search framework for the identification of interferon-responsive elements in DNA sequences—a case study with ISRE and GAS. Comput. Biol. Chem. 2006;30:134–147. doi: 10.1016/j.compbiolchem.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 55.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]