Systematic Discovery of In Vivo Phosphorylation Networks (original) (raw)
. Author manuscript; available in PMC: 2009 Jun 8.
Summary
Protein kinases control cellular decision processes by phosphorylating specific substrates. Proteome-wide mapping has identified thousands of in vivo phosphorylation sites. However, systematically resolving which kinase targets each site is presently infeasible, due to the limited specificity of consensus motifs and the potential influence of contextual factors, such as protein scaffolds, localisation and expression, on cellular substrate specificity. We have therefore developed a computational method, NetworKIN, that augments motifs with context for kinases and phosphoproteins. This can pinpoint individual kinases responsible for specific in vivo phosphorylation events and yields a 2.5-fold improvement in the accuracy with which phosphorylation networks can be constructed. We show that context provides 60–80% of the computational capability to assign in vivo substrate specificity. Applying this approach to a DNA damage signalling network, we extend its cell-cycle regulation by showing that 53BP1 is a CDK1 substrate, show that Rad50 is phosphorylated by ATM kinase under genotoxic stress, and suggest novel roles of ATM in apoptosis. Finally, we present a scalable strategy to validate our predictions and use it to support the prediction that BCLAF1 is a GSK3 substrate.
Introduction
The dynamic behaviour and decision processes of eukaryotic cells are controlled by post-translational modifications such as protein phosphorylation. These, in turn, can modify protein function by inducing conformational changes, or by creating binding sites for protein interaction domains (for example SH2 or BRCT) that selectively recognise phosphorylated linear motifs (Seet et al., 2006).
Decades of targeted biochemical studies and recent experiments employing mass spectrometry (MS) techniques have identified thousands of in vivo phosphorylation sites (Aebersold and Mann, 2003). These are collected in the Phospho.ELM database, which currently contains 7207 phosphorylation sites in 2540 human proteins (Diella et al., 2004). However, which of the approximately 518 human protein kinases (Manning et al., 2002) is responsible for each of these phosphorylation events is only known for just over a third of sites identified thus far (35% (Diella et al., 2004)), and this fraction is decreasing in the wake of additional proteome-wide studies. As a consequence, there is an ever-widening gap in our understanding of in vivo phosphorylation networks, which is difficult to close in a systematic way by current experimental methods, despite advances in high-throughput in vitro assays (Ptacek et al., 2005) and selective kinase inhibitors (Bain et al., 2003). Our understanding of phosphorylation-dependent signalling networks is therefore still fragmentary.
The desire to map phosphorylation networks has motivated the development of computational methods to predict the substrate specificities of protein kinases, based on experimental identification of the consensus sequence motifs recognised by the active site of kinase catalytic domains (Hjerrild et al., 2004; Obenauer et al., 2003; Puntervoll et al., 2003). However, these motifs often lack sufficient information to uniquely identify the physiological substrates of specific kinases. For example, the sites phosphorylated by different kinases from the CDK or Src families cannot be distinguished by their sequences, although consensus motifs of these kinases have been determined by in vitro experiments (Manke et al., 2005). Thus, the recognition properties of the active site alone are typically insufficient to reproduce the substrate specificities of protein kinases observed in living cells (Dar et al., 2005). Specificity in protein kinase signalling is also achieved through additional effects such as subcellular compartmentalisation, co-localisation via anchoring proteins and scaffolds (e.g. A-Kinase Anchoring Proteins and Ste5 (Bhattacharyya et al., 2006)), substrate capture by non-catalytic interaction domains (e.g. SH2 domains), temporal and cell-type specific co-expression, kinase docking motifs within substrates (e.g. for MAP kinases (Reményi et al., 2005)) and regulatory subunits (e.g. cyclins). Such information, which we term contextual, may therefore enhance the accuracy with which the in vivo substrates of protein kinases can be predicted.
Results
The NetworKIN methodology
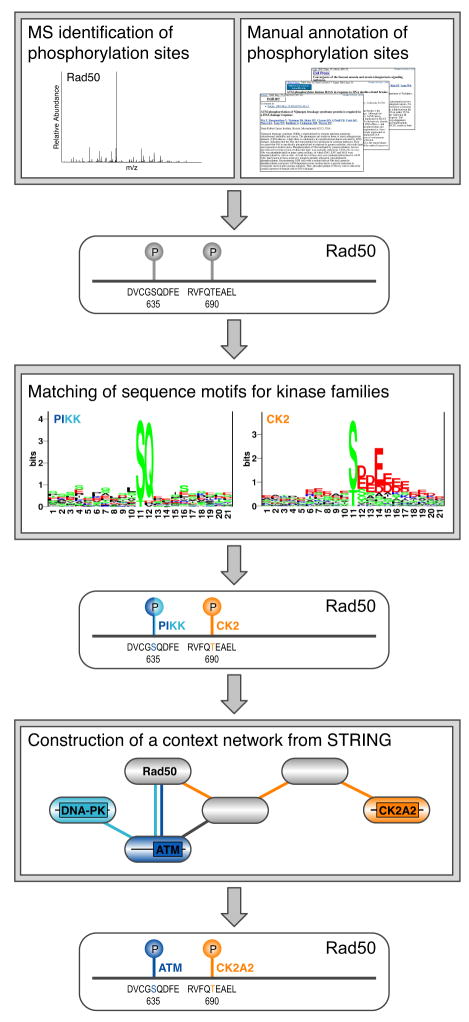
To explore the possibility of using context to enhance the identification of kinase substrates, we developed an integrative computational approach, NetworKIN. This combines consensus sequence motifs and protein association networks to predict which protein kinases target experimentally identified phosphorylation sites in vivo (Figure 1). The algorithm consists of two stages. In the first step we use neural networks and position-specific scoring matrices to assign each phosphorylation site to one or more kinase families, based on the intrinsic preference of kinases for consensus substrate motifs (Hjerrild et al., 2004; Obenauer et al., 2003). In the second stage, the context for each substrate is represented by a probabilistic protein network extracted from the STRING database (von Mering et al., 2005), which integrates information from curated pathway databases, co-occurrence in abstracts, physical protein interaction assays, mRNA expression studies, and genomic context (see Experimental Procedures). Within this context network, kinases of the appropriate families are identified through sequence similarity searches against an annotated database of kinase domain sequences (Figure 1). This approach captures both direct and indirect interactions; for example, phosphorylation events mediated by scaffolds are predicted, as the scaffolding protein provides a path in the probabilistic network between the substrate and kinase. The use of indirect links between kinases and their substrates enables non-obvious predictions that would be very difficult to spot by manually inspecting the available evidence.
Figure 1. Overview of the NetworKIN algorithm.
Phosphorylation sites determined experimentally (for example, by mass spectrometry) are mapped to a protein sequence (in this case Rad50). The kinase family likely to be responsible for phosphorylation of a site is predicted by consensus motifs that model the known sequence preferences of kinase catalytic domains (Hjerrild et al., 2004; Obenauer et al., 2003). Secondly, STRING is used to construct a context network for each substrate based on interaction and pathway databases, literature mining, mRNA expression studies and genomic co-occurrence evidence (von Mering et al., 2005). Within this network the nearest member of the relevant kinase family is identified for each phosphorylation site; for example, between members of the PIKK kinase family predicted by motifs, ATM is chosen over DNA-PK, as its path to Rad50 is shorter (see Experimental Procedures). However, a direct interaction between a kinase and a substrate is not a requirement, as illustrated by CK2A2 (CK2_α_′).
Benchmarking against known in vivo phosphorylation data
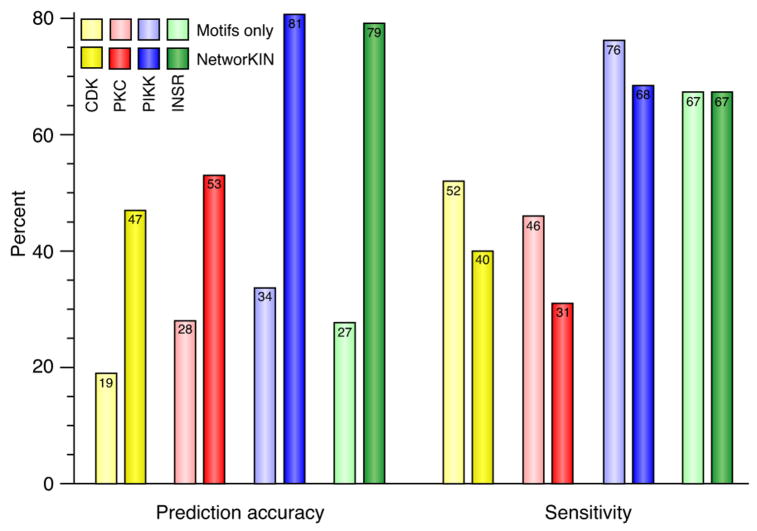
To test the method, we analysed the ability of NetworKIN to correctly predict which kinases are responsible for modifying each of 282 known in vivo phosphorylation sites from four well-studied kinase families (Figure 2, Figure S2, Table S1), and compared this with the results of purely motif-based methods (Hjerrild et al., 2004; Obenauer et al., 2003). We measured prediction accuracy (the fraction of predictions known to be correct), and sensitivity (the fraction of known sites that are correctly predicted). Using only consensus motifs we obtained a prediction accuracy of 25% (173 out of 693 predictions identified the correct kinase family) and a sensitivity of 61% (the correct kinase family was found for 176 of 282 sites). Although the kinase families used for benchmarking have by necessity been studied more than most kinases, the predictive power of the consensus sequence motifs for CDK, PKC, PIKK and INSR are representative for many other kinase families (Figure S1). By incorporating contextual information the prediction accuracy more than doubled to 64% (148/233 predictions) with only a slight drop in sensitivity to 52% (148/282 sites). Moreover, of the 148 correct kinase family predictions made when contextual information was included, the specific kinase gene product could be pinpointed in 72 cases (49%). In contrast, motif-based methods alone are generally not able to go beyond predicting the family of the responsible kinase (see Supplemental Experimental Procedures). These results highlight the importance of including contextual information in predicting kinase-substrate relationships.
Figure 2. Effects of including substrate context.
Manually curated datasets of CDK, PKC, PIKK and INSR in vivo phosphorylation sites were used to assess the prediction accuracy (the fraction of predictions that are known to be correct) and sensitivity (the fraction of known sites that are correctly predicted) of NetworKIN and solely motif-based methods (NetPhosK and Scansite, (see Supplemental Experimental Procedures)). This shows that including the cellular context (in the form of a protein association network) leads to a significant improvement in accuracy. Notably, the accuracy of NetworKIN predictions is likely to be an underestimate since not all the kinases that target each phosphorylation site in the set of test proteins may currently be known from experiments.
Context enhances cellular substrate specificity
The probabilistic networks around substrates permit us to estimate the degree to which context contributes to in vivo specificity from a computational perspective. Since sequence motifs alone give only 20% accuracy for well-characterised kinases (Figure 2), up to 80% of the ability to predict substrate specificity could come from contextual information. Conversely, the 2.5-fold increase in accuracy when including context (Figure 2) shows that the context must account for at least 60% ((0.64–0.25)/0.64 = 0.6) of the molecular information yielding precise predictions. By inference, the association of kinases and substrates in cells, for example by co-expression or through scaffolding proteins or regulatory subunits, plays a major role in delimiting the sites that are actually phosphorylated by kinase catalytic domains in vivo.
The human phosphorylation network
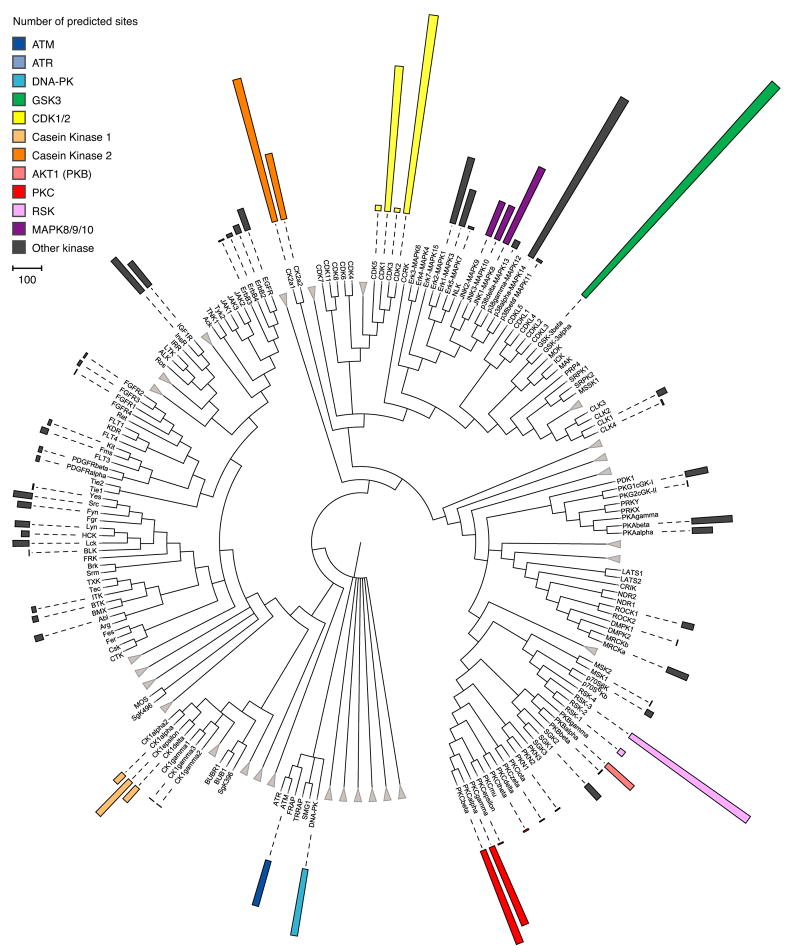
The algorithm currently covers 112 human kinases from 20 families (Figure 3). We applied it to the complete curated (see Supplemental Experimental Procedures) in vivo human phosphoproteome in Phospho.ELM (in total 7207 sites on 2540 proteins, including large-scale mass spectrometry data (Diella et al., 2004)). This resulted in a human phosphorylation network (HPN) consisting of 7143 site-specific kinase–substrate interactions (see Figure S3) between 1759 substrates and 68 kinases with predictions for 4488 phosphorylation sites (average 70 substrates per kinase; Figure S4). These data can be accessed at http://networkin.info and in Supplemental Data.
Figure 3. Number of predictions in the human kinome.
The human kinome consists of approximately 518 kinases (leafs) in a number of families (Manning et al., 2002). NetworKIN currently covers 20 of these families encompassing 112 individual kinases. Groups of kinases for which we do not have predictions are shown as collapsed branches (triangles). Using the complete Phospho.ELM database results in 7143 site-specific predicted kinase–substrate interactions (coloured bars indicate number of predicted phosphorylation sites) for 68 kinases. The figure was prepared with iTOL (Letunic and Bork, 2007)
A topological analysis suggests that networks predicted by NetworKIN are non-random in nature (see Figure S5 and Supplemental Experimental Procedures). The resulting clustering coefficients and topology coefficients indicate that the HPN bears more resemblance to physical protein-protein interaction networks (see Supplemental Data) than the context network used for prediction. This was even more evident if the same comparison was performed using only high-throughput data (data not shown). Transient interactions can be under-represented in high-throughput protein interaction screens (Aloy and Russell, 2002), thus one might not a priori expect the HPN and protein interaction networks to share topology. Further, the topology of the HPN does not simply reflect that of the context network on which it is based, which gives more confidence in the algorithm.
Co-localisation of kinases and their predicted substrates
Protein phosphorylation can control intracellular translocation and trafficking of proteins (Seet et al., 2006). Considerable progress has been made in determining subcellular localisation of proteins both on a systematic level (Foster et al., 2006) and individually. As the STRING database does not currently use localisation data for predicting protein associations, we can independently test if the computed phosphorylation networks are consistent with the localisation of the kinases and their predicted substrates.
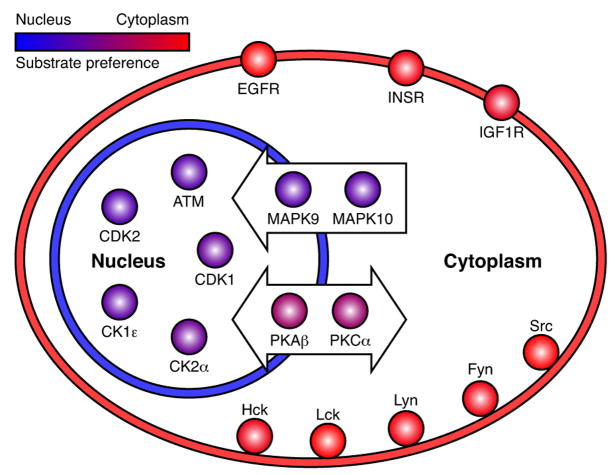
We mapped localisations from SwissProt to the HPN which resulted in 1005 phosphoproteins that are described as localising to either the cytoplasm or the nucleus (but not both). Since each of these proteins may contain multiple phosphorylation sites that are targeted by different kinases, NetworKIN predicted 2287 kinase–substrate interactions. Based on these interactions, we found 17 kinases that show a statistically significant preference for either cytoplasmic or nuclear substrates (Figure 4 and Table S5).
Figure 4. Subcellular localisation of kinases and their substrates.
A set of 960 cytoplasmic and 1327 nuclear phosphoproteins was extracted from the HPN based on subcellular localisation information from SwissProt. Among the predicted kinases we identified 17 kinases that showed a statistically significant preference for either cytoplasmic or nuclear substrates. These kinases were colour coded according to this preference and placed in the schematic figure according to their primary subcellular localisation; the kinases placed within the arrows are known to translocate to the nucleus upon activation or shuttle between the nucleus and the cytoplasm. Transmembrane and membrane-associated kinases are correctly predicted to selectively phosphorylate cytoplasmic substrates, whereas the kinases that are active only in the nucleus all show clear preference for nuclear substrates.
For membrane associated kinases (such as EGFR, INSR, and the SRC family) we almost exclusively predict cytoplasmic substrates. Although receptor tyrosine kinases (RTKs) can occasionally translocate to the nucleus, we predict very few nuclear substrates. However, we cannot exclude the possibility that the available phosphorylation data sets do not currently cover the cellular states where RTKs are active in the nucleus. In contrast, we find no kinases which are predicted to exclusively phosphorylate nuclear proteins. For the kinases that are primarily localised to the nucleus (ATM, CDK1, CDK2, CK1_ε_ and CK2_α_), we predict only 2–3 fold more nuclear than cytoplasmic targets (Table S5). There are at least three possible explanations for this: all nuclear kinases are synthesised in the cytosol and may phosphorylate cytosolic proteins prior to entering the nucleus, nuclear kinases may have access to cytosolic substrates during mitosis when the nuclear membrane is absent and finally many kinases may shuttle between the nucleus and the cytosol. This is exemplified by MAPK9 (Jnk2) and MAPK10 (Jnk1), which upon activation translocate from the cytosol to the nucleus or the perinuclear region (Mizukami et al., 1997; Whitmarsh et al., 2001). Consistent with this, we predict 3 fold more nuclear MAPK9/10 substrates than cytoplasmic ones. Conversely, PKA_β_ and PKC_α_ are both fairly pleiotropic kinases, which in the phosphorylation network show a weak preference for cytoplasmic substrates.
Despite the caveats of possible biases in the various datasets, the predicted kinase–substrate interactions are consistent with localisation data for the substrates and kinases. The cell membrane linked kinases show clear preference for cytoplasmic substrates, the predominantly nuclear kinases are biased towards nuclear substrates, and the kinases that shuttle between the cytosol and the nucleus exhibit a more even distribution of substrates.
Constructing a DNA damage response network
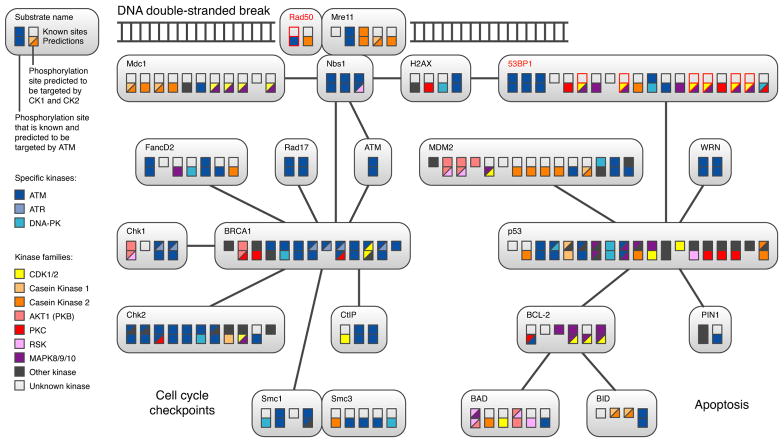
Using the HPN as a resource, we investigated protein phosphorylation within the DNA damage response (DDR) network of human cells (Figure 5 and Figure S6). An effective DNA damage response is critical for maintaining genomic integrity and preventing malignant transformation. The response to DNA double stranded breaks (DSBs) is primarily regulated by the kinase Ataxia-Telangectasia Mutated (ATM), a member of the PIKK family (Shiloh, 2006). Of the 45 sites in 16 known ATM substrates, we correctly predicted 39. In addition, we predicted 12 new ATM sites in DSB response and apoptosis related proteins (Figure 5), eight of which are found in six proteins not previously known to be ATM substrates. One of these novel predicted ATM substrates is Rad50, a component of the MRN complex that helps to stabilise broken chromosomes, and together with Nbs1 and Mre11, acts as a sensor of DNA damage, contributes to ATM recruitment to DSBs and activation of ATM by autophosphorylation (Shiloh, 2006). Serine 635 of Rad50 was shown to be phosphorylated in a large-scale mass spectrometry study (Beausoleil et al., 2004). The NetworKIN algorithm predicts it to be a potential ATM substrate site because it matches the consensus sequence of ATM and its relatives, and because the context network links Rad50 and ATM by primary experimental evidence, manually curated pathways and literature mining (Figure 1). The latter reflects the knowledge that ATM interacts with the MRN complex and co-purifies with Rad50 as part of the BASC super complex (Wang et al., 2000), a sensor of DNA damage.
Figure 5. Phosphorylation in the DNA damage response.
We modelled the primary DNA damage response and the apoptosis-related signalling by applying NetworKIN to in vivo phosphorylation sites (Diella et al., 2004). Only proteins that are known or predicted to be targeted by ATM are included. Boxes within each protein denote known phosphorylation sites, and are colour coded based on which kinases or kinase families are known (upper rows) or predicted (lower rows) to phosphorylate the site. In cases with multiple kinases predicted for a site, two kinases are shown as slashed boxes. A more comprehensive network (DDR+ subnetwork) containing additional proteins is included as an interaction map (Figure S6).
ATM kinase phosphorylates Rad50 in response to genotoxic stress
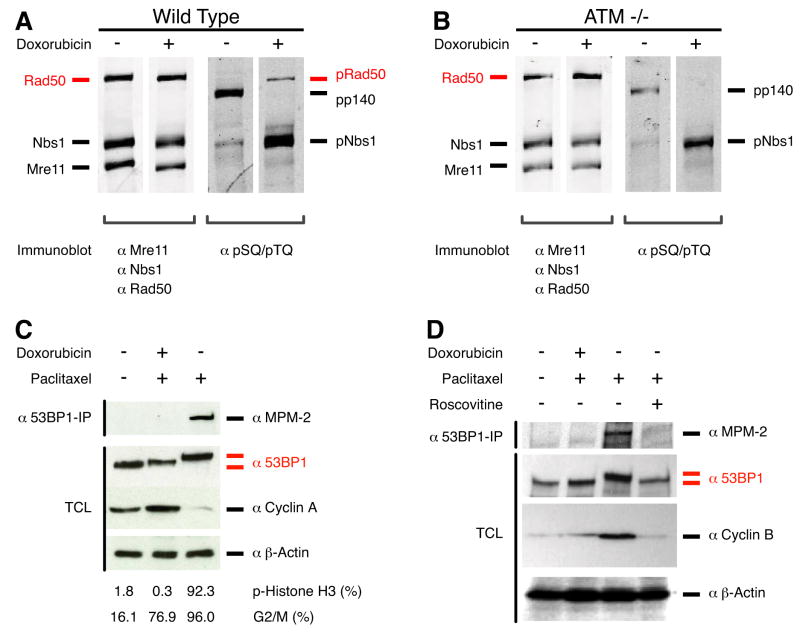
To test this prediction, we assayed the phosphorylation of Rad50 in response to DNA damage in wild type (ATM_wt_/wt, Figure 6A) and ATM null (ATM−/−, Figure 6B) lymphoblast cells using a phospho-S/T-Q-specific antibody which recognises the ATM phosphorylation sites. Treatment with the topoisomerase inhibitor doxorubicin induced the phosphorylation of Rad50 at the ATM-specific phosphorylation site in wild type cells but not in ATM null cells (Figure 6), which indicates that ATM is required for Rad50 phosphorylation induced by DNA damage as predicted. We also immunoprecipitated Rad50 from doxorubicin-treated wild type cells and analysed its phosphorylation by mass spectrometry. The MS/MS fragmentation spectra (Figure S7) confirmed the phosphorylation of Rad50 at the predicted ATM substrate site, S635, in agreement with published data (Beausoleil et al., 2004). Together, these data validate the effectiveness of the algorithm in predicting specific kinase–substrate relationships in vivo.
Figure 6. Phosphorylation of Rad50 and 53BP1.
A, Rad50 was immunoprecipitated from EBV-transformed human ATM_wt_/wt or B, ATM−/− lymphoblasts. The immunoprecipates were separated by SDS-PAGE and immunoblotted for Rad50 and co-associating proteins, Mre11 and Nbs1. These same immunoprecipitates were also probed with a phospho-S/T-Q specific antibody that recognises ATM/ATR motifs (two right panels). Rad50 was phosphorylated in the wild-type cells but not in the ATM null cells in response to DNA damage as predicted. Nbs1 phosphorylation was reduced, but not eliminated, in the ATM null cells, suggesting that other PIKK kinases are also active in these lymphoblast cell lines, but that these are not responsible for Rad50 phosphorylation. When probing with the phospho-S/T-Q specific antibody, the Nbs1 band is stronger than the Rad50 band due to the presence of three ATM sites in Nbs1 but only a single site in Rad50. An unidentified protein, p140 was recognised by the phospho-S/T-Q antibody. C,D Human osteosarcoma U2OS cells were left untreated or treated with paclitaxel/doxorubicin (G2/M checkpoint arrest), paclitaxel (mitotic arrest), or paclitaxel with the CDK-inhibitor Roscovitine. Subsequently, cells were harvested and analysed in parallel by immunoblotting and FACS (C panel only for FACS). Percentages of mitotic cells (phospho-Histone H3 staining) and G2/M cells (propidium iodide) as determined by FACS analyses are shown below panel C. Immunoblotting of total cell lysates (TCL) or 53BP1 immunoprecipitations was performed with antibodies indicated to the right of the panels (see Supplemental Experimental Procedures). Only 53BP1 immunoprecipitated from mitotic cells was recognised by the phospho-specific antibody MPM-2, indicating that CDK1 is responsible for 53BP1 phosporylation in vivo.
Multiple predicted ATM roles and targets
We predicted additional novel ATM targets including components of the NF-κ_B pathway (NF-κ_B2, IKK_β and NEMO) and regulators of apoptosis (Bcl-2, BAD and PIN1)(Figure 5) and the cell-cycle (Cdc25A). The predictions that ATM phosphorylates NF-κ_B-related proteins are far from obvious; in the context network, these proteins are only linked to ATM indirectly via two other proteins, namely I_κ_B_α and p53. While this manuscript was in preparation, it was demonstrated that ATM (located in the nucleus) indeed phosphorylates the site (S85) predicted by NetworKIN on the NEMO protein, and that following phosphorylation, both ATM and NEMO translocate to the cytosol (Wu et al., 2006). This intriguing observation provides some independent support for other predictions that ATM may phosphorylate cytosolic substrates (e.g. Cdc25A and IKK_β). ATM is known to contribute to cell-cycle arrest and to indirectly regulate apoptosis by activating the pro-apoptotic transcriptional programme of p53 and the anti-apoptotic transcriptional programme of NF-_κ_B (Rashi-Elkeles et al., 2006). We predict (Figure 5) that ATM contributes directly to the regulation of apoptosis by phosphorylating the pro-apoptotic protein BAD and the anti-apoptotic protein Bcl-2. Consistent with a role for ATM in apoptosis, ATM phosphorylates the pro-apoptotic Bcl-2 family member BID, which contributes to the S-phase checkpoint after DNA damage (Kamer et al., 2005; Zinkel et al., 2005). A direct role for ATM in apoptosis would constitute a significant expansion of ATMs regulatory activity. For an annotated list of ATM predictions covering 72 target proteins see Table S5.
The cell cycle-kinase CDK1 phosphorylates 53BP1 during mitosis
Although focused on ATM, Figure 5 also contains novel predictions for other kinases. For instance, we predict several kinases for the 53BP1 protein that has been shown to accumulate at DNA damage-induced foci and to be hyperphosphorlylated in an ATM-dependent manner (Anderson et al., 2001; Rappold et al., 2001; Schultz et al., 2000). Deletion of 53BP1 abrogates the G2 DNA damage checkpoint and compromises tumor suppression, classifying 53BP1 as a bona fide DNA damage checkpoint protein (Ward et al., 2003). DNA damage checkpoints inhibit cell-cycle progression by targeting the core proteins that drive cell-cycle progression; cyclin-dependent kinases (CDKs). Thus, the prediction of 53BP1 as a substrate for CDK1 and/or CDK2 (Figure 5) is interesting, as CDKs have previously been implicated in the DNA repair response by experiments showing that inhibition of CDKs in budding yeast results in defects in DNA damage induced arrest, and in homologous recombination repair pathways (Ira et al., 2004). To assess phosphorylation by CDKs, we analysed 53BP1 in human U2OS osteosarcoma cell cultures that had been subjected to stimuli, such as mitotic-arrest, that differentially modify the levels of CDK1 and CDK2 activity. As seen in Figure 6C, cells arrested in prometaphase with paclitaxel showed a slower migrating 53BP1, implying increased phosphorylation. These cells are inactive for CDK2 as reflected by the degradation of cyclin A, the most prominent cyclin partner of CDK2 in G2/M cells (Figure 6C, lane 3). To investigate whether cyclin B/CDK1 is responsible for mitotic 53BP1 phosphorylation, we employed the MPM-2 antibody, which recognises phosphorylated CDK1/2 consensus sites. 53BP1 immunoprecipitated from mitotic cells was recognised by MPM-2, indicating that CDK1 is responsible for 53BP1 phosporylation in vivo (Figure 6C and D, lane 3). Consistent with this view 53BP1 was phosphorylated by cyclin B/CDK1 in vitro (data not shown). To further investigate the involvement of CDK1 in mitotic phosphorylation of 53BP1, we utilised the CDK inhibitor roscovitine. Following treatment with roscovitine, cells exit from mitosis, as shown by the degradation of cyclin B (Figure 6D, lane 4). Incubation of mitotic cells with roscovitine efficiently reversed both the mobility shift and MPM-2 reactivity of 53BP1, indicating that CDK activity is required for the phosphorylation of 53BP1 that results in its altered mobility and recognition by the MPM-2 antibody (Figure 6D, lane 4). As controls, we analysed unsynchronised cells and cells that were prevented from entering mitosis by treatment with doxorubicin prior to paclitaxel to activate the G2/M-DNA damage checkpoint (Figure 6C,D lanes 1 and 2, respectively). In both these instances 53BP1 was not recognised by the anti-MPM2 antibody. The decreased mobility of 53PB1 in DNA checkpoint arrested cells is likely due to phosphorylation by the ATM kinase. These data indicate that 53BP1 is a substrate for CDK1 in mitotic cells as predicted by NetworKIN.
Additional potential cell-cycle regulation of the DDR network
Several other predictions in the DNA damage response network Figure 5 are of potential interest. For example, we predict that CDK1 and/or CDK2 phosphorylates CtIP(RBBP8), a potential tumor suppressor, on S327, suggesting that CtIP can be phosphorylated during M and S phase. CtIP was indeed recently shown to be involved in G2/M checkpoint control through BRCA1-dependent ubiquitination in response to S327 phosphorylation (Yu et al., 2006), and to counteract Rb-mediated G1 restraint (Chen et al., 2005). Similarly, we predict CDK1 and/or CDK2 to phosphorylate the SMC4 subunit of condensin, a protein complex which is known to be subject to cell cycle-dependent phosphorylation in both human cells and cells of other species (Takemoto et al., 2004). The complete set of predictions is available on the supplemental website (http://networkin.info) and in Supplemental Data.
Quantitative perturbations of phosphorylation networks
The examples provided by Rad50 and 53BP1 (Figure 6) illustrate the importance of reliable predictions, as even with such hypotheses in hand it takes considerable effort to validate individual in vivo kinase–substrate relationships using conventional techniques. An approach that, together with Networkin, could potentially accelerate this process is to perturb the activity of a specific kinase, for example by siRNA or selective inihibitors, and to monitor the phosphorylation of a predicted substrate by quantitative mass spectrometry. In particular a mass-spectrometric scan mode termed multiple reaction monitoring (MRM) is well suited to quantify the extent to which a given site is phosphorylated, by measuring specific fragment ions related to selected peptides (Anderson and Hunter, 2006; Ong and Mann, 2005; Schmelzle and White, 2006). MRM methods are increasingly being used to assay for phosphorylated peptides (Wolf-Yadlin et al., 2007).
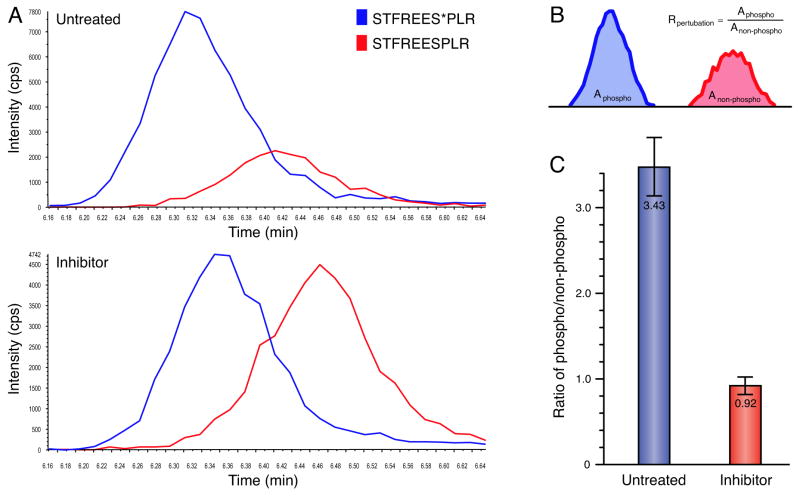
To test this approach, we have investigated the Bcl-2 interacting transcriptional repressor BCLAF1, which has previously been implicated in apoptosis and cancers (Kasof et al., 1999). BCLAF1 contains more than 30 phosphorylation sites identified by mass-spectrometry studies (Diella et al., 2004). Since little is known about the kinases that target this protein, we decided to track one of the several predicted GSK3_β_ sites by MRM (see Experimental Procedures). We employed an ABI/Sciex 4000QTRAP to assay the S531 phosphorylation site of BCLAF1 (Figure 7). The extracted ion currents indicate the relative amounts of phosphorylated/non-phosphorylated peptides in an untreated sample (Figure 7A, upper panel) or a sample treated with the GSK3 inhibitor lithium (Figure 7A, lower panel). In order to calculate the relative change in phosphorylation of S531 the peak integral was calculated (Figure 7B). The results based on two biological repeats are shown as the ratio of phospho/non-phospho peptide (Figure 7C), which indicates a 3.7-fold decrease upon lithium treatment. This observation, together with the NetworKIN prediction that GSK3_β_ directly phosphorylates BCLAF1, suggests that BCLAF1 is a novel target of this kinase. The approach presented here does not give definitive proof that a predicted kinases is the relevant in vivo enzyme, as it cannot rule out phosphorylation through another kinase. This problem will diminish as more kinase consensus motifs are added to the algorithm, as it will then be possible to use exclusion principles. However, given the prediction and observed perturbations we argue it is the most parsimonious explanation. Given the accuracy (Figure 2) of the NetworKIN method we think this is a reasonable approach to a large scale mapping of human phosphorylation networks.
Figure 7. Quantitative measurement of GSK3-dependent phosphorylation on BCLAF1.
A, Multiple reaction monitoring of S531 on BCLAF1. Human embryonic kidney (HEK) 293 cells were left untreated (upper panel) or treated (lower panel) with the GSK3 inhibitor lithium. Each curve (extracted ion currents) represent a MRM elution profile corresponding to the phosphorylated (blue, STFREEsPLR) and non-phosphorylated (red, STFREESPLR) peptides (see Experimental Procedures). B, The calculation of phosphorylation levels is given by the ratio of the integrated ion-currents. C, Treatment with the GSK3 inhibitor lithium results in a 3.7 fold decrease of phosphorylation of BCLAF1 at S531. The error bars show standard deviations.
Discussion
Context essential for modelling phosphorylation networks
The method presented here is designed to link experimentally identified phosphorylation sites to protein kinases. The algorithm relies on the fact that signalling proteins are modular, in the sense that they contain domains (catalytic or interaction) and linear motifs (phosphorylation or binding sites), which mediate interactions between proteins (Seet et al., 2006). It also exploits both the inherent propensity of kinase catalytic domains to phosphorylate particular sequence motifs, and contextual information regarding the physical association, co-occurrence in the genome and literature and co-expression of kinases and substrates. The improved predictive power gained from using context underlines the importance of extra-catalytic kinase–substrate interactions in the specificity of protein phosphorylation within cells. We would also suggest that this underlines the utility of network data in modelling molecular and cellular events.
Errors in phosphorylation networks
Although including contextual data markedly increases the accuracy of kinase–substrate relationship predictions, this method is obviously still prone to error. The HPN presented in this work offers insight into phosphorylation driven interaction networks and represents thousands of hypotheses, which can be further investigated. However, it is a predicted network and thus will contain errors, which may arise primarily three sources:
A fraction of experimentally determined phosphorylation sites will be false positives and some phosphorylation sites will have been missed, the latter resulting in incomplete networks. Identifying the position of phosphorylation sites within phospho-peptides identified with mass-spectroscopy is still a difficult problem and a source of error (Beausoleil et al., 2006). Whereas error rates can be determined in proteomics studies this is often not possible for low-throughput data. By inspecting the phosphorylation sites in the benchmark set that NetworKIN failed to predict, we were able to identify and correct the annotation in Phospho.ELM for 5 ATM and 5 DNA-PK sites, which had been reported in the literature, but which are likely phosphorylated by other kinases acting downstream of ATM and DNA-PK.
The consensus sequence motifs used for predicting kinase families can suffer from errors as well. The motifs may be biased relative to in vivo sites due to specific in vitro techniques. The lack of comprehensively determined in vivo substrate sites for many kinases means that the datasets available for training and testing motif predictors are limited, and thus the resulting predictors can be biased (Hjerrild et al., 2004). The false assignment of kinases to substrates due to indirect phosphorylation events can also lead to overly degenerate sequence motifs due to erroneous training data.
Finally, as we use a probabilistic association network to describe the context of kinases and substrates, errors in this network can lead to false couplings of kinases and substrates. However, this error rate is quantified and is easily controlled through benchmarking and weighting of the data (von Mering et al., 2005).
Coverage of the human phosphorylation network
It may seem contradictory that a method covering 22% (112/518) of the kinome should be able to make predictions for 62% (4488/7207) of all phosphorylation sites. An obvious explanation would be that NetworKIN overpredicts; however, this is not the case since the prediction accuracy is higher than the sensitivity (64% vs. 52%) and the method thus slightly underpredicts. The reason may rather be that most of the pleiotropic kinase families, which are responsible for the phosphorylation of a very large number of sites, are covered by the method, whereas the missing kinases tend to be more specific (or selectively expressed) and thus account for the phosphorylation of relatively few sites. Moreover, many sites are likely phosphorylated by multiple kinases in vivo (Bullock et al., 2005), which further increases the likelihood that at least one kinase for any given site is currently included in the algorithm.
Perspectives
Despite these potential sources of errors, combining multiple data types (i.e. consensus motifs, phosphorylation sites and association networks) is essential for constructing phosphorylation networks and is, as we show, also sufficiently accurate to allow meaningful theoretical and experimental investigations. Moreover, as experimental approaches such as MS-based phosphoproteomics improve (Olsen et al., 2006), these errors will continuously be diminished.
As the backlog of phosphorylation sites is reduced, and sites are entered into the Phospho.ELM database, the basis for deriving new and more accurate motifs will be improved accordingly. New techniques for determining kinase specificity are also improving, allowing the coverage of consensus motifs to be extended to the whole kinome (Hutti et al., 2004). Finally, as more data on protein interactions are generated and integrated with expression and other data in the STRING database this will result in more accurate association networks, improving the contextual information employed by the NetworKIN algorithm.
The availability of new data will also help extend the algorithm to include other post-translational modifications (for example, acetylation or methylation) to their relevant enzymes, and incorporate information regarding docking motifs and modification-dependent interaction modules (for example, Bromo and SH2 domains), thus enabling the construction of entire event-based signalling networks.
Our results clearly indicate that kinases and their substrates form complex and dynamic interaction networks. As we learn more about network mediated kinase specificity one can envision deployment of mixtures of kinase inhibitors to target the network rather than the individual kinases, for example for therapeutic purposes. Accurate and systematic pairing of post-translational modifications with the enzymes responsible for marking specific sites will ultimately provide critical information on the dynamics of signal propagation and processing in complex biological systems.
Experimental Procedures
The NetworKIN algorithm
Given a phosphorylation site, a set of possible kinase families is predicted using NetPhosK and Scansite (see below), and the set of candidate kinase families is given by a BLAST search in a database of kinase domains. The best candidate kinases within these families are identified from a protein network of functional associations (generated using the STRING database (von Mering et al., 2005)) by calculating the proximity to the substrate for all kinases, defined as the probability of the most probable path connecting them (Floyd-Warshall algorithm). Because it is not always possible, or even meaningful, to predict a single kinase for a site, all kinases with a functional association probability up to 0.01 worse than the best scoring one are suggested as candidates. The algorithm is implemented in ANSI-C and Python.
Integration of contextual data
To capture the biological context of a substrate, we use a probabilistic network of functional associations extracted from the STRING database (von Mering et al., 2005) (http://string.embl.de). This network is based on four fundamentally different types of evidence: genomic context (gene fusion, gene neighbourhood, and phylogentic profiles), primary experimental evidence (physical protein interactions and gene co-expression), manually curated pathway databases, and automatic literature mining. Table S3 shows that the three latter evidence types are of comparable importance, whereas genomic context methods contribute very little towards the predictions made by NetworKIN. As the curated pathway databases generally contain few errors, a confidence score of 0.9 is assigned to this type of evidence.
Physical protein interactions play the dominant role among the primary experimental data, whereas gene co-expression contributes only very little. Physical protein interactions were imported and merged from numerous repositories, and the reliability of each individual interaction was assessed based on the promiscuity of the interaction partners using a scoring schemes described elsewhere (von Mering et al., 2005). Gene co-expression was measured by calculating the Pearson correlation coefficient between two genes across all datasets in the GEO repository for the organism in question.
Automatic literature mining plays an important role as the majority of the accumulated knowledge on molecular biology is only available in the form of scientific papers. We thus extracted relations between proteins based on Medline abstracts, OMIM monographs, and gene summary paragraphs from model organism databases. To identify gene and protein names in these texts, a comprehensive synonyms list was compiled for each organism based on Ensembl, SwissProt, and various model organism databases. The vast majority of relations are extracted using a statistical co-occurrence method that counts the number of abstracts in which two proteins co-occur and compares this to the random expectation given how often each protein occurs. To obtain a high score, two proteins must thus repeatedly be mentioned together in the scientific literature. In addition, we extracted specific types of relations such as physical protein interactions using a natural language processing pipeline described elsewhere (Saric et al., 2006).
As a functional association implies that two proteins function in the same process, all the scoring schemes for all evidence types were benchmarked and calibrated on metabolic and signaling pathways from the KEGG database (Kanehisa et al., 2006), resulting in probabilistic scores for all evidence types. Since these scores are directly comparable and specify the reliability of each piece of evidence, no weighting scheme for different evidence types is needed. We subsequently transferred associations to orthologous proteins in other organisms, and combined multiple lines of evidence for a given binary association into a single confidence score using a Bayesian scoring scheme (von Mering et al., 2005).
Detection of Rad50 phosphorylation
EBV-transformed lymphoblasts from ATM_wt_/wt and ATM−/− individuals were treated with doxorubicin, lysed and immunoprecipitated proteins were isolated using SDS-PAGE and visualised by immunoblot analysis (see Supplemental Experimental Procedures). For mass spectrometry analysis, the experiment was repeated and the immunoprecipate was separated by SDS-PAGE. Excised gel bands containing Rad50 protein were digested with Trypsin (Promega, Madison, WI) according to the protocol described in (Houthaeve et al., 1995). After lyophilisation, tryptic peptides were analysed by liquid chromatography-mass spectrometry (see Supplemental Experimental Procedures).
Quantification of BCLAF1 phosphorylation
Human embryonic kidney (HEK) 293 cells were treated with 20 mM LiCl prior to cell lysis. BCLAF1 was immunoprecipated from the cell lysate and the immunoprecipitate was separated by SDS-PAGE (see Supplemental Experimental Procedures). Gel bands were excised and trypsinated as previously described (see Supplemental Experimental Procedures). The tryptic peptides were analysed by liquid chromatography-mass spectrometry utilizing an ABI/Sciex Tempo 1Dplus LC (Applied Biosystems, Foster City, California) into an ABI/Sciex QSTAR Elite mass spectrometer (ABI/Sciex, Foster City, CA). One clearly identified peptide containing a predicted GSK3_β_ site, STFREEsPLR was selected for further quantitative assay. Briefly, a targeted MRM analysis of the selected S531 peptide STFREESPLR and the corresponding phosphopeptide (STFREEsPLR) was performed using the most predominant product ion fragment identified from the full scan MS/MS spectra (data not shown). An MRM assay for the parent ion masses and proline promoted fragment ions of the unmodified (b7) and phosphorylated (b7–98) peptide was performed using an ABI/Sciex Tempo 1Dplus LC into an ABI/Sciex 4000QTRAP. The extracted ion currents (XIC) of the BCLAF1 peptides were integrated using Analyst 2.0 software (ABI/Sciex) and the ratio of phosphorylated/non-phosphorylated peptide amounts were calculated and compared to the same ratio calculated after inhibition of GSK3 with LiCl (Figure 7). The experiment was repeated twice on different days on different cell populations.
Supplementary Material
01
Acknowledgments
Thanks to Sara Quirk, Jeff Wrana, John Scott and Kresten Lindorff-Larsen for commenting on this manuscript. We are further indebted to Giselle Wiggin, Rizaldy Scott, Ivica Letunic, Jennifer Logan, Christian von Mering, Stephen A. Tate (ABI/Sciex) and Andrei Starostine for technical help and advice. This project was supported by the BioSapiens Network of Excellence (LSHG-CT-2003-503265), the EMBRACE project (LHSG-CT-2004-512092) and the GeneFun project (LSHG-CT-2004-503567) all funded by the European Commision FP6 Programme, the Danish Research Council for the Natural Sciences, the Lundbeck Foundation, Genome Canada through Ontario Genomics Institute and the NIH Integrative Cancer Biology Program grant U54-CA112967-03. R.L. is a Human Frontier Science Program Fellow. M.V.V. is the recipient of a VENI grant from the Dutch Organisation for Scientic Research. G.J.O. is the recipient of a Womens Excalibur Postdoctoral Fellowship (PF-06-094-01-CCG) from the American Cancer Society New England Division. R.L., L.J.J. and F.D. conceived and designed the computational strategy. L.J.J. and R.L. implemented the algorithm. F.D. curated the MS datasets. R.B.R. collected domain sets. R.L., G.J.O., K.C., L.T., C.J., M.V.V., J.R.W., J.J., P.M., T.P. and M.B.Y. conceived and designed the experiments. G.J.O., V.N., L.T., I.M.M., C.J., M.V.V., J.J., R.L., K.E., K.C. and P.M. performed the experiments. L.D.S. contributed reagents. R.L., A.P. and J.G.P. developed the website. R.L., L.J.J., G.J.O., M.V.V., K.C., R.B.R., P.B., M.B.Y. and T.P. wrote the paper.
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Aloy P, Russell RB. The third dimension for protein interactions and complexes. Trends Biochem Sci. 2002;27:633–8. doi: 10.1016/s0968-0004(02)02204-1. [DOI] [PubMed] [Google Scholar]
- Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villén J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Reményi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- Bullock AN, Debreczeni J, Amos AL, Knapp S, Turk BE. Structure and substrate specificity of the Pim-1 kinase. J Biol Chem. 2005;280:41675–41682. doi: 10.1074/jbc.M510711200. [DOI] [PubMed] [Google Scholar]
- Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EYHP, Lee WH. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Diella F, Cameron S, Gemünd C, Linding R, Via A, Kuster B, Sicheritz-Pontén T, Blom N, Gibson TJ. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Hjerrild M, Stensballe A, Rasmussen TE, Kofoed CB, Blom N, Sicheritz-Ponten T, Larsén MR, Brunak S, Jensen ON, Gammeltoft S. Identification of phosphorylation sites in protein kinase A substrates using artificial neural networks and mass spectrometry. J Proteome Res. 2004;3:426–33. doi: 10.1021/pr0341033. [DOI] [PubMed] [Google Scholar]
- Houthaeve T, Gausepohl H, Mann M, Ashman K. Automation of micro-preparation and enzymatic cleavage of gel electrophoretically separated proteins. FEBS Lett. 1995;376:91–94. doi: 10.1016/0014-5793(95)01242-7. [DOI] [PubMed] [Google Scholar]
- Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L, Haimovich G, Lerenthal Y, Marcellus RC, Gross A. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell. 2005;122:593–603. doi: 10.1016/j.cell.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasof GM, Goyal L, White E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol. 1999;19:4390–4404. doi: 10.1128/mcb.19.6.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AEH, Yaffe MB. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell. 2005;17:37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Yoshioka K, Morimoto S, Yoshida K. A novel mechanism of JNK1 activation. nuclear translocation and activation of JNK1 during ischemia and reperfusion. J Biol Chem. 1997;272:16657–16662. doi: 10.1074/jbc.272.26.16657. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–41. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemünd C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DMA, Ausiello G, Brannetti B, Costantini A, et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–30. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashi-Elkeles S, Elkon R, Weizman N, Linhart C, Amariglio N, Sternberg G, Rechavi G, Barzilai A, Shamir R, Shiloh Y. Parallel induction of ATM-dependent pro- and antiapoptotic signals in response to ionizing radiation in murine lymphoid tissue. Oncogene. 2006;25:1584–1592. doi: 10.1038/sj.onc.1209189. [DOI] [PubMed] [Google Scholar]
- Reményi A, Good MC, Bhattacharyya RP, Lim WA. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol Cell. 2005;20:951–962. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from medline. Bioinformatics. 2006;22:645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- Schmelzle K, White FM. Phosphoproteomic approaches to elucidate cellular signaling networks. Curr Opin Biotechnol. 2006;17:406–414. doi: 10.1016/j.copbio.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to dna double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Takemoto A, Kimura K, Yokoyama S, Hanaoka F. Cell cycle-dependent phosphorylation, nuclear localization, and activation of human condensin. J Biol Chem. 2004;279:4551–4559. doi: 10.1074/jbc.M310925200. [DOI] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res . 2005;33(Database Issue):D433–7. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh AJ, Kuan CY, Kennedy NJ, Kelkar N, Haydar TF, Mordes JP, Appel M, Rossini AA, Jones SN, Flavell RA, et al. Requirement of the JIP1 scaffold protein for stress-induced JNK activation. Genes Dev. 2001;15:2421–2432. doi: 10.1101/gad.922801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer SJ. A role for proapoptotic bid in the DNA-damage response. Cell. 2005;122:579–591. doi: 10.1016/j.cell.2005.06.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01