igr Genes and Mycobacterium tuberculosis Cholesterol Metabolism (original) (raw)
Abstract
Recently, cholesterol was identified as a physiologically important nutrient for Mycobacterium tuberculosis survival in chronically infected mice. However, it remained unclear precisely when cholesterol is available to the bacterium and what additional bacterial functions are required for its metabolism. Here, we show that the igr locus, which we previously found to be essential for intracellular growth and virulence of M. tuberculosis, is required for cholesterol metabolism. While igr_-deficient strains grow identically to the wild type in the presence of short- and long-chain fatty acids, the growth of these bacteria is completely inhibited in the presence of cholesterol. Interestingly, this mutant is still able to respire under cholesterol-dependent growth inhibition, suggesting that the bacteria can metabolize other carbon sources during cholesterol toxicity. Consistent with this hypothesis, we found that the growth-inhibitory effect of cholesterol in vitro depends on cholesterol import, as mutation of the mce4 sterol uptake system partially suppresses this effect. In addition, the Δ_igr mutant growth defect during the early phase of disease is completely suppressed by mutating mce4, implicating cholesterol intoxication as the primary mechanism of attenuation. We conclude that M. tuberculosis metabolizes cholesterol throughout infection.
Mycobacterium tuberculosis is an exquisitely adapted human pathogen that infects roughly a third of the world's population. Like many other intracellular parasites, M. tuberculosis resides largely within a modified phagosomal compartment of the host macrophage during infection. While there are obvious advantages to this intracellular life-style, survival and replication within a host-derived membrane also pose several particular challenges. Perhaps the most fundamental of these is the need to acquire nutrients, for example, carbon, from the host cell. While all vacuolar pathogens face this challenge, it remains unclear which carbon sources are readily available and how they are extracted from the cell.
Host lipids have long been implicated as nutrient sources for M. tuberculosis during intracellular growth and chronic infection (4, 13). This was first suggested by the observation that fatty acids but not carbohydrates stimulate respiration of M. tuberculosis isolated from the mouse lung (1). Subsequently, sequencing of the M. tuberculosis genome revealed at least 250 genes potentially involved in lipid metabolism (4). Many of these genes are transcriptionally induced during intracellular growth, and a few are known to be required for infection (2, 3, 11, 14, 18). However, the complexity of M. tuberculosis lipid metabolism has made it difficult to determine if any individual gene is genuinely required for host lipid catabolism, as opposed to the synthesis or modification of an endogenous bacterial lipid.
We previously identified a region of the M. tuberculosis genome (Rv3545c to Rv3540c) that is important for growth in macrophages and in mice (3) and therefore was renamed igr for _i_ntracellular _gr_owth. A number of other studies have also indicated that these genes are important in vivo (16-18). The convergence of these studies on this single locus highlights its potential importance, yet little was known about the mechanism by which these genes contribute to mycobacterial survival in the host. The igr locus consists of a single operon containing genes for a putative cytochrome P450 (igrA), two acyl coenzyme A dehydrogenases (igrBC), a conserved hypothetical protein with a hot dog domain (igrD), a putative enoyl coenzyme A hydratase with a hot dog domain (igrE), and a lipid carrier protein (igrF). Based on these homologies, we predicted that the igr locus was involved in lipid metabolism, but its precise function remained unclear.
Recently, cholesterol has been implicated as significant for M. tuberculosis during chronic infection. This was first hypothesized based on synteny between a cholesterol-catabolic locus of Rhodococcus strain RHA1, an environmental relative of M. tuberculosis, and a region of the mycobacterial chromosome known to be critical for bacterial growth in vivo (19). Included in this region is the mce4 operon, which encodes an ABC-like transporter that represents the major cholesterol uptake system of the bacterium (12, 15). Mutants lacking the ability to import cholesterol have a specific defect in survival during the chronic phase of infection in mice and in the gamma interferon-activated macrophages that characterize this stage of disease (15). Thus, cholesterol appears to be an important nutrient during chronic M. tuberculosis infection; however, it remains unclear if cholesterol is a constituent of the mycobacterial diet throughout infection and what other nutrients might be available at earlier time points.
Several lines of evidence indicated the igr genes might be important for cholesterol metabolism. First of all, a homologous operon is present in Rhodococcus strain RHA1 and is transcriptionally induced during growth on cholesterol (19). In addition, a genome-wide genetic interaction screen predicted a functional association between the Mce4 transporter and products of the igr genes (8). Finally, the predicted functions of several of these genes are consistent with a role in degrading either the side chain or sterol rings of the cholesterol molecule (19).
In this work, we demonstrate that the H37Rv:Δ_igr_ mutant is unable to grow in the presence of cholesterol, although ATP generation and electron transport are unaffected. Thus, cholesterol addition appears to inhibit the growth of the mutant, likely via the accumulation of a stable catabolic intermediate. This hypothesis is supported by the ability of a mce4 mutation to reverse this toxicity, presumably by preventing the uptake of cholesterol. This effect is even more dramatic in vivo, where the Δ_mce4_ mutation completely suppresses the Δ_igr_-encoded phenotype. Detailed analysis of the replication rate of the Δ_igr_ mutant in vivo confirmed that this strain divides more slowly than the wild type during infection. However, in contrast to previous studies that monitored only CFU counts, we show here that the slow replication rate of this strain remains constant throughout infection and results in a reduced cumulative bacterial burden. Taken together, the results of this work demonstrate that the igr genes are essential for growth on cholesterol as a carbon source and that cholesterol is metabolized by M. tuberculosis throughout infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Mycobacteria were grown in Middlebrook 7H9 medium containing 0.05% Tween 80 and albumin-dextrose-catalase supplement (Becton Dickinson) or on Middlebrook 7H10 agar supplemented with 0.06% oleic acid and albumin-dextrose-catalase at 37°C, unless otherwise indicated. Kanamycin and hygromycin were used at 33 and 50 μg/ml, respectively. Growth on cholesterol was assessed by using a minimal medium (MM) first described in reference 19. The MM used contains, per liter, 0.5 g asparagine, 1 g KH2PO4, 1.5 g Na2HPO4, 10 mg MgSO4 · 7H2O, 0.5 mg CaCl2, 0.1 mg ZnSO4, 50 mg ferric ammonium citrate, and 1 ml 1:1 (vol/vol) tyloxapol-ethanol, pH 7.2. A 100 mM cholesterol stock was obtained by dissolving the sterol in 1:1 (vol/vol) tyloxapol-ethanol with vortexing, followed by heating for 30 min at 65°C. For growth curves, log-phase bacteria were washed with MM and diluted to an optical density at 600 nm (OD600) of 0.1. Cholesterol-containing media were prepared by diluting a freshly prepared 100 mM stock to 0.2 mM in MM. Equal volumes of diluted bacteria and 0.2 mM cholesterol medium were mixed to obtain a final concentration of 0.1 mM cholesterol at an OD600 of ∼0.05. Glycerol or dextrose was added to MM to a final concentration of 0.1%, resulting in MM+CG. Growth was assessed by monitoring OD600.
Genetic manipulation of strains.
Strain H37Rv:Δ_igr_Δ_mce4_ was generated by a phage-mediated allelic exchange in which yrbE4a, the first gene of the mce4 operon, was replaced with a hygromycin resistance gene (8). Briefly, ∼1.5e10 mid-log-phase cells were washed with MP phage buffer and incubated with 10-fold excess phage for 3 h at 37°C. The cell-phage mixture was pelleted, resuspended in 7H9, plated on 7H10 plus hygromycin, and returned to the incubator for 3 to 4 weeks. Putative double mutants were screened by PCR for the absence of yrbE4a (sense, 5′-TTGATCCAACAACTTGCG; antisense, 5′-AATGGACACCAGCAACGTC) and confirmed by Southern blotting (data not shown).
Unstable plasmid pBP10 was electroporated into strain H37Rv:Δ_igr_. Transformants were selected on 7H10 plus kanamycin and maintained in the presence of this antibiotic until the start of each experiment.
Metabolic assays.
Mycobacteria were grown in Middlebrook 7H9, MM, MM supplemented with glycerol (MM+G), or MM+CG. Log-phase bacteria were washed with MM and diluted to an OD600 of 0.1. Bacteria were then aliquoted into 12- or 24-well plates with the appropriate medium to a final starting OD600 of 0.05. After 3 days, OD600 was measured with a FluoStar Omega (BMG LABTECH). ATP levels were measured with BactiterGlo (Promega) in accordance with the manufacturer's instructions. Briefly, 25 μl of BactiterGlo reagent was added to 25 μl of cell culture and incubated in the dark for 15 min in a 96-well plate (LUMITRAC 200; Greiner). Luminescence was then read by the FluoStar Omega. Relative luminescence units (RLU) represent levels of ATP present in cell culture. For the number of RLU per unit of OD600, the number of RLU was divided by the OD600 of that sample. alamarBlue was added as 10% of the bacterial culture volume after 3 days of growth in appropriate medium and incubated in 96-well plates for 48 h before fluorescence was read at 544/590 nm.
Macrophage infections.
THP-1 infections were carried out as previously described (10). Briefly, for each infection, bacterial stocks were grown to mid-log phase in 7H9 medium, diluted in warm RPMI 1640 medium plus 10% fetal calf serum, and added to each well of a 24-well plate containing 5 × 105 cells at a multiplicity of infection of 1. Cells were incubated at 37°C in 5% CO2. After 4 h, extracellular bacteria were removed by three washes with warm RPMI medium and the cells received fresh medium (RPMI 1640 medium plus 10% fetal calf serum). At each time point, triplicate wells were processed for each bacterial strain. alamarBlue reagent at 10% in RPMI 1640 medium was added to the wells, and after 6 h of incubation, a sample was transferred to a 96-well plate and fluorescence was measured at 544/590 nm.
Mouse infections.
Mice were infected with pBP10-carrying bacteria as described previously (3). Briefly, female C57BL/6 mice (Jackson Laboratories) were housed in a biosafety level 3 animal facility according to University of Washington Institutional Animal Care and Use Committee protocols. For the infection, a frozen stock was thawed, sonicated, diluted to ∼10e07 CFU/ml, and then nebulized with an inhalation exposure system (Glas-Col). The infectious dose and initial plasmid carriage were determined by plating lung homogenates on both 7H10 and 7H10 plus kanamycin. At each time point, five mice per group were killed and their lungs were homogenized and plated as serial dilutions. CFU counts were assessed after incubation at 37°C for 2 to 5 weeks. r, δ, and cumulative bacterial burden values were derived by using a recently developed mathematical model (6; see the supplemental material).
For mouse experiments involving the Δ_igr_ Δ_mce4_ mutant, C57BL/6 mice were infected with strain H37Rv or Δ_igr_ or Δ_igr_ Δ_mce4_ mutant bacteria. Log-phase cultures of different strains were washed twice and resuspended in phosphate-buffered saline (PBS)-0.05% Tween 80 buffer. The cultures were then sonicated for 90 s in a cup horn Sonifier (Branson Ultrasonics Corporation), and the mice were infected with a Glas-Col inhalation exposure system. At the indicated time points, groups of four mice were killed, their lungs were homogenized in PBS-0.05% Tween 80, and dilutions were plated on 7H10 agar to enumerate CFU.
Cholesterol catabolism assays.
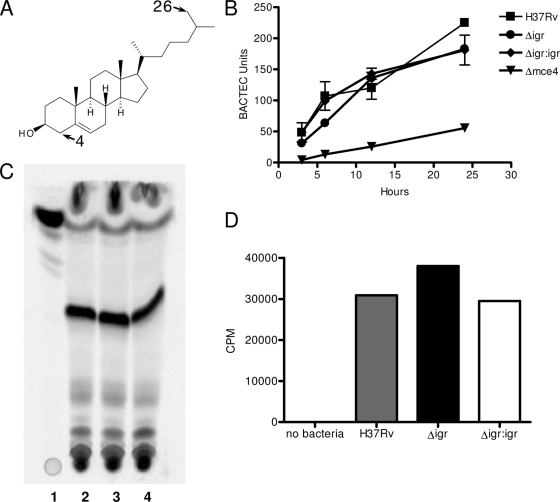
A-ring degradation of cholesterol was assayed by quantifying the release of 14CO2 after growing the strains in [4-14C]cholesterol while the degradation of the side chain was analyzed by quantifying the amount of 14C incorporated into the lipid fraction of different strains grown in [26-14C]cholesterol (Fig. 3A). Log-phase cultures of M. tuberculosis strains grown in 7H9 medium were washed twice with PBS and resuspended in MM (0.5 g/liter asparagine, 1 g/liter KH2PO4, 2.5 g/liter Na2HPO4, 50 mg/liter ferric ammonium citrate, 0.5 g/liter MgSO4 · 7H2O, 0.5 mg/liter CaCl2, 0.1 mg/liter ZnSO4) containing 0.1 μCi/ml [4-14C]cholesterol or [26-14C]cholesterol to a final OD600 of 0.5. To analyze ring degradation, 5-ml volumes of the cultures resuspended in [4-14C]cholesterol were transferred into sealed vials and incubated at 37°C. Release of 14CO2 was quantified by using a BACTEC TB-460 Instrument at various time points.
FIG. 3.
H37Rv:Δ_igr_ can import and partially degrade cholesterol. (A) The cholesterol molecule depicting C-4 and C-26. (B) CO2 production measured by BACTEC for H37Rv and the Δ_igr_, Δ_igr_:igr, and Δ_mce4_ mutant strains when incubated with 14C-4-radiolabeled cholesterol. (C, D) Total lipid fractions measuring incorporation of 14C-26-radiolabeled cholesterol by TLC (C) and counts per minute (D). (C) Lane 1, 14C-26-labeled cholesterol; lane 2, H37Rv; lane 3, Δ_igr_ mutant; lane 4, Δ_igr_:igr mutant. Major band is sulfolipid 1 (identified using a purified standard and mass spectrometry by S. Gillmore).
In order to analyze the degradation of the side chain, 10-ml volumes of the cultures resuspended in [26-14C]cholesterol were further incubated for 24 h at 37°C. Cell pellets were then washed twice and extracted with chloroform-methanol (2:1) for the total lipids. Total radiolabeled lipids were quantified by using a scintillation counter.
For thin-layer chromatography (TLC), the extracts were then dried under nitrogen and resuspended in the same solvent. Total lipid was resolved by TLC with glass-backed 250-μm-thick silica gel plates (Whatman) and single development of chloroform-methanol-water (60:12:1). The total lipid on the TLC plates was than detected with a PhosphorImager.
RESULTS
H37Rv:Δ_igr_ cholesterol metabolism.
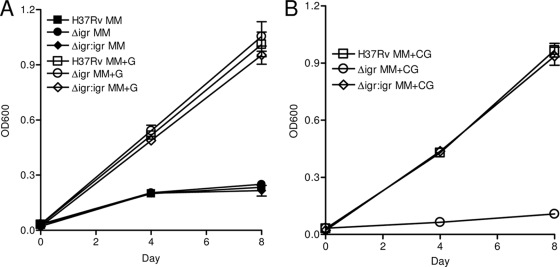
Deletion of the igr locus did not affect M. tuberculosis growth on a myriad of carbon sources, including pyruvate, valerate, isovalerate, proprionate, palmitate, dodecanoate, glycerol, Tween, or dextrose (3). Because an orthologue of the igr locus is conserved in the Rhodococcus cholesterol-catabolic region, we hypothesized that these genes might be involved in cholesterol utilization by M. tuberculosis. To investigate this, log-phase H37Rv and H37Rv:Δ_igr_ bacteria were inoculated into MM supplemented with various carbon sources. The two strains grew identically in MM or MM+G (Fig. 1A). Wild-type growth was nearly identical after the addition of 0.1 mM cholesterol (MM+CG) (Fig. 1A and B). In striking contrast, strain H37Rv:Δ_igr_ was unable to grow in cholesterol-containing medium (Fig. 1B). Examination of CFU counts over time demonstrated a less-than-twofold change in the number of viable bacteria, suggesting that cholesterol is inhibitory but does not kill H37Rv:Δ_igr_ under these conditions (data not shown).
FIG. 1.
H37Rv:Δ_igr_ is unable to grow in the presence of cholesterol. Growth of H37Rv (▪), H37Rv:Δ_igr_ (•), and the complemented H37Rv:Δ_igr_:igr strain (⧫) in MM (filled symbols in panel A) or in MM+G or MM+CG (open symbols in panels A and B, respectively). The data shown are the mean of triplicate cultures ± the standard deviation. For all panels, the data shown are from one of at least two experiments.
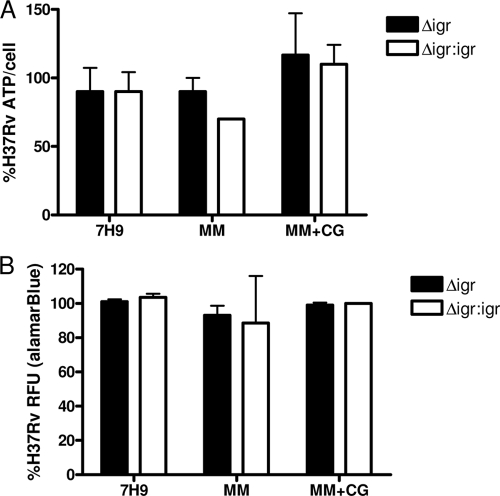
No change in ATP production and alamarBlue reduction by H37Rv:Δ_igr_ when exposed to cholesterol.
To understand better why deletion of the igr region causes growth arrest in medium containing cholesterol but not other carbon sources, the intracellular ATP concentrations of mutant and wild-type bacteria were compared during growth in various media. Surprisingly, the Δ_igr_ mutant showed no defect in ATP production in the presence of cholesterol (Fig. 2A). We observed the same phenomenon when measuring the metabolic activity of these strains by using alamarBlue, a dye responsive to electron transport (20). While there was a marked decrease in the growth of H37Rv:Δ_igr_ exposed to cholesterol (Fig. 1B), levels of alamarBlue reduction equaled those of wild-type cells (Fig. 2B).
FIG. 2.
H37Rv:Δ_igr_ is not attenuated for ATP production or alamarBlue reduction in the presence of cholesterol. (A) Amount of ATP per cell as a percentage of H37Rv after 3 days in 7H9, MM, or MM+CG. (B) alamarBlue reduction as a percentage of H37Rv after 48 h. Representative data from one of three experiments are shown as the mean of triplicate cultures ± the standard deviation.
H37Rv:Δ_igr_ can import and partially degrade cholesterol.
Because ATP production and electron transport were not diminished in the H37Rv:Δ_igr_ mutant after exposure to cholesterol, we hypothesized that deletion of the igr locus resulted in the accumulation of a cholesterol catabolite that is toxic to M. tuberculosis. This hypothesis requires that cholesterol be imported and at least partially degraded. To test this, we assessed the ability of the mutant to perform the initial steps of cholesterol catabolism. Cholesterol degradation occurs from both the A-sterol ring and the end of the side chain (Fig. 3A) (15). Previous work has shown that degradation of the A ring can be quantified by the conversion of 14C at the 4 position of cholesterol to 14CO2, and the initiation of side chain degradation can be monitored by the incorporation of 14C at the 26 position into branched-chain lipids (15). Upon incubation with these specifically labeled cholesterol species, we found no difference between the wild-type and H37Rv:Δ_igr_ strains in the ability to convert either the C-4 carbon to CO2 or the C-26 carbon into mycobacterial lipid (Fig. 3B to D). Thus, H37Rv:Δ_igr_ is able to import cholesterol and perform at least the initial steps of degradation, consistent with the hypothesis that a toxic intermediate accumulates in the mutant and is responsible for the growth-inhibitory effect of cholesterol.
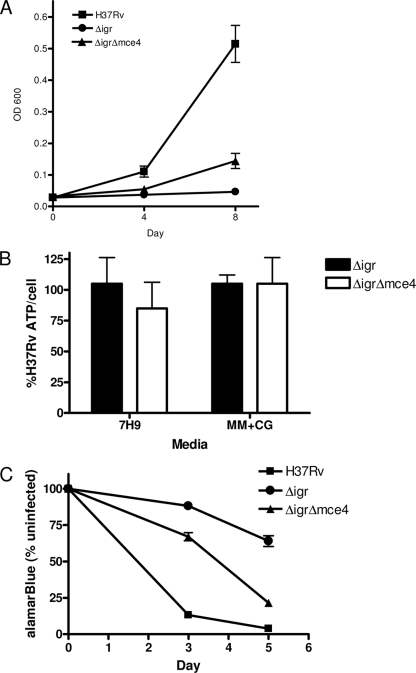
Suppression of H37Rv:Δ_igr_ phenotype by Mce4 disruption.
If the H37Rv:Δ_igr_ growth defect on cholesterol was due to the production of an inhibitory catabolite, we predicted that disruption of the mce4 cholesterol transport system would rescue H37Rv:Δ_igr_ growth in the presence of this sterol. Specialized phage transduction was used to generate H37Rv:Δ_igr_Δ_mce4_, in which yrbE4a, the first gene of the mce4 operon, is replaced with a hygromycin resistance gene in the H37Rv:Δ_igr_ background (17). Putative double mutants were identified by PCR, and yrbE4a deletion was confirmed by Southern blotting (data not shown). The wild-type and H37Rv:Δ_igr_ and H37Rv:Δ_igr_Δ_mce4_ mutant strains were tested for the ability to grow in cholesterol-containing medium. As predicted, the double-knockout strain grew better than H37Rv:Δ_igr_ although complementation was incomplete (Fig. 4A). Consistent with these growth data, deleting mce4 in the H37Rv:Δ_igr_ strain background did not alter the ATP levels per cell in the presence of cholesterol (Fig. 4B). From these in vitro data, we predicted that deletion of mce4 would rescue the attenuation phenotype initially caused by Δ_igr_ in a macrophage model of infection. THP-1 cells were infected with wild-type, Δ_igr_, and Δ_igr_ Δ_mce4_ bacteria, and macrophage survival was monitored by alamarBlue staining. As seen previously, Δ_igr_ was unable to kill THP-1 cells while wild-type M. tuberculosis efficiently killed the cells (Fig. 4C and reference 3). The Δ_igr_ Δ_mce4_ double mutant regained the ability to kill THP-1 cells (Fig. 4C). These data are consistent with the model in which Mce4 facilitates cholesterol entry into bacterial cells, where igr is required for full cholesterol metabolism.
FIG. 4.
Deletion of mce4 partially rescues cholesterol growth inhibition of Δ_igr_. (A) Growth of H37Rv (▪), H37Rv:Δ_igr_ (•), and H37Rv:Δ_igr_Δ_mce4_ (▴) strains in MM+CG. (B) Amount of ATP per cell as a percentage of H37Rv. (C) Killing of THP-1 cells by H37Rv (▪), H37Rv:Δ_igr_ (•), and H37Rv:Δ_igr_Δ_mce4_ (▴) strains monitored by alamarBlue staining. Data shown are the mean and standard deviation of triplicate cultures. For all panels, data are representative of at least duplicate experiments.
Slower H37Rv:Δ_igr_ replication in vivo.
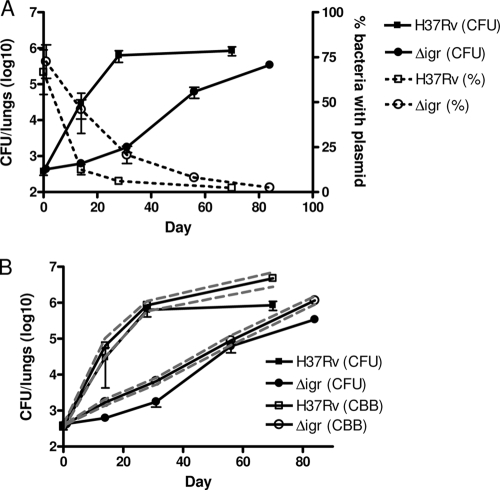
Infection of mice with H37Rv:Δ_igr_ yields little or no increase in bacterial numbers for the first 2 weeks. However, from 2 weeks onward, we observed a steady increase in CFU counts until 12 weeks, when the bacterial burden in the lungs was comparable to that of mice infected with wild-type bacteria (3). To understand better the replication dynamics of this strain early in infection, we infected mice with H37Rv:Δ_igr_ carrying unstable plasmid pBP10 (6). Since this plasmid is lost at a stable, quantifiable rate in the absence of antibiotics, the loss of pBP10 can serve as a molecular replication clock, allowing us to differentiate between slower in vivo replication of the mutant and enhanced host-mediated bacterial killing (6).
Figure 5A compares the proportion of plasmid-carrying bacteria to the total CFU count over time for both H37Rv and H37Rv:Δ_igr_. Consistent with the exponential increase in bacterial numbers observed during initial infection with H37Rv, plasmid loss in this strain was rapid; the proportion of cells carrying pBP10 dropped ∼80% by week 2. In contrast, the proportion of H37Rv:Δ_igr_ cells carrying the plasmid was reduced by only ∼40%. Slower plasmid loss in the mutant indicates that the modest CFU count increase of this strain in mouse lungs is due to slower replication in vivo, as opposed to a heightened susceptibility to host-mediated bacterial killing.
FIG. 5.
Replication dynamics of H37Rv:Δ_igr_ in mice. (A). The fraction of the bacterial population carrying unstable plasmid pBP10 confirms that mutant strain H37Rv:Δ_igr_ replicates more slowly than the wild type in C57BL/6 mice. H37Rv (▪), H37Rv:Δ_igr_ (•). The number of CFU per lung (filled line, closed symbols) and the percentage of bacteria carrying plasmid (dashed line, open symbols) are shown. (B) Mice infected with strain H37Rv:Δ_igr_ have a lower cumulative bacterial burden than those infected with the wild type. The number of CFU per lung (filled line, filled symbols), the cumulative bacterial burden (CBB) (filled line, open symbols), and the 95% confidence interval (dashed line) are shown. The data shown are from a single experiment, the average from five infected mice per time point with the standard deviation, and the data from H37Rv-infected mice previously appeared in reference 6.
We recently developed a mathematical model relating the probability of plasmid loss by log-phase bacteria (described by the segregation constant s) and the bacterial replication rate r. These values were experimentally derived simply by determining the number of live bacteria and the percentage carrying the plasmid at any time point (6). The segregation constant s must be determined independently for each bacterial strain. We derived s for H37Rv:Δ_igr_ and found that it is nearly identical to that of H37Rv (data not shown). We then calculated the bacterial replication rate r and death rate δ of H37Rv:Δ_igr_ during different intervals of infection (Table 1). In contrast to wild-type bacteria, r for H37Rv:Δ_igr_ was modest (0.23, corresponding to a doubling time of 72 h) and remained relatively constant through the course of the experiment. However, the bacterial death rate δ was also modest, never exceeding 0.2 at any point during the experiment. The mathematical model also allows us to estimate the cumulative bacterial burden, the sum of all bacteria live, dead, and removed, during infection. Figure 5B illustrates that even after 12 weeks of infection, the cumulative bacterial burden of H37Rv:Δ_igr_-infected mice is approximately 10-fold lower than that of mice infected with wild-type bacteria.
TABLE 1.
Estimates from bootstrap analysis performed on the in vivo mathematical modela
| Strain and day | Standard plating (CFU/ml) | Plasmid frequency (%) | Value/day | Cumulative no. of bacterial CFU/ml (95% CI)b | |
|---|---|---|---|---|---|
| r | δ | ||||
| H37Rv | |||||
| 1 | 3.78 × 102 | 66.68 | 3.78 × 102 (3.03 × 102-4.20 × 102) | ||
| 14 | 3.03 × 104 | 12.46 | 0.78 | 0.41 | 6.39 × 104 (3.18 × 104-1.04 × 105) |
| 28 | 6.31 × 105 | 6.06 | 0.31 | 0.07 | 8.56 × 105 (5.79 × 105-1.12 × 106) |
| 70 | 8.55 × 105 | 2.33 | 0.12 | 0.12 | 4.78 × 106 (2.79 × 106-7.01 × 106) |
| H37Rv:Δ_igr_ | |||||
| 1 | 4.32 × 102 | 72.57 | 4.32 × 102 (4.13 × 102-4.52 × 102) | ||
| 14 | 6.20 × 102 | 45.97 | 0.20 | 0.17 | 1.70 × 103 (1.39 × 103-2.08 × 103) |
| 31 | 1.77 × 103 | 20.70 | 0.26 | 0.20 | 6.58 × 103 (5.32 × 103-8.13 × 103) |
| 56 | 6.14 × 104 | 8.05 | 0.21 | 0.07 | 8.98 × 104 (6.60 × 104-1.22 × 105) |
| 84 | 3.42 × 105 | 2.47 | 0.23 | 0.17 | 1.16 × 106 (8.96 × 105-1.50 × 106) |
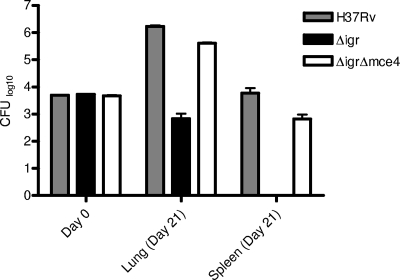
Mce4 deletion suppresses the in vivo growth defect of the Δ_igr_ mutant.
Taken together, these in vitro and in vivo data suggest that the slow growth of the Δ_igr_ mutant during infection might be due to continual cholesterol metabolism and the consequent accumulation of a toxic catabolic intermediate. To investigate this possibility, we determined whether the Δ_igr_ growth defect depended on the ability to acquire cholesterol from the host. Δ_mce4_ mutants grow similarly to the wild type for the first 3 to 4 weeks of infection and only show an obvious growth or survival defect after 6 to 8 weeks (15). This allowed us to ask whether the Δ_mce4_ mutation could suppress the early in vivo growth defect of the Δ_igr_ strain, which is readily observed within 3 weeks. We determined that the Δ_igr_ Δ_mce4_ mutant strain grew significantly faster than the Δ_igr_ strain in the mouse lung (Fig. 6). In contrast to the partial suppression seen in vitro, we found that the mce4 mutation almost completely suppressed the phenotype of the Δ_igr_ mutant strain at 21 days postinfection and restored both the growth rate in the lung and the ability to disseminate to the spleen.
FIG. 6.
Mce4 deletion suppresses the in vivo growth defect of the Δ_igr_ mutant. CFU were counted at 21 days postinfection in both the lungs and spleens of mice infected with wild-type H37Rv or the Δ_igr_ or Δ_igr_ Δ_mce4_ mutant strain. The results for the Δ_igr_ mutant are statistically significantly different from those for the Δ_igr_ Δ_mce4_ mutant in both the lung (P = 0.0002) and the spleen (P = 0.05).
DISCUSSION
The intimate relationship between mycobacteria and host cholesterol has been recognized for decades, and cholesterol is important at multiple stages of pathogenesis, including mycobacterial uptake (5) and persistence during chronic infection (8, 15). Mycobacterium-dependent accumulation of cholesterol esters in macrophages has been observed (9), and recently M. tuberculosis genes were shown to encode cholesterol-modifying activities that contribute to virulence (2, 19, 21, 22).
Deletion of the igr locus resulted in a profound growth defect in the presence of cholesterol. However, growth inhibition did not have an effect on the cells’ ability to make ATP or reduce alamarBlue. These data are consistent with a model in which the H37Rv:Δ_igr_ mutant cannot fully metabolize cholesterol, resulting in the build-up of a toxic metabolite, while cells are able to fully oxidize glycerol and glucose through unrelated metabolic pathways.
What does M. tuberculosis do with imported cholesterol? While the biochemical role of each of the genes in the igr locus is unknown, two have homology to enzymes involved in β oxidation and may be involved in the degradation of this steroid. Because H37Rv:Δ_igr_ is able to produce CO2 from C-4 of cholesterol as efficiently as wild-type cells (Fig. 3), the igr gene products are likely responsible for cholesterol degradation downstream of these steps. We do not yet have tools to study the localization of other carbons from the cholesterol molecule, but we are currently investigating this area. Due to homology with testosterone-metabolizing genes from Comamonas testosteroni, Van der Geize et al. speculated that the igr operon is responsible for the degradation of the C and D rings of cholesterol (19). However, the exact genes and degradation pathway of the C and D rings of testosterone from C. testosteroni are not known (7), and the igr genes could also be responsible for metabolism of the fatty acyl tail of cholesterol, a structure that is not common to testosterone. Deciphering the mechanism of action of the igr operon in cholesterol metabolism is currently under way in our laboratory.
The role of cholesterol import and metabolism in tuberculosis pathogenesis is also a matter of substantial interest, and in this respect the phenotypes of the Δ_igr_ and Δ_mce4_ mutants may be particularly informative. Loss of the Mce4 transporter results in rescue of the Δ_igr_ mutant sensitivity to cholesterol in culture and in macrophages (Fig. 4). However, rescue is incomplete, presumably because the mce4 mutation reduces but does not eliminate uptake of cholesterol (15). Sensitivity of the igr mutant to cholesterol in vitro argues that it may act as a biosensor of cholesterol availability in vivo. The Δ_igr_ mutant strain grows slowly throughout infection before establishing a chronic infection with much-reduced virulence. The slow but steady replication rate of H37Rv:Δ_igr_ in vivo (Fig. 5) argues that M. tuberculosis is exposed to and imports cholesterol throughout infection.
The behavior of the igr mutant in vivo differs from that of the wild type in another key respect. Replication is initially slower than that of the wild type, but bacterial numbers continue to increase for 12 weeks (Fig. 5 and reference 3), long after host immunity has contained a wild-type infection. Failure of the host immune system to control expansion of the mutant in this time may be due to the substantially reduced cumulative bacterial burden following infection with the mutant. Furthermore, the low cumulative bacterial burden of H37Rv:Δ_igr_ may explain why histopathology in H37Rv:Δ_igr_-infected mice is much less severe, even after 24 weeks of infection (3), and why these mice remained healthy to week 39, when a time-to-morbidity experiment was terminated (data not shown), while aerosol H37Rv-infected mice typically succumb to infection by weeks 28 to 30 (10).
We propose that the in vivo attenuation of H37Rv:Δ_igr_ is most likely due to its inability to metabolize cholesterol fully. This hypothesis is supported both by the inability of Δ_igr_ to grow in the presence of cholesterol in vitro and by the suppression of this defect by disruption of the mce4 cholesterol transport system. The inability of the mce4 deletion to rescue H37Rv:Δ_igr_ fully in vitro is likely due to an incomplete block of cholesterol import under these conditions (15). Interestingly, Δ_mce4_ suppression of the Δ_igr_ phenotype was almost complete in vivo (Fig. 6), suggesting that mce4 is particularly important for cholesterol import during infection. In addition, a recent study showed guinea pigs infected with a strain defective in the cholesterol-metabolizing dioxygenase HsaC resulted in reduced pathology in the lungs and spleens of infected animals (21). These studies, taken together, highlight the importance of cholesterol during M. tuberculosis infection, not only for host protection against disease but also for full virulence of the bacterium.
Cholesterol contributes to human health and disease in a myriad of ways. Our work is beginning to uncover new and unexpected ways that cholesterol plays a role in tuberculosis pathogenesis. We hope that understanding these processes better will lead to new interventions against this long-standing infectious scourge.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank members of the Sherman laboratory for helpful discussions, Kristi Guinn for expert help with the manuscript, and Sarah Gillmore for generous help with TLC and mass spectrometry assays.
This work was supported in part by Paul G. Allen Family Foundation grant 8999 to D.R.S. and by NIH award AI064282 to C.M.S.
Footnotes
▿
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Bloch, H., and W. Segal. 1956. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72**:**132-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brzostek, A., B. Dziadek, A. Rumijowska-Galewicz, J. Pawelczyk, and J. Dziadek. 2007. Cholesterol oxidase is required for virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 275**:**106-112. [DOI] [PubMed] [Google Scholar]
- 3.Chang, J. C., N. S. Harik, R. P. Liao, and D. R. Sherman. 2007. Identification of mycobacterial genes that alter growth and pathology in macrophages and in mice. J. Infect. Dis. 196**:**788-795. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitheead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393**:**537-544. [DOI] [PubMed] [Google Scholar]
- 5.Gatfield, J., and J. Pieters. 2000. Essential role for cholesterol in entry of mycobacteria into macrophages. Science 288**:**1647-1650. [DOI] [PubMed] [Google Scholar]
- 6.Gill, W., N. Harik, M. Whiddon, R. Liao, J. Mittler, and D. R. Sherman. 2009. A replication clock for Mycobacterium tuberculosis. Nat. Med. 15**:**211-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horinouchi, M., T. Kurita, T. Yamamoto, E. Hatori, T. Hayashi, and T. Kudo. 2004. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem. Biophys. Res. Commun. 324**:**597-604. [DOI] [PubMed] [Google Scholar]
- 8.Joshi, S. M., A. K. Pandey, N. Capite, S. M. Fortune, E. J. Rubin, and C. M. Sassetti. 2006. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc. Natl. Acad. Sci. USA 103**:**11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo, E., and K. Kanai. 1976. Accumulation of cholesterol esters in macrophages incubated with mycobacteria in vitro. Jpn. J. Med. Sci. Biol. 29**:**123-137. [DOI] [PubMed] [Google Scholar]
- 10.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R. Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 187**:**117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinney, J. D., K. Honer zu Bentrup, E. J. Muñoz-Elías, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406**:**735-738. [DOI] [PubMed] [Google Scholar]
- 12.Mohn, W. W., R. van der Geize, G. R. Stewart, S. Okamoto, J. Liu, L. Dijkhuizen, and L. D. Eltis. 2008. The actinobacterial Mce4 locus encodes a steroid transporter. J. Biol. Chem. 283**:**35368-35374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muñoz-Elías, E. J. M., and J. D. McKinney. 2006. Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8**:**10-22. [DOI] [PubMed] [Google Scholar]
- 14.Muñoz-Elías, E. J. M., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11**:**638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandey, A. K., and C. M. Sassetti. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 105**:**4376-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rengarajan, J. B., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 102**:**8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48**:**77-84. [DOI] [PubMed] [Google Scholar]
- 18.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198**:**693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Geize, R., K. Yam, T. Heuser, M. H. Wilbrink, H. Hara, M. C. Anderton, E. Sim, L. Dijkhuizen, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. USA 104**:**1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yajko, D. M., J. J. Madej, M. V. Lancaster, C. A. Sanders, V. L. Cawthon, B. Gee, A. Babst, and W. K. Hadley. 1995. Colorimetric method for determining MICs of antimicrobial agents for Mycobacterium tuberculosis. J. Clin. Microbiol. 33**:**2324-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yam, K. C., I. D'Angelo, R. Kalscheuer, H. Zhu, J. X. Wang, V. Snieckus, L. H. Ly, P. J. Converse, W. R. Jacobs, Jr., N. Strynadka, and L. D. Eltis. 2009. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 5**:**e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, X., E. Dubnau, I. Smith, and N. S. Sampson. 2007. Rv1106c from Mycobacterium tuberculosis is a 3β-hydroxysteroid dehydrogenase. Biochemistry 46**:**9058-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]