PTIP Associates with MLL3- and MLL4-containing Histone H3 Lysine 4 Methyltransferase Complex (original) (raw)
. Author manuscript; available in PMC: 2009 Aug 20.
Published in final edited form as: J Biol Chem. 2007 May 11;282(28):20395–20406. doi: 10.1074/jbc.M701574200
Abstract
PTIP, a protein with tandem BRCT domains, has been implicated in DNA damage response. However, its normal cellular functions remain unclear. Here we show that while ectopically expressed PTIP is capable of interacting with DNA damage response proteins including 53BP1, endogenous PTIP, and a novel protein PA1 are both components of a Set1-like histone methyltransferase (HMT) complex that also contains ASH2L, RBBP5, WDR5, hDPY-30, NCOA6, SET domain-containing HMTs MLL3 and MLL4, and substoichiometric amount of JmjC domain-containing putative histone demethylase UTX. PTIP complex carries robust HMT activity and specifically methylates lysine 4 (K4) on histone H3. Furthermore, PA1 binds PTIP directly and requires PTIP for interaction with the rest of the complex. Moreover, we show that hDPY-30 binds ASH2L directly. The evolutionarily conserved hDPY-30, ASH2L, RBBP5, and WDR5 likely constitute a subcomplex that is shared by all human Set1-like HMT complexes. In contrast, PTIP, PA1, and UTX specifically associate with the PTIP complex. Thus, in cells without DNA damage agent treatment, the endogenous PTIP associates with a Set1-like HMT complex of unique subunit composition. As histone H3 K4 methylation associates with active genes, our study suggests a potential role of PTIP in the regulation of gene expression.
Transcriptional regulation at the chromatin level requires concerted action of multiple chromatin remodeling complexes and histone-modifying complexes. Covalent modifications of histones, such as acetylation, methylation, phosphorylation, and ubiquitination, have been shown to play important roles in the regulation of both global and tissue- and developmental stage-specific gene expression. Histone lysine methylation is involved in both gene activation and repression, depending upon the specific lysine residue that gets methylated. For example, methylation at histone H3 lysine 27 is associated with gene repression, while di- and trimethylations at histone H3 lysine 4 (K4) are associated with gene activation (1, 2).
In yeast, a multisubunit complex (also known as COMPASS and Set1 complex) containing Drosophila Trithorax-related protein Set1 has been shown to be responsible for mono-, di-, and trimethylation of histone H3 K4 (3-7). In humans, multiple Set1-like complexes with robust histone H3 K4 methyltransferase activities have been isolated (8). Each of these complexes contains one or two SET domain-containing homologs of yeast Set1 and Drosophila Trithorax, such as hSet1 (human Set1) (9), MLL (mixed-lineage leukemia, also known as MLL1, HRX, ALL1) (10-13), MLL2 (mixed-lineage leukemia 2) (8, 10, 14-17), MLL3 (mixed-lineage leukemia 3), and MLL4 (mixed-lineage leukemia 4, also known as ALR) (15, 18, 19), which carry the enzymatic activity for the associated complexes. ASH2L, RBBP5, and WDR5, which are homologs of yeast COMPASS/Set1 complex subunits Bre2, Swd1, and Swd3, respectively, are shared by various human Set1-like histone methyltransferase (HMT)3 complexes. In addition, each of these complexes contains distinct but overlapping subunits (8, 10). For example, hSwd2 and CXXC1 are homologs of the Swd2 and Spp1 subunits of yeast COMPASS/Set1 complex and have been shown to associate with hSet1 complex (20). Menin, a protein with no homology with any of the yeast COMPASS/Set1 complex components, has been shown to be present in both MLL and MLL2 complexes (8, 10). MLL and MLL2 complexes have been shown to regulate expression of a subset of transcriptionally active genes, including clustered homeobox (Hox) genes (8, 10, 15, 21).
PTIP (Pax transactivation domain-interacting protein) is a ubiquitously expressed nuclear protein and was first identified, through yeast two-hybrid assays, as a Pax2 transactivation domain-interacting protein (22). PTIP carries six tandem BRCT (BRCA1 carboxyl-terminal) domains that are predominantly found in proteins involved in DNA damage response (Fig. 1_A_). Deletion of PTIP in mice leads to early embryonic lethality at E9.5 due to widespread cell death (23). In vitro, tandem BRCT domains in PTIP function as phosphopeptide binding modules that recognize substrates phosphorylated by DNA damage activated ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia- and RAD3-related (ATR) protein kinases. Upon ionizing radiation, ectopically expressed PTIP shows significantly increased association with DNA damage checkpoint protein 53BP1, translocates to DNA damage-induced foci, and co-localizes with phosphorylated histone H2AX, a marker for DNA double strand break (24, 25). However, the function of endogenous PTIP in cells without DNA damage agent treatment has remained unclear.
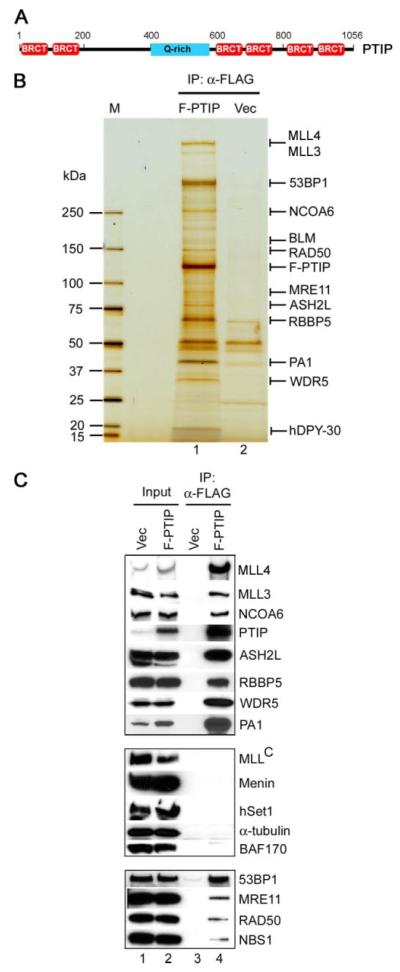
FIGURE 1. Ectopically expressed PTIP associates with two functionally distinct groups of proteins in cells.
A, schematic representation of full-length mouse PTIP protein containing 1,056 amino acids. The six BRCT domains are marked in red. The glutamine-rich (Q-rich) region is marked in blue. B, silver staining of affinity-purified PTIP-associated proteins. Nuclear extracts prepared from HeLaS cells stably expressing FLAG-tagged PTIP (F-PTIP) or vector only (Vec) were incubated with anti-FLAG M2-agarose. After extensive washing, bound proteins were resolved on 4–15% SDS-PAGE and analyzed by mass spectrometry as described under “Experimental Procedures.” IP, immunoprecipitation. M, protein marker. C, Western blot confirmation of mass spectrometry results. PTIP-associated proteins purified as in B were analyzed by Western blot with antibodies indicated on the right.
To understand the normal cellular function of PTIP, we affinity-purified PTIP-associated proteins from nuclear extracts prepared from HeLaS cells that were grown in the absence of DNA damage agent treatment. Mass spectrometry analysis and subsequent purification demonstrated that PTIP associated with ASH2L, RBBP5, WDR5, hDPY-30, NCOA6, a novel protein PA1, SET domain-containing histone methyltransferases MLL3 and MLL4, and substoichiometric amount of JmjC domain-containing putative histone demethylase UTX in a Set1-like complex that carried robust histone H3 lysine 4 (K4) methyltransferase activity.
EXPERIMENTAL PROCEDURES
Plasmids
Mammalian Gene Collection (MGC) clones obtained from ATCC were used as templates for PCR amplification of full-length cDNAs of mouse PTIP (GenBank™ accession number BC066014), human PA1 (GenBank™ accession number BC003640), UTX (GenBank™ accession number BC093868), RBBP5 (GenBank™ accession number BC053856), and WDR5 (GenBank™ accession number BC016103). Full-length ASH2L cDNA was amplified by reverse transcription-PCR from HeLaS cells using primers 5′-TAT AGG ATC CTG GTG ATG GCG GCG GCA GGA G-3′ and 5′-CAG CGA CTC GAG TCA GGG TTC CCA TGG GGG AC-3′. FLAG-tagged hDPY-30 was cloned by PCR from HeLaS cDNA using primers 5′-C AGT AGG ATC CTG GCC ACC ATG GAC TAC AAG GAC GAC GAT GAC AAG ATG GAG CCA GAG CAG ATG CTG G-3′ and 5′-TCG CTG CTC GAG TCA GTT TCG ATC TTC AAA CTG TGC-3′. cDNAs for the SET domain-containing COOH-terminal fragments of MLL3 (3,993–4,911 amino acids, GenBank™ accession number NP_733751) and MLL4 (4,289–5,262 amino acids, NP_003473) were amplified by reverse transcription-PCR from HeLaS cells. cDNAs were cloned into pcDNA3 (Invitrogen) for transient expression or as templates for in vitro transcription/translation (Promega). For glutathione _S_-transferase (GST) pull-down assays, cDNAs were cloned into pGEX-5X1 (Amersham Biosciences). For retrovirus-mediated gene transfer into HeLaS cells, FLAG-tagged PTIP, PA1, UTX, and hDPY-30 were cloned into pWZLneo, which carries neomycin resistance gene. For retrovirus-mediated gene transfer into immortalized mouse embryonic fibroblasts (MEFs), FLAG-tagged PA1 was also cloned into pMSCVpuro and pMSCVhygro (Clontech), which carry puromycin and hygromycin resistance genes, respectively. Cre recombinase gene was subcloned from pBabeCREpuro (gift of Thomas Blankenstein) into pMSCVpuro. All PCR products and plasmids were verified by DNA sequencing.
Antibodies
Rabbit anti-MLL3, anti-MLL4, anti-PA1, and anti-WDR5 antibodies were generated against and affinity-purified by the following peptides: DKRPRGRPRKDGASPFQ and TDKIKKRYRKRKNKLEET (human MLL3 residues 33– 49 and mouse MLL3 residues 1334 –1351, respectively), FVHSKSSQYRRLRTEWKN and EGDEKKKQQRRGRKRSKL (human MLL4 residues 5105–5122 and 1472–1489, respectively), RSQKREARLDKVLSD (human PA1 residues 193–207), and MATEEKKPETEAARAQP (human WDR5 residues 1–17). Rabbit anti-PTIP antibody was generated against GST-fused mouse PTIP residues 1–316 as described (22) and purified on protein A-agarose. Mouse IgG (catalog number sc-2025), anti-BAF170 (sc-10757), and anti-_α_-tubulin (sc-5286) were from Santa Cruz Biotechnology. Rabbit IgG (I-5006), anti-FLAG (F3165), and anti-FLAG M2-agarose (A2220) were from Sigma. Anti-53BP1 (A300–272A), anti-Menin (A300–105A), anti-human NCOA6 (A300–410A), anti-ASH2L (A300–107A), anti-RBBP5 (A300–109A), and anti-hSet1 (A300–289A) were from Bethel Laboratories. Anti-MLL COOH-terminal (05-765) was from Upstate Biotechnology. Anti-MRE11 (GTX70212) and anti-RAD50 (GTX70228) were from GeneTex. Anti-NBS1 (611870) and anti-CXXC1 (612334) were from BD Biosciences.
Cell Culture and Retrovirus-mediated Gene Transfer
All cells were routinely cultured in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum. Primary _PTIP_flox/flox MEFs were isolated from E13.5 embryos of PTIP conditional knock-out mice (26) and immortalized by transfection with pcDNA3 expressing SV40T following 3T3 protocol (27). Retrovirus infection of immortalized MEFs was performed as described (28). HeLaS cells stably expressing FLAG-tagged full-length mouse PTIP, human PA1 and hDPY-30 (F-PTIP, F-PA1, and F-hDPY-30, respectively) were established using retrovirus-mediated gene transfer. Briefly, Phoenix amphotropic retrovirus packaging cells were seeded at a density of 1.1 × 106 cells in 4 ml of medium per 6-cm dish. After overnight incubation, cells were transfected with 8 _μ_g of pWZLneo-based plasmids by lipofectAmine2000 (Invitrogen). 48 h later, the cells were changed to fresh medium. Virus-containing supernatant was collected 12 h later, filtered through 0.45-mm membrane, and supplemented with 8 _μ_g/ml polybrene and was added to HeLaS cells that were plated at a density of 3.5 × 105 cells per 6-cm dish 1 day before. Two days later, cells were serial diluted in medium containing 1 mg/ml G418 (Invitrogen). Fourteen days later, colonies were expanded, and those expressing FLAG-tagged PTIP were identified by Western blot with anti-FLAG antibody.
Purification of PTIP HMT Complex
HeLaS cells expressing FLAG-tagged PTIP or vector only were grown to confluence on 200 × 15 cm dishes in Dulbecco’s modified Eagle’s medium containing 8% bovine serum and 2% fetal bovine serum. To isolate PTIP-associated proteins, 300 mg of nuclear extracts prepared as described (28) were precleared twice with 0.6 ml of mouse IgG-agarose (Sigma) for 2 h in buffer A (20 mM HEPES (pH 7.9), 180 mM KCl, 0.2 mM EGTA, 1.5 mM MgCl2, 20% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.1% Nonidet P-40) supplemented with 1 _μ_g/ml aprotinin, 2 _μ_g/ml leupeptin, and 0.7 _μ_g/ml pepstatin (Roche Applied Science), followed by overnight immunoprecipitation with 0.5 ml anti-FLAG M2-agarose at 4 °C. After washing with buffer A for 12 times, bound proteins were eluted twice with 0.25 mg/ml FLAG peptide (Sigma), concentrated with microcon column (Millipore), resolved on 4–15% SDS-PAGE (Bio-Rad), and analyzed by mass spectrometry. To purify PTIP HMT complex, anti-RBBP5 antibody was cross-linked to protein A-agarose (Amersham Biosciences) using disuccinimidyl suberate (Pierce). PTIP-associated proteins purified as above were incubated with immobilized anti-RBBP5 at 4 °C overnight. After washing with buffer A containing 300 mM KCl five times, bound proteins were eluted twice with 100 mM triethylamine (pH 11.5) for 10 min, neutralized with 1 M Tris (pH 7.5), concentrated with microcon column, and resolved on 4–15% SDS-PAGE. For one-step purification of PTIP HMT complex, nuclear extracts prepared from HeLaS cells expression FLAG-tagged PA1 were precleared twice with mouse IgG-agarose in buffer A, followed by overnight immunoprecipitation with anti-FLAG M2-agarose at 4 °C. After extensive washing with buffer A, bound proteins were eluted twice with 0.25 mg/ml FLAG peptide.
Mass Spectrometric Protein Identification
For mass spectrometric protein identification, excised gel bands were placed in V-shaped 96-well microtiter plate (Greiner) with needle-punctured bottoms and robotically processed on a Proteam 100 Genesis liquid handling system (Tecan). The robotic procedure encompassed gel destaining in 50% acetonitrile, 100 mM ammonium bicarbonate, protein reduction in 10 mM Tris (carboxyethylphosphine) (Pierce), 50 mM ammonium bicarbonate, alkylation with iodoacetamide, and digestion with bovine trypsin (Roche Applied Science) at 37 °C for 8 h. The gel-released peptides were captured on Poros 20 R 2 beads (Applied Biosystems), transferred into ZipTips (Millipore), serving as frits, and eluted onto stainless steel sample plates using 60% acetonitrile, 20% methanol, in 0.1% aqueous trifluoroacetic acid and co-crystallized with the MALDI matrix _α_-cyano-4-hydroxy cinnamic acid. Single-stage mass spectrometric analyses were performed on a Protof2000 MALDI-quadrupole time-of-flight instrument (PerkinElmer/Sciex) and multistage mass spectrometric fragmentation spectra were obtained on a self-built MALDI-quadrupole ion trap as described previously (29). Spectra were analyzed by data base searching using Xproteo, Profound, and TheGPM X! Tandem.
Co-immunoprecipitation, Western Blot, and GST Pull-down Assay
For co-immunoprecipitation, 1–5 mg of HeLaS nuclear extracts were incubated with 2–5 _μ_g of antibodies. The precipitates were washed five times with buffer A. Western blot and GST pull down of in vitro translated proteins were done essentially as described (28).
HMT Assay and Edman Degradation
HMT assay and Edman degradation to map methylation site on histone H3 were done as described (8).
RESULTS
Affinity Purification of PTIP-associated Proteins
To understand the normal cellular function of PTIP, we sought to isolate PTIP-associated proteins from cells cultured in the absence of DNA damage agent treatment. For this purpose, a HeLaS cell line stably expressing FLAG-tagged full-length mouse PTIP (F-PTIP) was established using retrovirus-mediated gene transfer. Nuclear extracts were prepared and were incubated with anti-FLAG M2-agarose. After extensive washing, bound proteins were eluted with FLAG peptide and resolved on 4–15% SDS-PAGE followed by mass spectrometry analysis (Fig. 1_B_ and supplemental Table S1). Consistent with previous reports of interaction between PTIP and 53BP1 (25), a high molecular mass band above 250 kDa was identified as 53BP1. A 159-kDa band was identified as BLM (Bloom Syndrome protein), a RecQ family DNA helicase that has been shown to associate with 53BP1 in response to DNA damage (30). In addition, two bands were identified as MRE11 and RAD50, components of the MRE11-RAD50-NBS1 (MRN) complex that plays key role in DNA damage detection and repair (31). Mass spectrometry results were confirmed by Western blot analysis (Fig. 1_C_, bottom panel).
Surprisingly, the remaining bands (Fig. 1_B_) were identified to be ASH2L, RBBP5, WDR5, hDPY-30, SET domain-containing histone methyltransferases MLL3 and MLL4, and NCOA6 (nuclear receptor coactivator 6, also known as NRC/AIB3/ASC-2/PRIP/TRBP/RAP250 (32)), proteins that have been reported to be subunits of various Set1-like HMT complexes (8-10, 33, 34). In addition, a 42-kDa band was identified to be a novel protein, which we named as PA1 (PTIP-associated 1). Polyclonal antibodies were raised against PA1, WDR5, MLL3, and MLL4. Western blot using available antibodies confirmed mass spectrometry results (Fig. 1_C_, top panel). Taken together, our affinity purification led to the identification of two functionally distinct groups of proteins that associated with ectopically expressed PTIP in cell nuclei: those involved in DNA damage response and repair (53BP1, BLM, MRE11, RAD50, and NBS1) and those involved in regulation of gene expression through chromatin methylation on histone H3 K4 (ASH2L, RBBP5, WDR5, hDPY-30, NCOA6, MLL3, and MLL4).
PTIP Associates with ASH2L, RBBP5, WDR5, hDPY-30, NCOA6, MLL3, MLL4, and PA1 in One Complex
PTIP-associated proteins isolated as above were adjusted to contain 300 mM KCl and fractionated on Superose 6 gel filtration column followed by Western blot analysis. As shown in Fig. 2_A_ bottom panel, a portion of PTIP co-eluted in high molecular mass fractions with both groups of PTIP-associated proteins, peaking at fractions with ∼1.8 MDa. Consistent with the presence of ASH2L and RBBP5, these PTIP-containing high molecular weight fractions were associated with robust HMT activity on unmodified histone H3 peptide (Fig. 2_A_, top panel). These data suggest PTIP and associated proteins may form high molecular weight protein complex(es) with HMT activity.
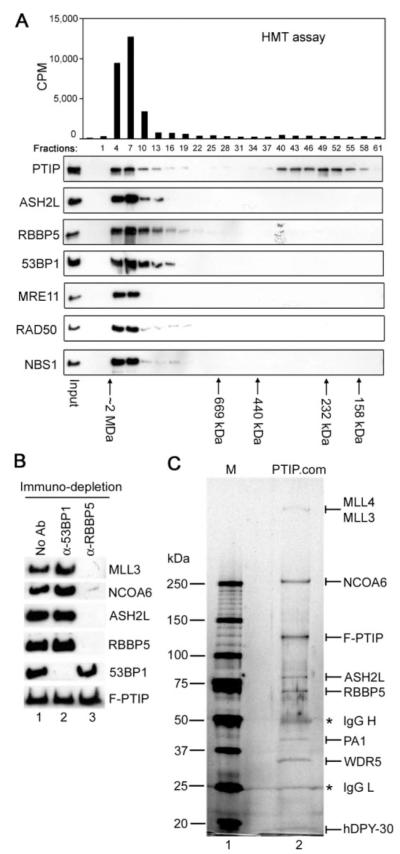
FIGURE 2. PTIP associates with ASH2L, RBBP5, WDR5, hDPY-30, NCOA6, MLL3, MLL4, and PA1 in one complex.
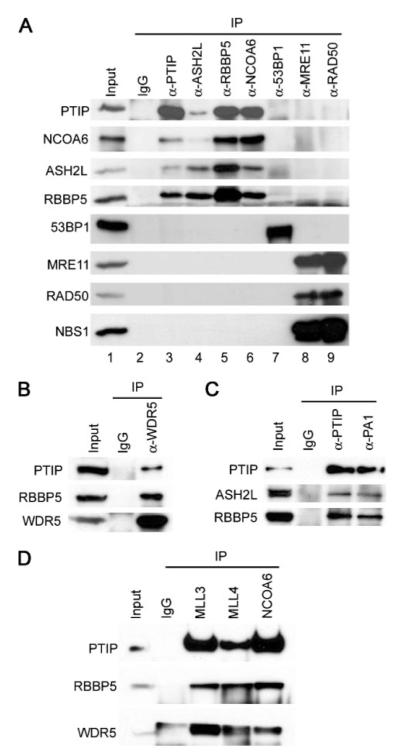
A, PTIP exists in high molecular weight complex(es) associated with HMT activity. PTIP-associated proteins isolated as in Fig. 1_B_ were fractionated on Superose 6 gel filtration column (Amersham Biosciences) in buffer A containing 300 mM KCl. Fractions were analyzed by Western blot with antibodies indicated on the left (lower panel) and by HMT assay on unmodified histone H3 peptide (residues 1–20) (upper panel). Positions of protein standards are indicated at the bottom. Note that PTIP protein alone purified by gel filtration did not possess any HMT activity. B, the two functionally distinct groups of PTIP-associated proteins do not exist in the same complex. PTIP-associated proteins purified as in Fig. 1B were subjected to immuno-depletion with anti-53BP1 or anti-RBBP5 antibodies, followed by Western blot with antibodies indicated on the right. No Ab, no antibody. C, purification of PTIP complex. PTIP-associated proteins isolated as in Fig. 1_B_ were incubated with immobilized anti-RBBP5 antibody at 4 °C overnight. After washing with buffer A containing 300 mM KCl, bound proteins were eluted with 100 mM triethylamine (pH 11.5), neutralized with 1 M Tris (pH 7.5), concentrated with microcon column, resolved on 4–15% SDS-PAGE, and stained with silver. Protein identities were confirmed by mass spectrometry and/or Western blot. PTIP.com, PTIP complex. IgG heavy chain (IgG H) and light chain (IgG L) are marked with asterisks.
To determine whether the two functionally distinct groups of PTIP-associated proteins co-existed in one complex, PTIP-associated proteins were subjected to immuno-depletion with either anti-53BP1 or anti-RBBP5 antibodies. As shown in Fig. 2_B_, anti-RBBP5 antibody almost completely depleted RBBP5, ASH2L, MLL3, and NCOA6, while it had minimal effect on 53BP1 level. Conversely, anti-53BP1 antibody depleted 53BP1 without affecting the levels of RBBP5, ASH2L, MLL3, and NCOA6, suggesting that the two functionally distinct groups of PTIP-associated proteins did not co-exist in the same complex. Neither anti-53BP1 nor anti-RBBP5 antibodies could significantly deplete the overexpressed PTIP as the majority of which existed as monomer or oligomer as revealed by gel filtration (Fig. 2_A_). While it remains to be determined how the DNA damage response and repair proteins associate with PTIP, data from gel filtration and immuno-depletion experiments strongly suggest that PTIP associates with RBBP5, ASH2L, MLL3, and NCOA6 in one high molecular weight complex.
To purify the PTIP complex associated with HMT activity, PTIP-associated proteins isolated through F-PTIP as described in Fig. 1_B_ were incubated with immobilized anti-RBBP5 antibody. After extensive washing with buffer A containing 300 mM KCl, bound proteins were eluted with 100 mM Triethylamine (pH 11.5) and resolved on SDS-PAGE (Fig. 2_C_). Silver staining revealed several prominent bands that migrated at a similar molecular mass as that of MLL4 (∼520–540 kDa), MLL3 (∼520–540 kDa), NCOA6 (250 kDa), PTIP (120 kDa), ASH2L (77 kDa), RBBP5 (67 kDa), PA1 (42 kDa), WDR5 (35 kDa), and hDPY-30 (15 kDa) shown in Fig. 1_B_. The identities of proteins in each prominent band were confirmed by mass spectrometry and/or Western blotting (data not shown). Consistent with immuno-depletion results (Fig. 2_B_), no detectable amount of 53BP1 (∼300 kDa) was precipitated by anti-RBBP5 antibody. Thus, PTIP associates with ASH2L, RBBP5, WDR5, hDPY-30, NCOA6, MLL3, MLL4, and PA1 in one complex.
One-step Purification of PTIP Complex through FLAG-tagged PA1
Human PA1 (GenBank™ accession number EF195235) contains 254 amino acids and has no recognizable conserved domains (Fig. 3_A_). Mouse PA1 protein (GenBank™ accession number AAH03932) contains 253 amino acids and shares 87% identity at the amino acid level with its human ortholog. The theoretical molecular mass of PA1 protein is 28 kDa. Probably due to its relatively low pI (isoelectric point) at 4.4, PA1 protein runs at around 42 kDa on SDS-PAGE (Figs. 1_B_,2_C_, and 3_C_). When ectopically expressed in HeLa cells, PA1 was localized in the nucleus (data not shown). In GST pull-down assay, GST-fused PA1 interacted directly with in vitro translated PTIP but not with ASH2L, RBBP5, WDR5, and NCOA6 (Fig. 3_B_).
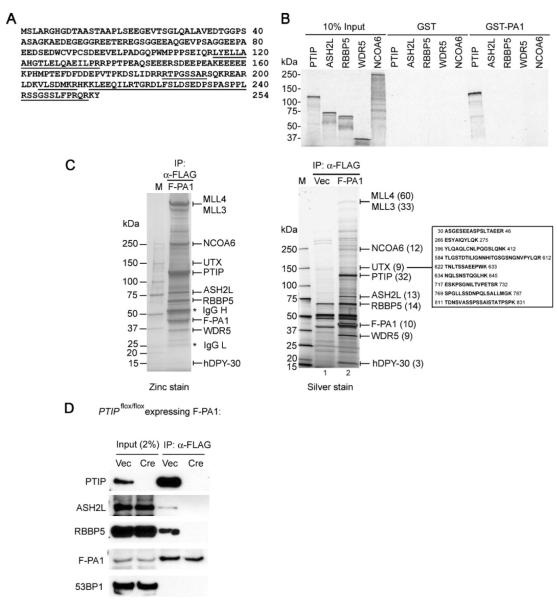
FIGURE 3. One-step purification of PTIP complex through FLAG-tagged PA1.
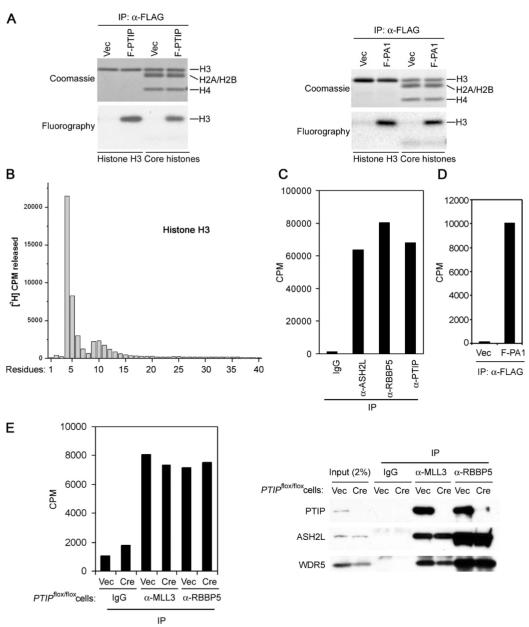
A, amino acid sequence of the novel human protein PA1. Peptide sequences identified by mass spectrometry analysis of PTIP-associated proteins as described in the legend Fig. 1B are underlined. B, PA1 binds directly to PTIP. GST or GST-PA1 was incubated with in vitro translated, [35S]methionine-labeled PTIP, ASH2L, RBBP5, WDR5, and NCOA6 in GST pull down assay. C, one-step purification of PTIP complex through FLAG-tagged PA1. Nuclear extracts prepared from HeLaS cells expressing FLAG-tagged PA1 (F-PA1)or vector only (Vec) were incubated with anti-FLAG M2-agarose. After extensive washing, bound proteins were resolved on 4–15% SDS-PAGE followed by silver staining (right panel) or by staining using Zinc Stain & Destain kit (Bio-Rad) (left panel). The identities of the proteins were determined by mass spectrometry analysis. The numbers of matching tryptic peptides are indicated in parentheses. The sequences and locations of tryptic peptides matching UTX are listed on the right side of the silver-stained gel. Note that the ∼540-kDa band containing both MLL3 and MLL4 could be well stained by zinc but not silver. D, PA1 requires PTIP for interaction with the rest of the PTIP complex. SV40T-immortalized PTIP conditional knock-out MEF cell line _PTIP_flox/flox was infected with a retroviral vector pMSCVhygro expressing F-PA1. After selection with hygromycin, cells were subsequently infected with retroviruses pMSCVpuro expressing vector (Vec) or Cre recombinase to delete PTIP gene. After selection with puromycin, cells were expanded, and nuclear extracts were prepared and subjected to immunoprecipitation with anti-FLAG M2-agarose. The immunoprecipitates were analyzed by Western blot with antibodies indicated on the left.
The direct interaction between PA1 and PTIP prompted us to test whether the PTIP complex could also be purified through PA1. For this purpose, a HeLaS cell line stably expressing FLAG-tagged PA1 (F-PA1) was established using retroviral gene transfer method. Nuclear extracts were prepared and subjected to immunoprecipitation using anti-FLAG M2-agarose. After extensive washing, bound proteins were eluted with FLAG peptide, concentrated, and resolved on 4–15% SDS-PAGE. The SDS-PAGE gels were stained separately with silver staining and the less sensitive zinc staining methods, as the high molecular mass protein bands over 200 kDa did not stain well with the silver staining method (compare left versus right panels in Fig. 3_C_). Mass spectrometry analysis revealed that F-PA1 pulled down the entire PTIP complex, including the SET domain-containing histone methyltransferases MLL3 and MLL4, NCOA6, PTIP, ASH2L, RBBP5, WDR5, and hDPY-30 (Fig. 3_C_ and data not shown). Thus, PTIP complex could be isolated through FLAG-tagged PA1 in one-step purification.
To find out how PA1 interacts with the PTIP complex, SV40T-immortalized PTIP conditional knock-out mouse embryonic fibroblast (MEF) cell line _PTIP_flox/flox was infected with a retroviral vector pMSCVhygro expressing F-PA1. After selection with hygromycin, cells were subsequently infected with retroviruses pMSCVpuro expressing Cre recombinase to delete PTIP gene. After selection with puromycin, cells were expanded and nuclear extracts were prepared and subjected to immunoprecipitation with anti-FLAG M2-agarose followed by Western blot analysis. As shown in Fig. 3_D_, Cre expression eliminated PTIP expression in _PTIP_flox/flox cells. F-PA1 was unable to pull down the PTIP complex components from PTIP gene-deleted cells, indicating the PA1 requires PTIP for interaction with the rest of the complex.
Interestingly, mass spectrometry analysis of PTIP complex isolated through F-PA1 also identified nine peptides that matched protein sequence of UTX, a JmjC domain-containing putative histone demethylase (35) (Fig. 3_C_). UTX is present at substoichiometric level in PTIP complex isolated from HeLaS cells, likely due to the relatively low level of UTX expression in HeLaS cells, which is only 9.6% of that in 293 cells as determined by quantitative reverse transcriptase PCR. The specific association between UTX and PTIP complex was confirmed by Western blot analysis (Fig. 6_B_).
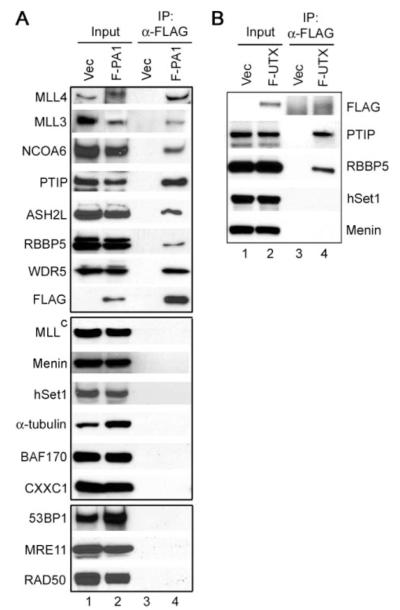
FIGURE 6. PA1 and the JmjC domain-containing putative histone demethylase UTX specifically associate with PTIP complex.
A, Western blot characterization of subunit composition of PTIP complex isolated through F-PA1. Nuclear extracts prepared from HeLaS cells stably expressing F-PA1 were incubated with anti-FLAG M2-agarose. Immunoprecipitates were analyzed by Western blot using antibodies indicated on the left. B, nuclear extracts prepared from HeLaS cells expressing FLAG-tagged UTX (F-UTX) were incubated with anti-FLAG M2-agarose. Immunoprecipitates were analyzed by Western blot against distinct subunits (hSet1, Menin, and PTIP) or the shared RBBP5 subunit of various human Set1-like complexes.
Endogenous PTIP Does Not Associate with DNA Damage Response Proteins in Cells without DNA Damage Agent Treatment
The interactions among endogenous PTIP complex subunits were confirmed by reciprocal co-immunoprecipitation from HeLaS nuclear extracts using antibodies against selected PTIP complex components (Fig. 4). Anti-PTIP antibody readily precipitated endogenous ASH2L, RBBP5, and NCOA6 (Fig. 4_A_, lane 3). Conversely, antibodies against ASH2L, RBBP5, WDR5, PA1, NCOA6, and the SET domain-containing MLL3 and MLL4 efficiently pulled down the endogenous PTIP (Fig. 4_A_, lanes 4–6, Fig. 4, B—D). In contrast, reciprocal co-immunoprecipitation failed to detect endogenous interaction between PTIP complex subunits (PTIP, ASH2L, RBBP5, and NCOA6) and DNA damage response proteins (53BP1, MRE11, and RAD50) (Fig. 4_A_, lanes 3–9). These data suggest that at least in cells cultured under normal condition without DNA damage agent treatment, endogenous PTIP does not associate significantly with DNA damage response proteins. However, such data do not exclude the possibility that in cells treated with DNA damage agent, endogenous PTIP, and potentially other subunits of PTIP complex, may interact with DNA damage response and repair proteins including 53BP1. Indeed, a recent report described increased association between endogenous PTIP and 53BP1 upon ionizing radiation (24).
FIGURE 4. In cells without DNA damage agent treatment, endogenous PTIP complex components do not associate with 53BP1 and other DNA damage response proteins.
A, rabbit IgG (as control), anti-PTIP, ASH2L, RBBP5, NCOA6, 53BP1, MRE11, and RAD50 antibodies were incubated with 1 mg of HeLaS nuclear extracts. The immunoprecipitates were subjected to Western blot with antibodies indicated on the left. B—D, endogenous interactions among PTIP complex components. Experiments were done as described for A.
Consistent with our observation that endogenous PTIP did not associate with DNA damage response proteins in cells without DNA damage agent treatment, PTIP complex isolated using FLAG-tagged PA1 did not contain significant amount of 53BP1 and other DNA damage response proteins (Fig. 3_C_ and Fig. 6_A_, bottom panel).
PTIP Complex Methylates Histone H3 on K4
Purified PTIP complex was analyzed in HMT assays using histone H3 or core histones as substrates. As shown in Fig. 5_A_, PTIP complex efficiently methylated histone H3 both in the isolated form and in the context of core histones. In contrast, PTIP protein alone purified by gel filtration did not possess any HMT activity (Fig. 2_A_). To determine which residue in histone H3 was methylated, recombinant histone H3 was radio-labeled in HMT assay of PTIP complex, followed by 40 cycles of Edman degradation and scintillation counting. As shown in Fig. 5_B_, PTIP complex specifically methylated K4 on histone H3. Consistent with this result, PTIP complex could methylate unmodified histone H3 peptide but not the one with trimethylation on K4 or the one with K4 mutated to Arginine (data not shown). Moreover, immunoprecipitates generated by anti-PTIP, ASH2L, and RBBP5 antibodies from HeLaS nuclear extracts carried strong HMT activities, indicating that endogenous PTIP complex was associated with robust HMT activity (Fig. 5_C_). Likewise, PA1 was associated with robust HMT activity in cells (Fig. 5_D_). Taken together, these data indicate that PTIP complex is a Set1-like HMT complex and carries robust histone H3 K4 methyltransferase activity both in vitro and in cells.
FIGURE 5. PTIP complex methylates histone H3 on K4.
A, PTIP complex methylates histone H3. Proteins immunoprecipitated by anti-FLAG M2-agarose from nuclear extracts of HeLaS cells expressing F-PTIP (left panel) or F-PA1 (right panel) were subjected to HMT assay using recombinant histone H3 or core histones as substrates. Assay products were separated on SDS-PAGE, stained with Coomassie Blue, followed by fluorography. B, PTIP complex methylates histone H3 on K4. Four _μ_g of histone H3 methylated as described for A was subjected to 40 cycles of Edman degradation and scintillation counting. Positions of histone H3 residues are indicated at the bottom. C, endogenous PTIP complex subunits are associated with robust HMT activity. HeLaS nuclear extracts were immunoprecipitated with indicated antibodies. Immunoprecipitates were subjected to HMT assay on unmodified histone H3 peptide (residues 1–20). D, nuclear extracts prepared from HeLaS cells expression F-PA1 or vector were immunoprecipitated with anti-FLAG M2-agarose followed by HMT assay. E, PTIP is not required for the enzymatic activity of the associated HMT complex. SV40T-immortalized _PTIP_flox/flox MEF cell line was infected with retroviruses expressing vector (Vec) or Cre to delete PTIP gene. Nuclear extracts were prepared and subjected to immunoprecipitation with indicated antibodies. The immunoprecipitates were subjected to HMT assay on unmodified histone H3 peptide (residues 1–20) (left panel) and Western blot analysis (right panel).
To investigate whether PTIP is required for the enzymatic activity of the associated MLL3/MLL4-containing HMT complex, anti-MLL3 antibody was used to immunoprecipitate from nuclear extracts prepared from wild type or PTIP-deficient cells. The immunoprecipitates were subjected to HMT assay and Western blot analysis. As shown in Fig. 5_E_, PTIP was not required for the histone H3 methyltransferase activity of the associated HMT complex. Anti-MLL3 antibody could still pull down endogenous RBBP5 and WDR5, suggesting that the remaining complex in the PTIP-deficient cells is largely intact. Similar results were obtained when anti-MLL4 antibody was used (data not shown). Such observation is consistent with the fact that only the MLL3 and MLL4 subunits of this complex contain the catalytic SET domain, which has been shown to carry the enzymatic activity for almost all histone lysine methyltransferases including yeast Set1 and human MLL and MLL3 (4, 13, 33).
PTIP HMT Complex Has Unique Subunit Composition
Besides the shared subunits ASH2L, RBBP5, and WDR5, and the unique SET domain-containing enzymatic subunit, each of the human Set1-like HMT complexes contains distinct subunits (Table 1) (8, 10). The subunit composition of the PTIP HMT complex was examined by Western blot analysis (Figs. 1_C_ and Fig. 6_A_). MLL, Menin, hSet1, CXXC1, _α_-tubulin, and BAF170 are unique subunits of the corresponding human Set1-like HMT complexes (8-10, 33, 34). However, none of them associated with PTIP or PA1 (Figs. 1_C_ and 6_A_, middle panels), indicating that PTIP and PA1 are unique to the PTIP HMT complex. Such result is also consistent with the absence of matching peptides of MLL, Menin, hSet1, and CXXC1 in the mass spectrometry analysis of F-PTIP- and F-PA1-associated proteins (supplemental Table S1 and data not shown).
TABLE 1.
Subunit compositions of human Set1-like histone H3 K4 methyltransferase complexes
| YeastCOMPASS/Set1complex | Human Set1-like histone H3 K4methyltransferase complexes | |||
|---|---|---|---|---|
| hSet1complex | MLLcomplex | MLL2complex | MLL3/MLL4complex | |
| Set1 | hSet1 | MLL | MLL2 | MLL3/MLL4 |
| Bre2 | ASH2L | ASH2L | ASH2L | ASH2L |
| Swd1 | RBBP5 | RBBP5 | RBBP5 | RBBP5 |
| Swd3 | WDR5 | WDR5 | WDR5 | WDR5 |
| Sdc1 | hDPY-30 | hDPY-30 | hDPY-30 | hDPY-30 |
| Swd2 | hSwd2 | |||
| Spp1 | CXXC1 | |||
| HCF1 | Menin | Menin | PTIP | |
| HCF1/HCF2 | PA1 | |||
| NCOA6 | ||||
| UTX |
To confirm the interaction between UTX and PTIP complex revealed by mass spectrometry analysis of F-PA1-associated proteins (Fig. 3_C_), and to find out whether UTX is also present in other Set1-like HMT complexes, a HeLaS cell line expressing FLAG-tagged UTX was established using retroviral gene transfer method. Nuclear extracts were prepared and subjected to immunoprecipitation using anti-FLAG M2-agarose followed by Western blot analysis. As shown in Fig. 6_B_, UTX specifically associated with PTIP but not hSet1 and Menin, suggesting that UTX is unique to the PTIP complex (Fig. 6_B_). Taken together, our data indicate that PTIP complex is distinct from any of the previously reported Set1-like HMT complexes and has unique subunit composition.
hDPY-30 Binds ASH2L Directly and Is a Common Subunit of All Human Set1-like HMT Complexes
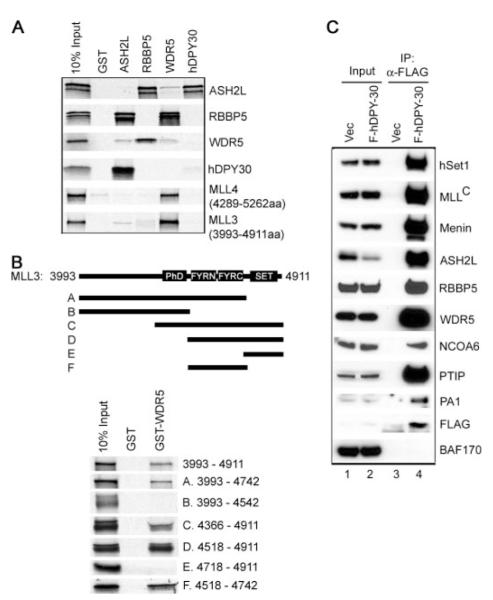
To investigate the protein-protein interactions among PTIP HMT complex components, GST pull-down assays were performed by incubating GST-fused ASH2L, RBBP5, WDR5, and hDPY-30 with in vitro translated selected subunits of the PTIP HMT complex (Fig. 7_A_). It has been shown previously that ASH2L, RBBP5, and WDR5 constitute a subcomplex that binds directly with the SET domain-containing COOH-terminal fragment of the enzymatic subunit MLL in the MLL complex (13). We confirmed direct interactions among ASH2L, RBBP5, and WDR5 and found that WDR5 bound directly with the COOH-terminal SET domain-containing fragments of MLL3 (3,993–4,911 amino acids) and MLL4 (4,289 –5,262 amino acids). Furthermore, WDR5 was found to interact with the FYRN and FYRC domain-containing region on MLL3 (Fig. 7_B_).
FIGURE 7. hDPY-30 binds ASH2L directly and is shared by all human Set1-like complexes.
A, hDPY-30 binds ASH2L directly. To characterize the direct interactions among PTIP complex components, GST or GST-ASH2L, -RBBP5, -WDR5, and -hDPY-30 were incubated with in vitro translated, [35S]methionine-labeled ASH2L, RBBP5, WDR5, hDPY-30, and the COOH-terminal SET domain-containing fragments of MLL3 and MLL4 in GST pull-down assay. B, WDR5 interacts with the FYRN- and FYRC-domain-containing region of MLL3. Top panel, schematic diagram of deletion mutations of the COOH-terminal 3,993– 4,911 amino acids of MLL3. Lower panel, GST or GST-WDR5 was incubated with in vitro translated, [35S]methionine-labeled deletion mutations of the COOH-terminal fragment of MLL3 in GST pull-down assay. C, hDPY-30 is shared by all human Set1-like complexes. Nuclear extracts prepared from HeLaS cells stably expressing FLAG-tagged hDPY-30 (F-hDPY-30) were incubated with anti-FLAG M2-agarose. Immunoprecipitates were analyzed by Western blot using antibodies against distinct subunits (hSet1, MLL, Menin, NCOA6, PTIP, and PA1) or shared subunits (ASH2L, RBBP5, and WDR5) of various human Set1-like complexes.
hDPY-30 is the human homolog of the Sdc1 subunit of yeast COMPASS/Set1 complex and has been shown to associate with the MLL2 complex (8). In GST pull-down assay, hDPY-30 directly interacts with ASH2L but not RBBP5 or WDR5, suggesting that hDPY-30 may be shared by multiple human Set1-like HMT complexes (Fig. 7_A_). To test this possibility, a HeLaS cell line stably expressing FLAG-tagged hDPY-30 was established using retroviral gene transfer method. Nuclear extracts were prepared and subjected to immunoprecipitation using anti-FLAG M2-agarose followed by Western blot analysis. As shown in Fig. 7_C_, hDPY-30 was found to associate with hSet1, MLL, Menin, PTIP, NCOA6, PA1 in addition to ASH2L, RBBP5, and WDR5 in cells, indicating that the evolutionarily conserved hDPY-30 is a shared subunit of all human Set1-like complexes.
DISCUSSION
As a first step toward the understanding of the normal cellular function of BRCT domain-containing protein PTIP, we isolated PTIP-associated proteins from cells cultured in the absence of DNA damage agent treatment. We show (i) that while ectopically expressed PTIP is capable of interacting with 53BP1 and other DNA damage response proteins, endogenous PTIP associates with MLL3- and MLL4-containing Set1-like complex; (ii) that PTIP complex carries robust HMT activity in vitro and in cells. By Edman degradation analysis, we show unequivocally that PTIP complex methylates K4 residue of histone H3; (iii) that a novel nuclear protein PA1 binds PTIP directly and PTIP HMT complex can be isolated in one step through FLAG-tagged PA1; (iv) that PTIP, PA1, and UTX, a JmjC domain-containing putative histone demethylase, associate specifically with the PTIP HMT complex; (v) that in addition to ASH2L, RBBP5, and WDR5, hDPY-30 is a common subunits shared by all human Set1-like HMT complexes, thus further establishing the evolutionary conservation between human Set1-like complexes and the yeast COMPASS/Set1 complex; (vi) that PTIP HMT complex has unique subunit composition. As histone H3 K4 methylation associates with active genes, and Xenopus PTIP has been implicated in transcriptional regulation (36), our data suggest that PTIP and associated Set1-like HMT complex may be involved in the regulation of gene expression through modulation of histone H3 K4 methylation.
Consistent with previous reports (24, 25), we show that ectopically expressed F-PTIP is capable of interacting with 53BP1 and other DNA damage response proteins in HeLaS cells cultured under normal condition (Fig. 1). However, such an interaction is clearly due to the overexpression of F-PTIP, as endogenous PTIP does not interact significantly with 53BP1 and other DNA damage response proteins in cells without DNA damage agent treatment (Fig. 4_A_). Consistent with this, PTIP HMT complex purified through FLAG-tagged PA1 contains little DNA damage response proteins (Fig. 6_A_, bottom panel). It will be interesting to determine whether the endogenous PTIP and other subunits of the PTIP HMT complex interact with 53BP1 in response to DNA damage agent treatment.
In yeast, a single COMPASS/Set1 complex is responsible for mono-, di-, and trimethylation on histone H3 K4 (4). However in humans, there exist multiple Set1-like histone H3 K4 methyltransferase complexes with overlapping but distinct subunit compositions (summarized in Table 1). Each of these human Set1-like HMT complexes contains one or two enzymatic subunits that carry the signature SET domain, such as hSet1, MLL, and MLL2. PTIP complex contains two SET domain-containing large proteins, MLL3 (4,911 residues) and MLL4 (5,262 residues), although the specific contributions of each of them to the HMT activity and functions of the entire complex remain to be determined. Recently, Lee et al. (19) showed data suggesting that MLL3 and MLL4 may exist in separate HMT complexes. We cannot rule out the possibility that PTIP complex consists of two HMT complexes with one contains MLL3 and the other contains MLL4. However, PTIP and PA1 clearly associate with both MLL3 and MLL4 in cells (Figs. 1 and 3).
PTIP complex also contains ASH2L, RBBP5 and WDR5, the three subunits shared by all human Set1-like complexes. The identification of hDPY-30, homolog of the Sdc1 subunit of the yeast COMPASS/Set1 complex, in PTIP complex further indicates that PTIP complex is a bona fide Set1-like complex. hDPY-30 binds directly with ASH2L (Fig. 7_A_), consistent with the observation that within the yeast COMPASS/Set1 complex, Sdc1 interacts directly with Bre2, which is the yeast homolog of ASH2L (37). ASH2L, RBBP5, and WDR5 have been shown to form a subcomplex interacting with the SET domain-containing COOH terminus of MLL (12, 13). We show hDPY-30 is also shared by various human Set1-like HMT complexes and WDR5 binds directly with the COOH-terminal SET domain-containing fragments of MLL3 and MLL4 (Fig. 7_A_). These data suggest that hDPY-30, ASH2L, RBBP5, and WDR5 may form a four-subunit subcomplex that directly interacts, likely through the WDR5 subunit, with the SET domain-containing histone methyltransferases of Set1-like complexes and regulates the enzymatic activities of these complexes.
Besides the SET domain-containing enzymatic subunits and the shared ASH2L, RBBP5, WDR5, and hDPY-30 subunits, each of the human Set1-like HMT complexes contains unique or distinct subunits (Table 1). For example, hSwd2 and CXXC1 have been shown to specifically associate with human Set1 (hSet1) complex, while Menin is shared by MLL and MLL2 complexes only (8, 10). PTIP, PA1, NCOA6, and UTX specifically associate with PTIP complex (Figs. 1_C_ and 6). MLL, hSet1 and CXXC1, Menin, _α_-tubulin, and BAF170, which are subunits unique to the previously reported human Set1-like HMT complexes (8-10, 20, 33, 34), are not present in PTIP complex, indicating that PTIP complex is distinct from any of the previously isolated Set1-like HMT complexes and has unique subunit composition (Fig. 6).
PTIP complex shares several subunits with the ASC-2 complex that also contains both MLL3 and MLL4 (33). However, there are clear distinctions between the two complexes. First, hDPY-30, PTIP, PA1, and UTX are present in PTIP complex but not in ASC-2 complex. Second, _α_- and _β_-tubulins are components of ASC-2 complex (33). However, they are clearly absent from PTIP complex. Last and most important, PTIP complex shows robust histone H3 K4 methyltransferase activity (Fig. 5). On the contrary, ASC-2 complex only shows very weak methyltransferase activity toward histone H3, and the residue being methylated remains undefined (33).
Recently, a partially purified complex that minimally contained MLL4, ASH2L, RBBP5, and WDR5 was found to interact with estrogen receptor α and regulate transcriptional activity of estrogen receptor α (38). It is unclear whether PTIP and other subunits of the PTIP HMT complex are present in this complex and whether this complex carries any HMT activity.
The roles of PTIP and its tandem BRCT domains in the complex remain to be investigated. PTIP is clearly not required for the enzymatic activity of the associated HMT complex. Also, the remaining complex in the absence of PTIP seems largely intact (Fig. 5_E_). PTIP was cloned as a Pax2 transactivation domain-interacting protein (22). Xenopus PTIP has been shown to physically interact with Smad2 via its BRCT domains and function as a transcriptional coactivator for SMAD2 (36). We speculate that PTIP may interact with transcription factors that bind directly to promoters and facilitate the recruitment of the PTIP HMT complex to target gene promoters. The tandem BRCT domains in PTIP has been shown to bind phosphopeptide (25). It is conceivable that PTIP may interact with other subunits within the complex or with other proteins that associate with the PTIP complex in a phosphorylation-dependent manner.
Considerable evidence suggests functional differences among human Set1-like HMT complexes. First, these complexes contain distinct subunits that are linked to diverse biological functions. However, it remains to be determined how the distinct subunits, either alone or in combination, help define the specific functions for the associated complex. Second, the SET domain-containing enzymatic subunits of these complexes carry distinct domains in addition to the COOH-terminal catalytic SET domain. For example, BROMO domain, which recognizes acetylated histone lysine residues, is only found in MLL protein, suggesting the potential link between histone acetylation event and MLL complex. Yeast Set1 protein contains RNA recognition motif that is required for histone H3 K4 trimethylation but not dimethylation (39). In human, RNA recognition motif is only present in hSet1 but not in MLL, MLL2, MLL3, and MLL4, suggesting these complexes may have different abilities in carrying out mono-, di-, and trimethylation on histone H3 K4.
Recently, yeast COMPASS/Set1 complex has also been implicated in the regulation of chromosome segregation during mitosis through methylation of kinetochore protein Dam1 (40), a function independent of histone methylation and transcriptional regulation. Ectopically expressed PTIP interacts with 53BP1, a master regulator of cellular response to DNA damage, and translocates to DNA double strand break foci upon ionizing radiation, suggesting that PTIP may be involved in DNA damage response and repair (24, 25). It will be interesting to test whether the endogenous PTIP complex and the associated HMT activity are involved in DNA damage response, a function distinct from its potential role in transcriptional regulation.
Supplementary Material
Supplementary data
Acknowledgments
We thank J. Reddy for NCOA6/PRIP cDNA, G. Nolan for Phoenix retrovirus packaging cell lines, S. Agarwal for sharing reagents, and T. Blankenstein for pBabeCREpuro. We thank D. Forrest and S. Simons for critical reading of the manuscript.
Footnotes
*
This work was supported by the National Institutes of Health and by the Intramural Research Program of the NIDDK (to K. G.).
3
The abbreviations used are:
HMT
histone methyltransferase
BRCT
BRCA1 carboxyl-terminal
GST
glutathione _S_-transferase
MEFs
mouse embryonic fibroblasts
PTIP
Pax transactivation domain-interacting protein
MALDI
matrix-assisted laser desorption ionization
◆
This article was selected as a Paper of the Week.
_Addendum_—Issaeva et al. (Mol. Cell. Biol. (2007) 27, 1903) recently reported that the MLL4 (ALR) HMT complex contains PTIP.
REFERENCES
- 1.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 3.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noma K, Grewal SI. Proc. Natl. Acad. Sci. U. S. A. 2002;99(Suppl 4):16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. Proc. Natl. Acad. Sci. U. S. A. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, Kay GF, Hayward NK, Hess JL, Meyerson M. Mol. Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 9.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Mol. Cell. Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 12.Steward MM, Lee J-S, O’Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 13.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 14.Sierra J, Yoshida T, Joazeiro CA, Jones KA. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Development (Camb.) 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 16.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 17.Huntsman DG, Chin SF, Muleris M, Batley SJ, Collins VP, Wiedemann LM, Aparicio S, Caldas C. Oncogene. 1999;18:7975–7984. doi: 10.1038/sj.onc.1203291. [DOI] [PubMed] [Google Scholar]
- 18.Prasad R, Zhadanov AB, Sedkov Y, Bullrich F, Druck T, Rallapalli R, Yano T, Alder H, Croce CM, Huebner K, Mazo A, Canaani E. Oncogene. 1997;15:549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Lee D-K, Dou Y, Lee J, Lee B, Kwak E, Kong Y-Y, Lee S-K, Roeder RG, Lee JW. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Skalnik DG. J. Biol. Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 21.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lechner MS, Levitan I, Dressler GR. Nucleic Acids Res. 2000;28:2741–2751. doi: 10.1093/nar/28.14.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho EA, Prindle MJ, Dressler GR. Mol. Cell. Biol. 2003;23:1666–1673. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jowsey PA, Doherty AJ, Rouse J. J. Biol. Chem. 2004;279:55562–55569. doi: 10.1074/jbc.M411021200. [DOI] [PubMed] [Google Scholar]
- 25.Manke IA, Lowery DM, Nguyen A, Yaffe MB. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 26.Kim D, Wang M, Cai Q, Brooks H, Dressler GR. J. Am. Soc. Nephrol. 2007;18:1458–1465. doi: 10.1681/ASN.2006060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todaro GJ, Green H. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 29.Kalkum M, Lyon GJ, Chait BT. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2795–2800. doi: 10.1073/pnas.0436605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, Halazonetis TD, Harris CC. J. Cell Biol. 2004;166:801–813. doi: 10.1083/jcb.200405128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Bosch M, Bree RT, Lowndes NF. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan MA, Samuels HH. Endocr. Rev. 2005;26:583–597. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- 33.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, Roeder RG, Azorsa DO, Meltzer PS, Suh PG, Song EJ, Lee KJ, Lee YC, Lee JW. Mol. Cell. Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. Mol. Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 35.Klose RJ, Kallin EM, Zhang Y. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu K, Bourillot P-Y, Nielsen SJ, Zorn AM, Gurdon JB. Mol. Cell. Biol. 2001;21:3901–3912. doi: 10.1128/MCB.21.12.3901-3912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dehe P-M, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, Shevchenko A, Rosaleny LE, Tordera V, Chavez S, Stewart AF, Geli V. J. Biol. Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 38.Mo R, Rao SM, Zhu Y-J. J. Biol. Chem. 2006;281:15714–15720. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 39.Schlichter A, Cairns BR. EMBO J. 2005;24:1222–1231. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data