SNPs in human miRNA genes affect biogenesis and function (original) (raw)
Abstract
MicroRNAs (miRNAs) are 21–25-nucleotide-long, noncoding RNAs that are involved in translational regulation. Most miRNAs derive from a two-step sequential processing: the generation of pre-miRNA from pri-miRNA by the Drosha/DGCR8 complex in the nucleus, and the generation of mature miRNAs from pre-miRNAs by the Dicer/TRBP complex in the cytoplasm. Sequence variation around the processing sites, and sequence variations in the mature miRNA, especially the seed sequence, may have profound affects on miRNA biogenesis and function. In the context of analyzing the roles of miRNAs in Schizophrenia and Autism, we defined at least 24 human X-linked miRNA variants. Functional assays were developed and performed on these variants. In this study we investigate the affects of single nucleotide polymorphisms (SNPs) on the generation of mature miRNAs and their function, and report that naturally occurring SNPs can impair or enhance miRNA processing as well as alter the sites of processing. Since miRNAs are small functional units, single base changes in both the precursor elements as well as the mature miRNA sequence may drive the evolution of new microRNAs by altering their biological function. Finally, the miRNAs examined in this study are X-linked, suggesting that the mutant alleles could be determinants in the etiology of diseases.
Keywords: microRNA, miRNA, SNP, miRNA-polymorphism, miR-SNP, miR-TS-SNP
INTRODUCTION
MicroRNAs (miRNAs) are emerging as one of the most interesting small regulatory, noncoding RNAs in molecular biology. They are single stranded, about 22-nucleotides (nt) long, evolutionarily conserved, and perform functions equivalent to those of transcription factors by switching off or fine tuning target gene expression. It is believed that they function largely, though not exclusively, through base pairing to the complementary sequences in the 3′ untranslated region (3′UTR) of target genes to suppress translation. Generally, they can repress translation by guiding the miRNA-mediated RNA-induced silencing complex (miRISC) binding to the 3′UTR of a complementary message, disrupting translation initiation or elongation (Filipowicz et al. 2008). Several lines of evidence suggest that they may perform their function in the RNA processing bodies (P-bodies) by sequestering target transcripts to P-bodies for storage, decapping, deadenylation, and degradation (Liu et al. 2005; Rehwinkel et al. 2005; Chu and Rana 2006).
miRNA genes are scattered among each of the chromosomes in humans, except for the Y chromosome. They primarily derive from intronic or exonic capped, polyadenylated RNA polymerase II transcripts, termed “primary” miRNAs (pri-miRNA) (Lee et al. 2002, 2003; Cai et al. 2004). Mature miRNAs are generated by a two-step processing mechanism (Supplemental Fig. S1). pri-miRNAs are first processed to “hairpin-like,” partially duplexed “precursor” miRNAs (pre-miRNA) in the nucleus. Aside from a small group of pre-miRNAs that are generated through mRNA splicing/debranching machinery termed the “miRtron pathway” (Berezikov et al. 2007; Okamura et al. 2007; Ruby et al. 2007), most pre-miRNAs are processed from pri-miRNAs by the nuclear ribonuclease (RNase) III Drosha, which partners with the RNA-binding protein DiGeorge syndrome critical region gene 8 (DGCR8) (Lee et al. 2003; Han et al. 2004). pre-miRNAs are typically 55–80 nt in length and are exported to the cytoplasm by exportin-5/RAN-GTP (Yi et al. 2003). The pre-miRNAs are processed into ∼21–22-nt long miRNA/miRNA* duplexes by RNase III Dicer, which partners with the RNA-binding protein transactivation response (TAR) element RNA-binding protein (TRBP) (Chendrimada et al. 2005; Haase et al. 2005). The production of miRNA/miRNA* duplexes is an essential step in miRNA biogenesis and precisely defines the ends of the mature miRNAs for preferential loading of the guide strand (Khvorova et al. 2003). The choice of the guide strand is dependent in part on the thermodynamic end properties of the duplex, with the least thermodynamically stable 5′ end usually being chosen as the guide strand, while the other strand, labeled miRNA*, is usually degraded (Ruvkun 2001; Hutvagner and Zamore 2002; Khvorova et al. 2003; Schwarz et al. 2003). Most recently, the fates of the miRNA guide and miRNA* strands have been shown to be tissue dependent (Ro et al. 2007), with both strands being functionally active under specific conditions (Okamura et al. 2008). Argonaute-mediated loading into the processing complex can increase the bias of strand loading (Seitz et al. 2008), and RNA-binding proteins can selectively block the processing of pri-miRNAs (Piskounova et al. 2008; Viswanathan et al. 2008).
The mature miRNAs are used to guide miRISC to the complementary sequences in the 3′UTR of targeted transcripts. The result is site-specific mRNA cleavage when the pairing is nearly complete (mostly in plants, rare in animals) or translational inhibition when imperfect base pairing occurs (mostly in animals) (Bartel 2004). For translational suppression, Watson–Crick base pairing between six or seven consecutive nucleotides in the target mRNA's 3′UTR and nucleotides 2–7 or 2–8 (the “seed sequence”) of the miRNA's 5′ end is required. The critical role played by the “seed sequence” in the majority of miRNA/mRNA interactions implies that a single nucleotide change in the seed sequence, or shift of the processing sites during biogenesis of the miRNA/miRNA* duplex could result in a novel miRNA with alternated target-spectra. Therefore, both the 5′ end of the mature miRNA that is generated from the 5′ arm of the pre-miRNA (5p) by Drosha, and the 5′ end of the mature miRNA that is produced by Dicer from the 3′ arm of the pre-miRNA (3p), will be under strong selective pressure to be highly conserved. The sequence preceding the 5′ end or trailing the 3′ end of the pre-miRNAs form an ∼11-base-pair (bp)-long imperfect stem that is recognized by DGCR8 as part of the required structure for Drosha cutting (Han et al. 2004, 2006). The terminal loop is also important for Dicer/TRBP complex binding (Zeng 2006), as well as for other protein binding (Viswanathan et al. 2008). Sequences outside of the seed in the mature miRNA sequence can also impact the strength of inhibition as well as the spectra of targeted transcripts.
The hairpin structure-guided miRNA processing, the thermodynamic influences on strand loading, and the base-pairing requirements for miRNA/mRNA interaction imply that single nucleotide polymorphisms (SNPs) in miRNA genes should affect miRNA biogenesis and function. Similarly, SNPs in the miRNA target also affect miRNA function. To clarify possible confusions in terminology with respect to SNPs, we use the terms “miR-SNP” to refer to the variation that occurs in the miRNA gene sequence, and “miR-TS-SNP” to refer to SNPs that occur in the miRNA target site (TS) or binding site. Since one miRNA can have multiple targets, miR-SNPs would be expected to exhibit more profound and broader biological effects than miR-TS-SNPs.
The roles that sequences flanking the pre-miRNA play in miRNA processing have been thoroughly studied (Han et al. 2004; Zeng an Cullen 2004, 2005; Zeng et al. 2005; Han et al. 2006). miR-SNPs in miR-125a and Kaposi's sarcoma-associated herpes virus-encoded miR-K5 were reported to impair miRNA processing by the Drosha/DGCR8 complex (Gottwein et al. 2006; Duan et al. 2007). miR-196a2-SNP (rs11614913) in the mature miR-196a2 was reported to be associated with a significantly decreased rate of survival in individuals with non-small cell lung cancer, and the investigators of this study also suggested an association of rs11614913 with enhanced processing of mature miR-196a (Hu et al. 2008a). miR-146a-SNP (rs2910164) within the pre-miR-146a sequence reduced both the amount of pre- and mature miR-146a, and apparently affected the Drosha/DGCR8 processing step (Hu et al. 2008b; Jazdzewski et al. 2008; Shen et al. 2008). miR-196a2-SNP, miR-146a-SNP, miR-149-SNP (rs2292832), and miR-499-SNP (rs3746444) are each associated with increased breast cancer risk (Hu et al. 2008b). miR-146a-SNP was associated with papillary thyroid carcinoma (Jazdzewski et al. 2008), breast/ovarian cancer (Shen et al. 2008), and hepatocellular carcinoma (Xu et al. 2008). The miR-146a-SNP and hepatocellular carcinoma report is somewhat controversial since it claims that the SNP enhanced miR-146a expression, while three other reports show reduced expression. Each of the above are examples of SNPs created by changes in DNA-coding sequences, but miRNAs can also be post-transcriptionally modified, such as by RNA editing via ADAR. A to I-edited pre-miR-151 blocks its processing by Dicer/TRBP (Kawahara et al. 2007a). ADAR-edited pri-miR-142 was more easily degraded by Tudor-SN (Yang et al. 2006). Edited miR-376a-5p within the middle of the “seed” region alters the set of targets regulated by this miRNA (Kawahara et al. 2007b). A survey of RNA editing of miRNAs from 10 human tissues implies RNA editing of miRNA happens quite often and it is a mechanism to increase the diversity of miRNAs and their targets (Blow et al. 2006).
The above examples show that mutant or post-transcriptionally edited miRNAs can result in alterations of processing and function. Hence, SNPs that occur in sequences downstream from or upstream of the pre-miRNA, sequences in the terminal loop of pre-miRNA, and sequences in the miRNA and miRNA* duplexes may also play important roles in miRNA biogenesis and function (Supplemental Fig. S1).
We have been analyzing X-linked miRNA genes from male patients with diagnosed schizophrenia or autism, and compared our findings with a gene pool analysis consisting of over 7000 chromosomes from normal individuals. The statistical correlation of SNPs in miRNAs with Schizophrenia is presented in a separate study (Feng et al. 2009). The present study represents the first large-scale, systematic genetic analysis of naturally occurring miR-SNPs. Twenty-four different point mutations have been determined in either the mature miRNA sequences or the precursor regions for sixteen different X-linked miRNA genes. Here, we address the effects on miRNA generation and function generated by SNPs in X-linked miRNAs.
Of the tested miR-SNPs, one variant resulted in elevated levels of the mature miRNA sequence, several variants resulted in reduced levels of the mature miRNA sequence relative to wild type, and another variant resulted in the generation of a novel miRNA due to an alteration in the Drosha and/or Dicer processing sites. This latter miRNA-SNP also had an alteration in the strand-loading bias relative to the wild-type version. Our results demonstrate that a single base alteration even outside of the mature miRNA sequence can have profound consequences on miRNA generation and function.
RESULTS
miRNA variants
A total of 59 microRNA genes on the X chromosome (covered in miRBase 8–10 when the experiments were conducted; the other 18 X-linked miRNAs in the current miRBase 12.0 were not available at the time of this study) were analyzed in 193 male schizophrenia patients and 191 male controls. Some variants were further screened by analyzing whether or not they were present in 7000 control chromosomes from normal individuals.
Twenty-four variants within pre-miRNAs and the immediate flanking regions were identified (Table 1). They are further characterized into four groups based on the location within the hairpin structure (Supplemental Fig. S1).
TABLE 1.
List of all miRNA gene variants that were found in the control population or the patient samplesa
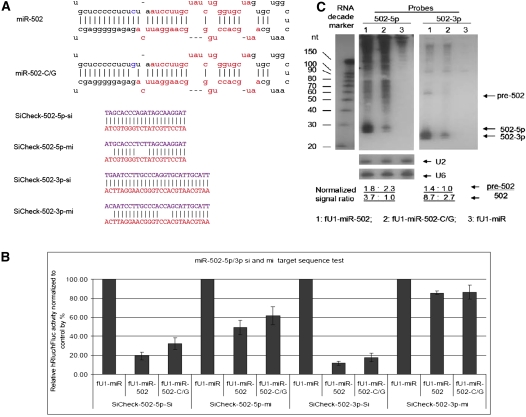
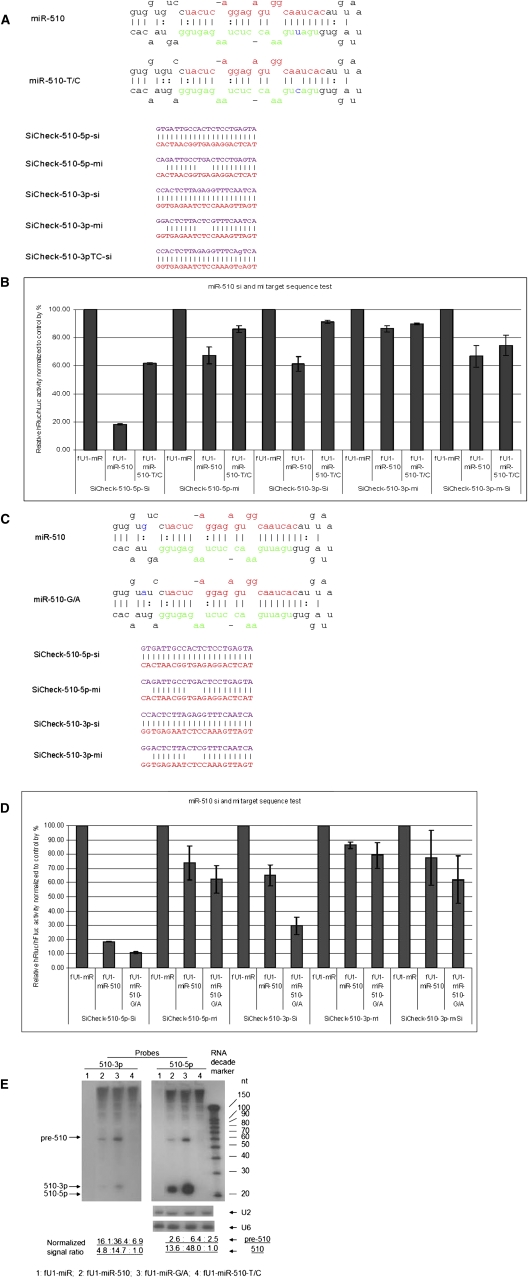
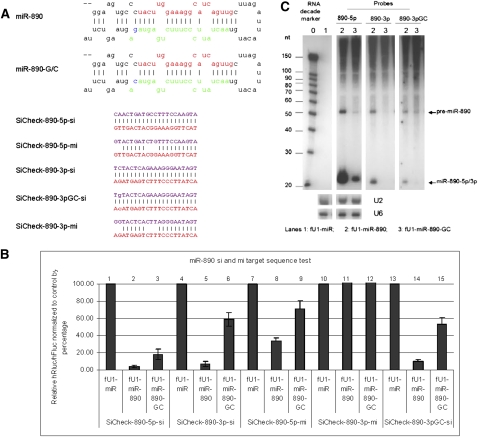
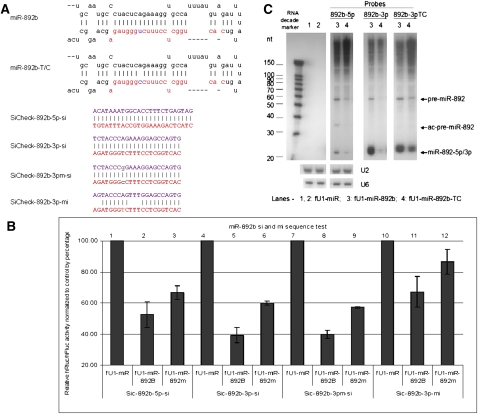
In order to evaluate the consequences of these point mutations, we have developed a facile assay to monitor the processing and function of both strands of the miRNAs by using both miRNA (seed sequence complementarity) and siRNA (fully complementary) assays. The functional assays were carried out in transient cotransfections of expressed pri-miRNA with target sequences in the 3′UTR of the Renilla luciferase encoding transcripts. At least six of the variants, miR-502-C/G (Fig. 1A, rare variant and associated with schizophrenia), miR-510-T/C (Fig. 2A, rare variant and associated with schizophrenia), miR-510-G/A (Fig. 2C), miR-890-C/G (Fig. 3A), miR-892b-T/C (Fig. 4A), and miR-934-T/G (Fig. 5A), each showed reduced or enhanced repression of the “si” and “mi” reporters in transient transfection assays. For each of these variants, Northern blotting was performed to detect the effects of the SNP on processing of the pre- and mature miRNAs. Mature miRNA cloning was performed on miR-510-G/A, miR-890-C/G, and miR-934-T/G to study the affect of the SNP on the maturation of the miRNAs. While all three SNPs apparently affect the miRNA processing, we observed that only the miR-934-T/G transversion also altered the Drosha or Dicer excision sites (Supplemental Table S1), which also resulted in changing the strand bias for RISC loading relative to the wild-type miRNA (Fig. 5).
FIGURE 1.
Functional test of miR-502 and miR-502-C/G. (A) Sequences of miRNAs and reporters. Stem–loop sequences of miR-502, miR-502-C/G, and the sequences inserted into the 3′UTR of Rluc for the “si” and “mi” reporters. (B) Cotransfection test results. The knockdown of 5p-si, 5p-mi, and 3p-si reporters is reduced in the mutant, whereas the expression of 3p-mi reporter is the same in the WT and Mutant. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (C) Northern blot results. Blot was hybridized with miR-502-3p probe (right) and a miR-502-5p probe (left). (Lane 1) RNAs from cells transfected with the WT miRNA construct; (lane 2) samples from cells transfected with the Mutant miRNA construct; (lane 3) RNAs from cells transfected with the miRNA expression vector fU1-miR. U2 and U6 snoRNA was used as an RNA loading control. Quantification of the signal was first normalized to U2, and then it was further normalized to the band with the lowest signal.
FIGURE 2.
Functional test of miR-510, miR-510-T/C, and miR-510-G/A. (A) Sequences of miRNAs and reporters. Stem–loop sequences of miR-510, miR-510-T/C, and the sequences inserted into the 3′UTR of Rluc for the “si” and “mi” reporters. (B) Transfection test results of T/C mutant. The knockdown of reporters for 5p-si, 5p-mi, 3p-si, and 3p-m-si (mutant form) from the mutant form are reduced. The repression for 3p-mi is approximately the same for both the WT and the mutant. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (C) Sequences of miRNAs and reporters. Stem–loop sequences of miR-510, miR-510-G/A, and the inserted sequences into the 3′UTR of Rluc for the “si” and “mi” reporters. (D) Transfection test results of miR-510-G/A. The knockdown of reporters for 5p-si, 5p-mi, 3p-si, and 3p-m-si (G/A mutant form) from the mutant are all increased. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (E) Northern blot results. Blot was probed with the miR-510-5p probe (right) and miR-510-3p probe (left); U2 and U6 snoRNAs were used as RNA sample loading controls. (Lane 1) Transfected with fU1-miR; (lane 2) transfected with miR-510 WT; (lane 3) transfected with the miR-510-T/C mutant; (lane 4) transfected with fU1-miR-510-G/A. Quantification of the signal was first normalized to U2, and then it was further normalized to the band with the lowest signal.
FIGURE 3.
Functional test of miR-890 and miR-890-G/C. (A) Sequences of miRNAs and reporters. Stem–loop sequences of miR-890, miR-890-G/C, and the sequences inserted into the 3′UTR of Rluc for the “si” and “mi” reporters. (B) Transfection test results. The knockdown of reporters for 5p-si, 5p-mi, 3p-si, and 3pGC-si (mutant form) from the mutant form are reduced. The expression of the 3p-mi reporter is approximately the same for both the WT and mutant. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (C) Northern blot results. Blot was probed with miR-890-5p, 3pGC, and 3p for samples transfected with fU1-miR (lane 1), fU1-miR-890 (lane 2), and fU1-miR-890-G/C (lane 3). U2 and U6 snoRNAs were used as RNA loading controls.
FIGURE 4.
Functional test of miR-892b and miR-892b-T/C. (A) Sequences of miRNAs and reporters. Stem–loop sequences of miR-892b, miR-892b-T/C, and the sequences inserted into the 3′UTR of Rluc for the “si” and “mi” reporters. (B) Transfection test results. The knockdown of reporters for 5p-si, 3p-si, 3pm-si (mutant form), and 3p-mi from the mutant form are reduced. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (C) Northern blot results. Blot was hybridized with probes for miR-892b-5p, 3p, and 3pTC (mutant form) using samples transfected with fU1-miR (lane 1), fU1-miR-892b (lane 2), and fU1-miR-892b-T/C (lane 3). U2 and U6 snoRNAs were used as RNA loading controls.
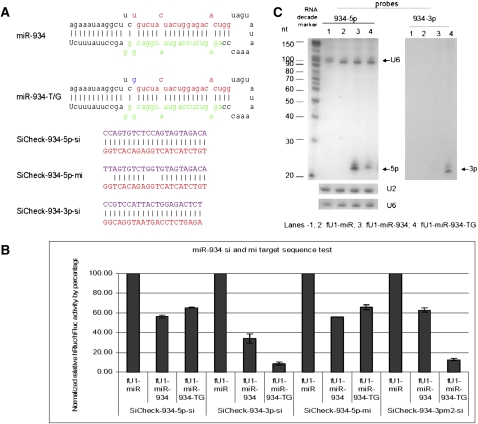
FIGURE 5.
Functional test of miR-934 and miR-934-T/G. (A) Sequences of miRNAs and reporters. Stem–loop sequences of miR-934, miR-934-T/G, and the sequences inserted into the 3′UTR of Rluc for the “si” and “mi” reporters. (B) Transfection test results. The knockdown of 5p-si and 5p-mi reporters by the mutant were reduced and strong knockdown of the 3p-si and 3pm2-si reporters are observed from the mutant miRNA. Each bar represents the average of at least three independent transfections, with duplicate determinations for each construct. Error bars represent the SD. (C) Northern blot results. Blot was probed with a miR-934-5p probe (left) and 3p probe (right). U2 and U6 snoRNAs were probed as RNA gel loading controls. (Lanes 1,2) From cells transfected with fU1-miR; (lane 3) from cells transfected with fU1-miR-934; (lane 4) from cells transfected with the fU1-miR-934-T/G.
Variant miRNAs impair miRNA processing
In our tests, we observed several examples in which miR-SNPs resulted in reduced processing. Four of the six observed miR-SNPs, miR-502-C/G, miR-510-T/C, miR-890-C/G, and miR-892b-T/C, produced less-detectable mature miRNAs than the wild-type versions of these miRNAs. While the 502-G/C SNP occurs 2 nt before the 5′ end of 502-5p, the other three SNPs all occur in the mature 3p products.
Variant miR-502-C/G: This variant produces a bulge that changes the structure of the stem of the precursor miRNA (Fig. 1A). Most likely, this structural change affects the site of Drosha cleavage in producing pre-miR-502. Reduced target knockdowns were observed in the transfection assays (Fig. 1B). The impaired functional activity of the variant was supported by Northern blot analysis, as the production of pre-miR-502 and mature 502-5p/3p was both reduced (Fig. 1C).
Variant miR-510-T/C: Currently there are no published reports to validate the informatic prediction of the miR-510-3p products. Our 3p reporter transfection assays show its ability to knockdown the corresponding “si” target sequence (Fig. 2B). The T/C transition produces a pre-miR-510 with much less activity for both 5p and 3p products (Fig. 2B). Therefore, this mutation affects the function of both strands of mature miR-510. Northern blot analyses confirmed that the production of both pre-miR-510 and miR-510-5p/3p are reduced (Fig. 2E).
Variant miR-890-C/G: Currently there are no published data to validate the predicted miR-890 3p products. Our 3p reporter transfection assays show its ability to knockdown the corresponding “si” target sequence. Transfection and Northern blotting data show that the C/G transversion in miR-890 affects the production of both the 3p and the 5p strands (Fig. 3B,C) with the production of both strands being reduced. Because the C/G transversion may be at the Drosha cleavage site, we wanted to define the exact sequence of its 3p products by miRNA cloning. The cloning data show that the cutting sites for both the 5p and 3p products were not altered by this miR-SNP (Supplemental Table S1). The 5p and 3p mature sequences in the cloning data were the same for both the wild-type and mutant. Dot blotting analyses (Supplemental Fig. S4b) also show that more clones of the miR-890 than miR-890-C/G for both the 3p and 5p probes were obtained, which is consistent with the transfection and Northern blot data (Fig. 3B,C).
Variant miR-892b-T/C: like miR-510-T/C and miR-890-C/G, this SNP occurs in the 3p of miR-892b, but miR-892b-3p is listed in miRBase. Although there are currently no published data that support the existence of miR-892b-5p products, our 5p reporter transfection assays show its ability to knockdown the corresponding “si” target sequence. Transfection and Northern blotting data show that the T/C transition in miR-892b affects the production of both 5p and 3p strands (Fig. 4B,C).
Variants in miRNA that enhance processing of mature miRNAs
We were surprised to find that a G/A transition in pri-miR-510 enhanced the production of miR-510-5p and -3p (-3p is miR-510*) (Fig. 2D,E). The G-to-A transition occurs at the fourth nucleotide upstream of the 5′ end of the mature miR-510-5p (Fig. 2C). Variants at this position may affect Drosha processing of this substrate, since it may provide a more stable stem preceding the mature miRNA sequence. The reporter assay data show that the siRNA activity of the mutant is markedly higher than the wild type (Fig. 2D). Northern blot data show that the production of both pre-miR-510 and mature miR-510-5p/3p are increased (Fig. 2E). Dot-blotting data also revealed more clones of the miR-510-G/A than miR-510 (Supplemental Fig. S4A). The miRNA cloning data show that the generation of the 5p product is the same for both the wild type and mutant, and apparently this SNP does not affect the Drosha cutting sites (Supplemental Table S1). However, we didn't observe any colonies hybridizing to the 3p probe in either the wild-type or SNP blots; therefore, the exact sequence of miR-510-3p is unknown.
Variant miRNA with altered mature miRNA processing sites and miRISC strand loading bias
It is reasonable that SNPs could alternate Drosha or Dicer excision sites, since their cutting sites are structure based and not sequence based. Variant miR-934-T/G occurs at the first nucleotide of the miR-934-5p (Fig. 5A), which is also the Drosha processing site. Because the variant occurs at the 5′ end and the base of the 5′end plays an important role in strand selection into miRISC, the T/G transversion of this variant is particularly interesting. Transfection and Northern blot results show that this SNP affects the production of both strands (Fig. 5B,C). First, the transfection assay shows that repression of the 5p reporter is reduced by the SNP, and Northern blots confirmed the reporter assay results (Fig. 5B). Second, the length of the 5p product seems to be increased in the Northern blot. The most dramatic changes are in the 3p product. Transfections show that repression of the 3p reporter by the SNP is increased, and Northern blots show that the variant produces more 3p than wild type. Thus, the guide strand and passenger strand in miRISC are inverted in the wild-type versus mutant miRNAs. Cloning also yielded more 5p wild-type clones and more mutant 3p clones (Supplemental Fig. S4c). The cloning data also show that the production of 3p is altered, with both the Drosha and Dicer cutting sites being offset by one nucleotide from the wild type, resulting in a different 3p product (Supplemental Table S1). This not only produced a novel miRNA, but it also affected the strand selection in miR-934/miR-934*. The wild-type miR-934-5p starts with a U and is most likely selected as the predominant guide strand due to the lower thermodynamic stability of the 5′ end. The U/G transversion changes the first nucleotide of the 5p product to “G<” which affected the Dicer cutting site, moving it back one nucleotide from the original “G” to an “A.” Thus, the 3p product in the mutant has a lower 5′ end thermodynamic stability, and this is probably responsible for altered guide strand selectivity, which is consistent with the reported requirements for asymmetric strand loading (Schwarz et al. 2003).
DISCUSSION
miRNAs play an essential role in various biological functions including development, cell differentiation, proliferation, viral pathogenesis, and progression of human diseases (Bartel 2004; Bushati and Cohen 2007). Spatial and temporal expression of individual miRNAs, plus the synergistic effects of miRNAs that bind to the same target, make miRNAs a family of regulatory factors that can subtly and precisely modulate target gene expression. The function of miRNAs can be compromised by variations in their sequences (Gottwein et al. 2006; Duan et al. 2007) or their target-site sequences (Kawahara et al. 2007b; Saunders et al. 2007). Our data are consistent with other published reports which show that there are more SNPs that impair miRNA generation than SNPs that enhance miRNA generation. It is of interest that some miR-SNP-related reports did not observe obvious aberrant processing of miR-SNPs, perhaps owing to the subtle changes that such SNPs can have on the processing reactions (Diederichs and Haber 2006; Chen et al. 2007). In contrast, changes in miRNA-binding sites (miR-TS-SNPs) are readily associated with loss of miRNA function (Abelson et al. 2005; Clop et al. 2006; Arisawa et al. 2007; Martin et al. 2007; Mishra et al. 2007; Sethupathy et al. 2007; Yu et al. 2007).
miR-SNPs may contribute to the evolution of miRNAs with new or altered functions. miR-SNPs may reveal a mechanism for the generation of clustered miRNAs, miRNA homologs, or family members of miRNAs during evolution. Most miR-SNPs we observed are located in clusters and some of them, such as mir-510 and miR-509, rapidly evolved in primates (Zhang et al. 2007). One clear example is the generation of miR-509-3-5p by the deletion of an “A” from miR-509-5p, which is processed from miR-509-1 or 2 (Supplemental Fig. S2). There are three copies of miR-509, miR-509-1, and miR-509-2 that produce the same mature miRNAs, while miR-509-3 produces a different 5p product. Most likely, the miR-509-3-5p was created by the deletion of an “A” from miR-509-5p. We also observed a high percentage of an ATG insertion in the 5′ end of miR-509-1-3p. This insertion may affect both 5p and 3p processing. Eventually, under selective pressure to target different mRNAs or to target with different specificities, the three copies of miR-509 may have developed into different family members with the same seed, like the let-7 family, or different miRNAs in the same cluster, like the miR-25-93-106b cluster. It appears to us that the miR-509 structure is more flexible, since we did not observe significant differences in the processing or function among three different miR-509-3 variants (Supplemental Fig. S3).
Many factors may contribute to differences in miRNA expression profiles, including transcriptional regulation, post-transcriptional miRNA processing, the stability of the pri-miRNA or pre-miRNA, and pre-miRNA export. The existence of miRNA targets may also result in miRNA stabilization because of engagement in miRISC. Our data clearly show different miRNA profiles as a consequence of subtle genetic changes in pre-miRNAs and their immediate flanking sequences.
Previous in silico studies from Bentwich et al. (2005) and Zhang et al. (2007) show that miRNA family expansion during primate evolution may have occurred through tandem duplication. Copy-number variations and high rates of gene conversion in the newly emerged miRNAs in primates may have resulted in production of novel miRNAs with more specialized functions. As a result, gene conversion may be a major mechanism in the biogenesis of miRNAs during evolution, especially in clusters of miRNAs, homologs, or miRNA families. Finally, some of the SNPs that we have characterized by altered processing or abundance may play significant roles in disease development and progression.
MATERIALS AND METHODS
Cell lines and plasmids
HEK293 cells were purchased from ATCC and were maintained in high glucose (4.5 g/L) DMEM supplemented with 2 mM glutamine, 10% FBS, and 2 mM Penicillin/Streptomycin. Transfections to HEK293 cells were performed with Lipofectamine 2000 (Invitrogen) in duplicate in 24-well plate formats with cells at 80% confluency.
Cell-based miRNA processing test
Primary miRNA expression plasmids and reporter constructs with either fully complementary targets or seed sequences complementary to the miRNAs were cotransfected into HEK293 cells. Dual-reporters (expressing both Firefly Luciferase [Fluc] and Renilla Luciferase [Rluc]) carrying the fully complementary miRNA targets (“si” reporters) in the 3′UTR of the Renilla transcript were constructed. These were used to validate the expression level and the ability of cloned primary miRNA expression plasmids to produce functional, mature miRNAs. Dual-reporters carrying the partially complementary sequence (“mi” reporters: sequences that are complementary to the mature miRNA sequence except for mismatches at positions 11–13 and the last two nucleotides in the miRNA/mRNA duplex) of a miRNA in the Rluc 3′UTR were used to quantitatively measure the translational repression from the corresponding miRNA.
In order to express the pri-miRNAs, we retrieved the predicted stem–loop sequences from miRBase (Griffiths-Jones et al. 2008). The stem–loop sequence plus flanking sequences extending at least 100 bases in both directions were PCR amplified from patient genomic DNAs when possible. An miRNA expression vector was constructed by cloning the human Pol II U1 promoter upstream of a multiple cloning site in the Bluescript SK plasmid to create the plasmid SK-U1. Secondly, the U1 transcriptional termination sequence was cloned downstream of the MCS of SK-U1 to create the fU1-miR miRNA expression vector. The primary miRNA sequence was cloned into the XhoI and BamHI sites of fU1-miR. miRNA variants were cloned in the same manner as the wild-type miRNAs from sample DNA when available. If samples were no longer available, the QuikChange Site-Directed Mutagenesis Kit II (Stratagene) was used to create mutants within the wild-type expression constructs.
All miRNAs and their homologous mutant “si/mi” reporters were generated by inserting annealed oligos into the psiCheck2.2 XhoI/SpeI sites or XhoI/NotI sites in the 3′UTR of the Rluc.
About 5 × 104 HEK293 cells per well in 500 μL of growth media were plated in 24-well plates 1 d prior to transfection. The cells were at 80% confluency at the time of transfection. Each well was transfected with 5 ng of reporter, 100 ng of miRNA expression constructs (1:20 reporter/miRNA ratio; 1:5, 1:1 or 5:1 ratio was used if the knock down of the “si” target was very efficient, e.g., >95%: 1:5 as 25 ng of miRNA expression plasmid and 75 ng of stuffer Blue-script SK) and 1 uL of Lipofectomine 2000. Forty eight hours post transfection, the cells were lysed with 100 μL of Passive Lysis Buffer (Promega) and Luciferase levels were analyzed from 20 μL of lysates using the Dual Luciferase reporter assay (50 μL of each substrate reagent, Promega) on a Veritas Microplate Luminometer (Turner Biosystems). Changes in expression of Rluc (target) were calculated relative to Fluc (internal control) and normalized to the miRNA expression vector control (fU1-miR).
Mutagenesis of miRNA target sites
When the DNA from patient samples containing the SNPs was not available, the appropriate changes were introduced into the wild-type sequences using the QuikChange Site-Directed Mutagenesis Kit II (Stratagene) following the protocol provided in the kit. Mutations were confirmed by sequencing.
RNA isolation, Northern blot, and mature miRNA cloning
RNA isolation, Northern blot, and small RNA cloning were carried out as previously reported (Sun et al. 2007). Briefly, RNA was isolated with RNA STAT-60 (Tel-Test, Inc.) and 20 μg of total RNA was loaded into a denaturing 12.5% SDS–polyacrylamide gel. A DNA oligonucleotide probe complementary to the mature miRNA sequence was labeled with [γ-32P]ATP. For Northern blots, at least two independent transfections with different preparation of plasmids were performed in HEK293 cells to detect processing of expressed pri-miRNAs. One transfection contained pri-miRNA expression constructs alone, while the other one was cotransfected with a control 25/27-mer dicer substrate siRNA that targets HIV Tat/Rev as transfection efficiency control (only one set of the data was showed in the figures). The hybridization was performed overnight in PerfectHyb Plus Hybridization Buffer from Sigma. The blots were washed once for 10–30 min with 6x SSPE/0.1% SDS, followed by two washings with 6x SSC/0.1% SDS for 10–30 min each. U2 or U6 snoRNA were used as RNA loading controls. For small RNA cloning, small RNAs below 40 nt were fractionized by a flashPAGE Fractionator System. Small RNAs were first polyadenylated, then ligated with a 5′ RNA adaptor. The 5′-adaptor-added polyadenylated small RNAs were RT-PCR amplified and the products cloned. Dot-blot hybridizations were used to identify positive clones. The positively hybridizing clones were sequenced to verify the processed mature miRNA sequences.
Dot blotting
Bio-Rad membranes were cut to the same size as the bottom of Petri-Dish plates. The membranes were laid on the colonies for 20 sec or until they were wet, then lifted and washed twice in 0.5N NaOH for 5 min each (the plates were put back into the 37°C incubator for 5–6 h to preserve the colonies). Next, the membranes were washed twice in 0.5M Tris-HCl (pH 7.5) for 5 min each. Then, the membranes were washed twice in 6x SSC/0.1% SDS for 5 min each. Finally, the membranes were washed in 95% EtOH for 5 min and dried between two sheets of Whatman paper. All washings were performed at room temperature. Just before hybridization, membranes were soaked in 6x SSPE/0.1% SDS twice for 5 min each. The probe and the temperature of hybridization and the washing condition were the same as those for the Northern blots above. The only difference was that the hybridization duration was 1 h. Usually the signal is strong enough to detect after the blots are exposed to film for 5–6 h. Positive colonies were located and plasmid DNAs were made for sequencing.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI29329 and HL07470 to J.J.R. We wish to thank Brain Luk for critical reading of this manuscript.
Footnotes
REFERENCES
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Hasegawa S, Takagi T, Wang FY, et al. A polymorphism of microRNA 27a genome region is associated with the development of gastric mucosal atrophy in Japanese male subjects. Dig Dis Sci. 2007;52:1691–1697. doi: 10.1007/s10620-006-9648-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-20060704-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jensen LR, Gecz J, Fryns JP, Moraine C, de Brouwer A, Chelly J, Moser B, Ropers HH, Kuss AW. Mutation screening of brain-expressed X-chromosomal miRNA genes in 464 patients with nonsyndromic X-linked mental retardation. Eur J Hum Genet. 2007;15:375–378. doi: 10.1038/sj.ejhg.5201758. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Sequence variations of microRNAs in human cancer: Alterations in predicted secondary structure do not affect processing. Cancer Res. 2006;66:6097–6104. doi: 10.1158/0008-5472.CAN-06-0537. [DOI] [PubMed] [Google Scholar]
- Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS. Evidence for X-chromosomal schizophrenia associated with microRNA alterations. PLoS One. 2009;4:e6121. doi: 10.1371/journal.pone.0006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Cai X, Cullen BR. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol. 2006;80:5321–5326. doi: 10.1128/JVI.02734-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008a;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Carcinogenesis. 2008b;29:2341–2346. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007a;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007b;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mishra PJ, Humeniuk R, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both microRNAs and their miRNA strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: A mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Ambrosone CB, Dicioccio R, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the mir-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- Sun G, Li H, Rossi JJ. Cloning and detecting signature microRNAs from mammalian cells. Methods Enzymol. 2007;427:123–138. doi: 10.1016/S0076-6879(07)27007-7. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, Su H, Zhuang SM. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, Wu M, Shen SH. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Peng Y, Wang W, Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]