3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy (original) (raw)
. Author manuscript; available in PMC: 2009 Sep 21.
Published in final edited form as: Nature. 2008 Apr 30;453(7193):415–419. doi: 10.1038/nature06893
Abstract
Cytoplasmic polyhedrosis virus (CPV) is unique within the Reoviridae family in having a turreted single-layer capsid contained within polyhedrin inclusion bodies, yet being fully capable of cell entry and endogenous RNA transcription1–4. Biochemical data have shown that the amino-terminal 79 residues of the CPV turret protein (TP) is sufficient to bring CPV or engineered proteins into the polyhedrin matrix for micro-encapsulation5,6. Here we report the three-dimensional structure of CPV at 3.88Å resolution using single-particle cryo-electron microscopy. Our map clearly shows the turns and deep grooves of α-helices, the strand separation in β-sheets, and densities for loops and many bulky side chains; thus permitting atomic model-building effort from cryoelectron microscopy maps. We observed a helix-to-β-hairpin conformational change between the two conformational states of the capsid shell protein in the region directly interacting with genomic RNA. We have also discovered a messenger RNA release hole coupled with the mRNA capping machinery unique to CPV. Furthermore, we have identified the polyhedrin-binding domain, a structure that has potential in nanobiotechnology applications.
Three-dimensional structures of several sub-viral particles in Reoviridae have been determined at atomic resolutions by X-ray crystallography7–9. Those structures have provided a wealth of information about the unity and diversity of viral organization, double-stranded (ds)RNA genome packing and transcription, and mRNA processing and releasing. For example, nascent mRNA of bluetongue viruses is capped within the capsid shell before release7,10,11, but orthoreoviruses release their nascent mRNA into a turret chamber, where it is subsequently capped8. Both of these viruses are thought to release their mRNA from the capsid shell through five-fold axial holes on icosahedral vertices. The orthoreovirus shell protein λ1 alone cannot assemble into icosahedral particles12. For CPV, however, previous studies have shown that the apical pentameric pore is blocked for mRNA release13 and the capsid shell protein (CSP) alone can assemble into virus-like particles in vitro14. In addition, there is a CPV-unique haemagglutinin-like protein (that is, the ‘A spike’) situated on the top of each turret4. In the absence of an atomic model of the full CPV virion, many fundamental issues have remained elusive, including the structural basis for RNA packing, transcription, processing and release, and the mechanism underlying CPV’s unique property of polyhedra embedding. In contrast to CPV polyhedrin protein, which forms diffraction-quality micro-crystals even in vivo6,15, CPV virions have eluded X-ray crystallographic efforts, probably due to the intrinsic flexibility of the haemagglutinin-like A spikes.
To address some of these issues, we attempted to push the resolution limit of single-particle cryo-electron microscopy. Focal pairs of 16-megapixel charge-coupled device (CCD) images of CPV were recorded at liquid-nitrogen temperature in a 300-kV FEI cryoelectron microscope (Supplementary Fig. 1a, b). Rigorous screening of images was carried out to select cryo-electron microscopy images with visible contrast transfer function (CTF) rings beyond 5Å and without noticeable specimen drift, charging, or image astigmatism. The final map (Fig. 1a and Supplementary Movie 1) was reconstructed by merging 12,814 particles from only the close-to-focus images with a B-factor16–18 of 35Å 2 and underfocus values ranging from 0.15 to 1.3 µm (Supplementary Table 1).
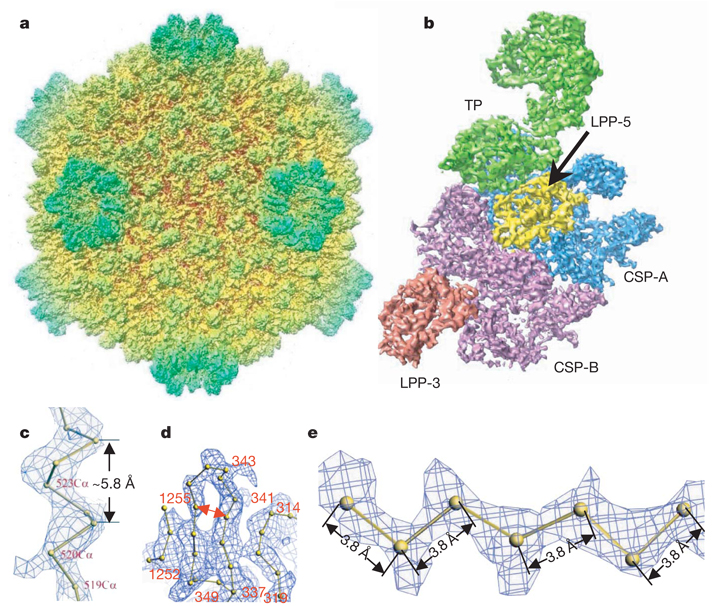
Figure 1. Overall structure of the CPV capsid.
a, Radially coloured, shaded surface view of the CPV reconstruction as viewed along a two-fold axis. b, An extracted asymmetric unit, colour coded by protein subunits, including one turret protein (TP) (green), two copies of the capsid shell proteins (CSP-A in blue and CSP-B in purple), and two copies of the large protrusion protein (LPP-5, near the five-fold axis, in yellow; LPP-3, near the three-fold axis, in brown). c, d, Views of density maps of one α-helix (c) and four β-strands (d) for CSP-B superimposed with the corresponding Cα model, showing the clear turn and deep groove of the α-helix with a pitch of ~5.8Å and the density for the bulky side chain of residue 520 in c, and the clear separation of β-strands in d (note also the densities for the bulky side chains of amino acids 341 and 343). e, Cα model of a β-strand of TP in the density map, showing zigzagging of the backbone and the clear densities for side chains. All the inter-consecutive Cα distances are ~3.8Å , which is a well established distance in polypeptide chains.
The effective resolution of our map was estimated to be better than 4Å by examining the structural features revealed by the map, such as clear turns and deep grooves of helices, strand separation in β-sheets, a zigzagging feature of polypeptide backbones in β-strands, and densities of bulky side chains (Fig. 1c– Fig. 1e, Supplementary Fig. 2 and Supplementary Movies 2 and 3), which were comparable to those revealed in crystallographic density maps at a similar resolution19. In addition to its resolving power, the quality of the map was also convincingly verified by its accuracy, such as the pitch in α-helices being ~5.8Å (Fig. 1c) and the inter-consecutive Cα distances in polypeptide chains being ~3.8Å (Fig. 1e). This assessment is consistent with the 3.88Å resolution, as estimated by the criterion of 0.5 Fourier shell correlation coefficient (Supplementary Fig. 1c). Finally, throughout the reconstruction, we only applied icosahedral averaging. Thus, the close similarities of the densities for CSP-A and CSP-B (see below), as well as the densities of the non-symmetry-related two large protrusion protein (LPP) molecules, both at an overall structural level and for a variety of detailed features, serve as an excellent internal control for the high quality of the density map (Fig. 1b and Supplementary Figs 3–5).
The 750-Å diameter CPV has a _T_=1 icosahedral capsid shell decorated with 12 turrets on the five-fold vertices (Fig. 1a). The asymmetric unit consists of two CSPs, two LPPs and one TP (Fig. 1b, Supplementary Fig. 3 and Supplementary Movie 4). As with other members of Reoviridae, the structures of which have been determined to atomic resolution by X-ray crystallography7–9, the CSPs of CPV have two conformational states: CSP-A and CSP-B. Although no sequence homology was detectable, our cryo-electron microscopy structures of CPV CSPs show an overall thin-plate-like shape similar to that of the orthoreovirus λ1 without obvious domain boundaries7,8, with the exception of the unique small protrusion domain (SPP) (Fig. 2a and Supplementary Figs 4 and 5). The identified secondary structures of CPV CSP (for example, the helices in the apical domain and the β-sheets in the dimerization domain) all have topological counterparts in λ1, although their detail orientations differ locally, indicating that CPV CSP has a fold similar to that of λ1. Using the crystallographic modelling tool O (ref. 20), we have built Cα models for the two CSPs without the SPP (Fig. 2a and Supplementary Figs 4 and 5), two LPPs (Supplementary Fig. 6), and two N-terminal domains of TP based on our density map. At the current resolution, accurate registration of residues remains challenging and, consequently, residue numberings were tentative.
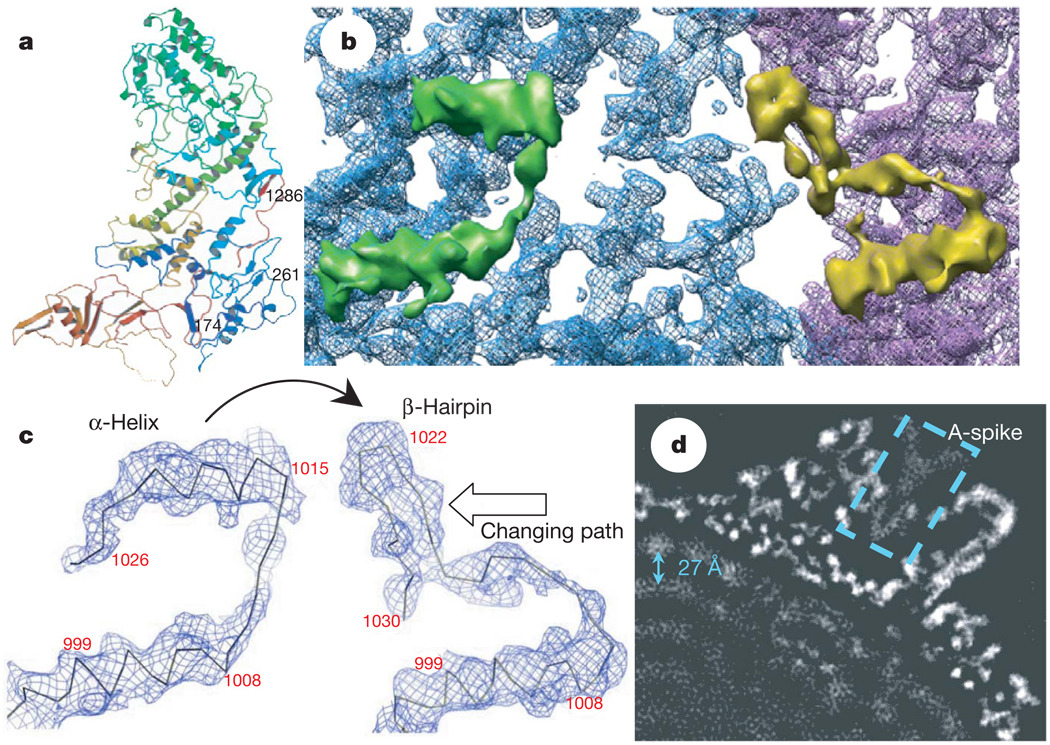
Figure 2. A conformational change between CSP-A and CSP-B: implication for packing and sliding of the dsRNA genome.
a, Atomic model of CSP-A, coloured from blue at the N terminus to red at the C terminus. b, Close-up view of the CSP-A (blue) and CSP-B (purple) density map, showing that an α-helix (the upper helix within the green density) in CSP-A transforms into part of a β-hairpin in CSP-B (the upper part in the yellow density). c, Density maps with Cα models, showing the conformational change between CSP-A (left) and CSP-B (right). One α-helix in CSP-A transforms into part of the β-hairpin in CSP-B (indicated by curved arrow). The changing path is also indicated by an empty arrow. d, A 10Å slab extracted from the two-fold map showing the ordered dsRNA genome with a ~27Å distance between the adjacent dsRNA strands (arrow). The dotted box indicates the A-spike plug all the way in the central chamber of the turret.
A marked local conformational change has been identified between the two conformers CSP-A and CSP-B in their regions interacting with viral RNA (Fig. 2), in addition to a conserved global conformational swing about a pivot in the middle7,8 (Supplementary Figs 4 and 5). Whereas residues 1014–1024 form an α-helix in CSP-A, the same fragment exists as part of a β-hairpin (residues 1017–1029) in CSP-B. The α-helix of CSP-A is connected to another approximately parallel α-helix (residues 996–1009) through a 4-residue loop (residues 1010–1013). In CSP-B, this loop is rotated about 45° clockwise (viewed from inside the capsid) and subsequently pushes the connected β-hairpin to rotate ~90° clockwise and backward ~20Å to the edge of the carapace domain to interact with its adjacent CSP-A (Fig. 2b, c). The genomic dsRNA is coiled and tightly packed within the capsid with a multilayer arrangement21. The outermost RNA layer interacts with the inner side of the capsid shell (Fig. 2d and Supplementary Fig. 7). The α-helix (residues 1014–1024) of CSP-A is the most inward component and most likely to be involved in the interaction with viral RNA. The functional implications of the observed conformational change remain unknown; however, the interactions with viral RNA genome suggest an association with RNA packaging, replication and transcription (See ‘Discussion’ in the Supplementary Information).
Uncapped nascent mRNA is unstable and degrades prematurely if released into the cytoplasm. Viruses in the Reoviridae overcome this problem by capping nascent mRNAs before releasing them. Whereas viruses with non-turreted cores cap their mRNA within the capsid shell, turreted viruses cap their mRNA in the turret outside the capsid shell. CPV TP differs from orthoreovirus λ2 despite sharing similar domain architectures. The λ2 pentamer of orthoreovirus is a hollow cylinder with five flaps forming a lid on the top8. The TP pentamer of CPV is a hollow cone with a wide opening on the top, as TP lacks the immunoglobulin fold flap domains of the orthoreovirus λ2. Instead, the central chamber of the CPV turret is plugged all the way down by its haemagglutinin-like protein A-spike (dotted box in Fig. 2d and yellow in Supplementary Fig. 8a, b and Supplementary Movie 6), which confers the CPV’s infectivity. The A-spike adopts a shape complementary to that of the inner space of the turret and totally blocks the central channel at the lower part of the turret, leaving only a peripheral space of ~20Å in the middle to upper regions of the turret (Fig. 2d, Supplementary Fig. 8a and Supplementary Movie 6). In keeping with this arrangement of the turret, the CSPs of CPV do not form a single hole at each five-fold axis for mRNA release, like orthoreovirus8, but instead form five holes surrounding the five-fold axis, similar to those seen in a low-resolution structure of the rotavirus22. In CPV, each of those holes is ~10Å in diameter, formed by the apical domains of two CSP-As and one CSP-B (Fig. 3a), and is immediately adjacent to the active-site cleft of the guanylyltransferase (GTase) of TP (Fig. 3b, c and Supplementary Fig. 8c). Notably, the narrowest region of each hole is formed by three loops (Fig. 3a).
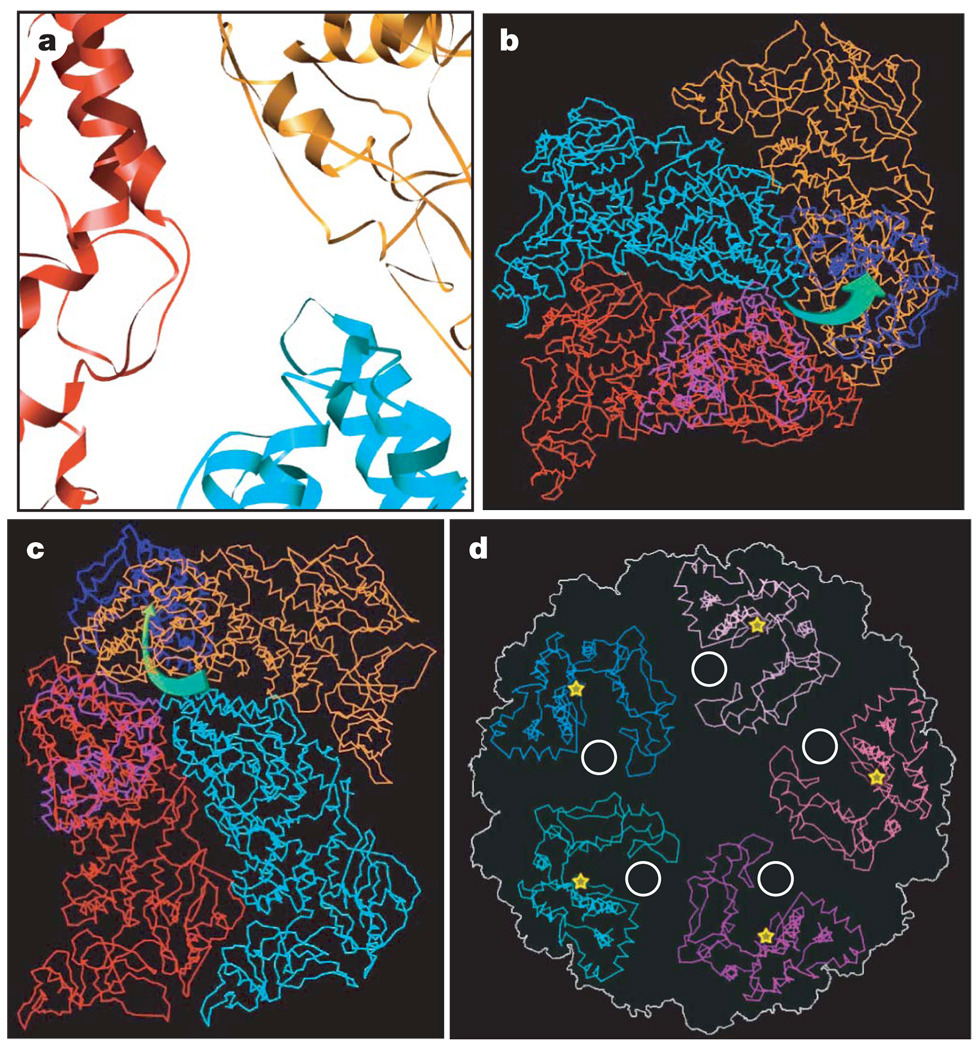
Figure 3. Nascent mRNA release hole coupled with the GTase active site of TP in a way unique to CPV.
a, Close-up view of the atomic model of CSP-A (two copies) and CSP-B. The mRNA release hole comprises loops contributed by the two copies of CSP-A (red and orange) and one CSP-B (cyan). b, c, Cα models of one CSP-B (cyan), two copies of CSP-A (red and orange) and two GTase domains (blue and light blue), showing the mRNA releasing and capping pathway, which is illustrated by green arrows in b (viewed from outside of the capsid) and c (viewed from inside of the capsid). d, Cα models of the GTase domains in a turret, showing that the active-site cleft (yellow stars) of GTase rotates away from the central chamber, facing sideways, thus coupled with the mRNA release hole (white circle).
Consistent with this difference in the location of the putative mRNA release hole, the structure of the CPV GTase domain, which catalyses the first step of RNA capping, also differs from that of orthoreovirus. Although CPV and orthoreovirus belong to the same family, the CPV GTase domain is rather like the GTase of the completely unrelated chlorella virus PBCV-1 (ref. 23) in having two sub-domains, forming a deep and narrow cleft (Fig. 3d and Supplementary Fig. 9). On the basis of the structural resemblance, we speculate that the active site of the CPV GTase is on the larger sub-domain, opening to the cleft. Rather than facing the central chamber, as for the orthoreovirus GTase, the active-site cleft of the CPV GTase rotates away from the central chamber, thus facing sideways towards the putative RNA release hole. This configuration of GTase allows it to be coupled with the mRNA release hole in CPV (Fig. 3b–d). This configuration is different from that of orthoreovirus as mentioned above, but still ensures that the nascent mRNA bumps into the active-site cleft of GTase immediately after it exits the capsid shell, resulting in immediate and efficient capping as in the orthoreovirus (Fig. 3b–d and Supplementary Movie 7). The PBCV-1 GTase exists in two conformations: the inactive open conformation and the active closed conformation23. Notably, the CPV GTase domain resembles the inactive open conformation more closely, consistent with the fact that our structure was obtained from quiescent, non-transcribing CPV virions.
Mutagenesis studies have shown that the N-terminal 79 residues of TP are required for specific interaction of the CPV capsid with polyhedrin trimer during polyhedra-embedding or micro-encapsulation into polyhedra5,6,24. Consistent with this observation, our density map of TP shows a CPV-unique domain anchored to the N-terminal end of the GTase domain through a clearly resolved linker (Fig. 4 and Supplementary Movies 8 and 9). This domain has a palm-shaped structure with a dimension of 15Å by 20Å consisting of four β-strands (Fig. 4d, e). Because the GTase domain in orthoreovirus is the N-terminal end of the capping complex λ2, the additional domain attached to the CPV GTase domain is probably the N-terminal domain of CPV TP (that is, the CPV-unique polyhedrin- binding domain (PBD)). Indeed, our model shows that this domain is β-strand rich, which is in agreement with the secondary structure prediction from the primary structure of the TP N-terminal 79 amino acids. This domain is located on the outer surface of the CPV turret, and is thus easily accessible by the polyhedrin trimer. Its sheet-like feature apparently provides a large interaction surface, which may be the structural basis for its strong and specific binding to the polyhedrin trimer6.
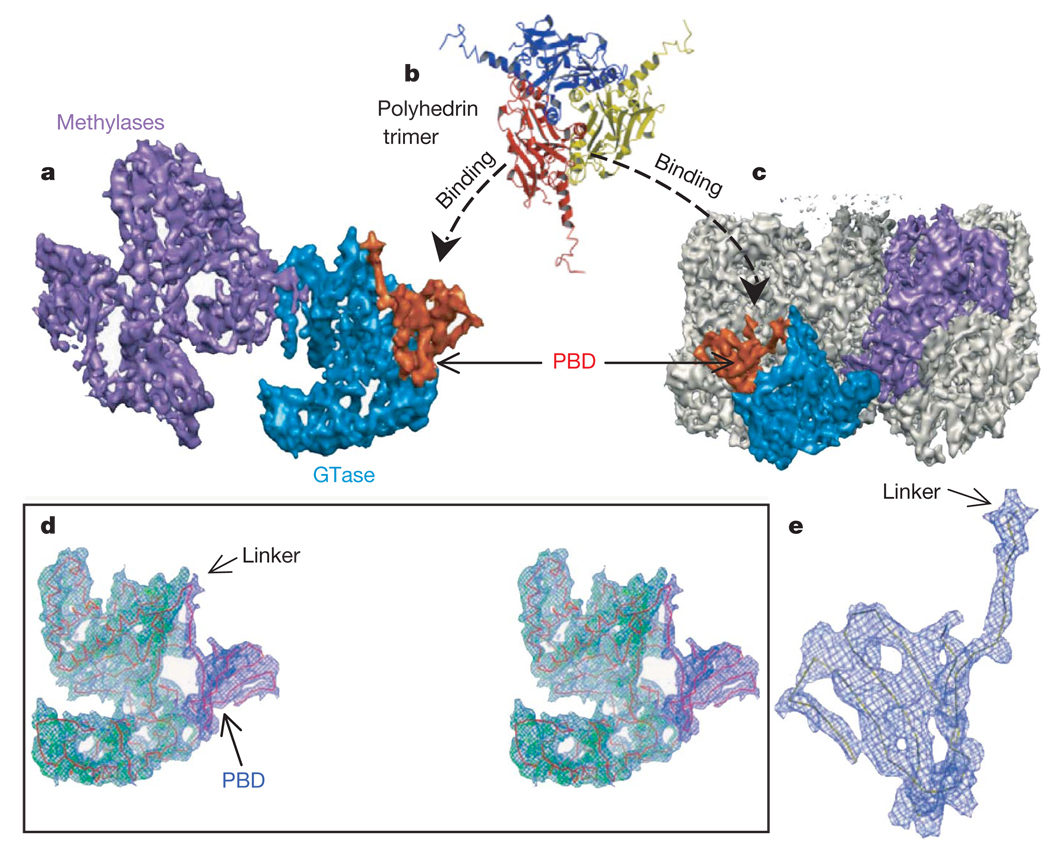
Figure 4. Location and structure of the unique polyhedrin-binding domain (PBD) of TP.
a, Shaded surface representation of TP viewed from inside, showing the two methylase domains (purple), GTase domain (blue), and CPV’s unique PBD (orange). The PBD is absent from other dsRNA viruses and consists of residues 1–79 of TP. b, Ribbon model of the CPV polyhedrin trimer, which is the building block of the crystalline polyhedra6. c, View of one entire turret, showing the orientation and position of one TP (coloured) in the turret. d, Stereo view of a cryo-electron microscopy density map superimposed with the Cα model of GTase domain (green) and PBD (blue), which are connected together through a linker. e, Blown-up view of the model of PBD superimposed with its cryo-electron microscopy density.
Because the PBD has the unique ability to bring proteins of various sizes into nano-scale inclusion bodies (polyhedra) with controllable disassembly behaviour, this would offer better systems for delivery and protein microarrays5. Our elucidation of the structural details of this domain opens doors for rational designs of such systems.
We have obtained a 3.88Å density map of CPV by single-particle cryo-electron microscopy, which is of sufficient quality for us to attempt atomic modelling and has led to three important findings (summarized in Supplementary Fig. 10). In the last decade there have been great efforts at sub-nanometre resolution in single-particle cryo-electron microscopy reconstructions16,25. Our structure was obtained from only ~13,000 particles recorded in a 300-kV microscope without an energy filter. With further advancement of cryoelectron microscopy instrumentation, the development of more sophisticated algorithms and ever-growing computational power, there is every reason to believe that we are entering the age of atomic resolution for single-particle cryo-electron microscopy, which will have profound impacts on biology, chemistry and medicine.
Note added in proof: Recently, three other papers have appeared describing near-atomic-resolution structures of the rotavirus double-shell particles26, the ε15 acteriophage27 and the GroEL complex 28. These advancements illustrate the potential towards atomicresolution structural determination by single-particle cryo-electron microscopy.
METHODS SUMMARY
The CPV virions were isolated and purified from infected Bombyx mori larvae using a previously described procedure4,13, with some modifications primarily to preserve the structural integrity for high-resolution structure determination. Focal-pair images were collected at liquid-nitrogen temperature on a 16-megapixel CCD camera (TVIPS) in an FEI Polara cryo-electron microscope operated at 300 kV with a dosage of ~20 e− per Å 2 at a magnification of ×154,380 (step size: 0.97Å per pixel). Determination of particle orientation, centre parameters and subsequent three-dimensional reconstruction were carried out using the IMIRS package29, using integrated strategies of common lines, projection matching30 and Fourier–Bessel synthesis methods31,32. Before the merging of particles for three-dimensional reconstruction, the Fourier transform values of individual images were corrected for the CTF with 15% amplitude contrast and a decay factor of 35Å 2. We estimated the effective resolution of the final reconstruction to be beyond 4Å using a structure-feature-based criterion commonly accepted in protein crystallography, such as visible turns and deep grooves for helices, strand separations in β-sheets and clear densities for bulky side chains. This is consistent with the 3.88 Å effective resolution based on the criterion of the 0.5 Fourier shell correlation coefficient between two reconstructions. Because CPV has no recognizable sequence homologies (that is,<,8% sequence identity) with orthoreovirus8, bluetongue virus7 and phytoreovirus9, experimental Cα-trace model building based on cryo-electron microscopy densities was necessary and was accomplished using O (ref. 20).
METHODS
Virus purification
In contrast to the stringent requirement on sample purity by conventional X-ray crystallography and nuclear magnetic resonance (NMR), we believe that sample integrity is more important than purity for high-resolution single-particle cryo-electron microscopy structure determination of viruses and large protein assemblies. In cryo-electron microscopy images, impurities or contaminations, if ever existing, can be distinguished from particles of interest either visually or by computational means, thus they will not have a severe impact on subsequent data processing. However, subtle structural deformation or flexibility of the complexes cannot be easily sorted out and can become a limiting factor towards high-resolution reconstruction. To avoid possible structural damages caused by multi-step sucrose-gradient centrifugation, we isolated the CPV virions from Bombyx mori larvae using a simplified protocol modified from a previous procedure4,13. Purified polyhedra were treated with an alkaline solution of 0.2M Na2CO3-NaHCO3 (pH 10.8) for 1 h. The suspension was centrifuged at 10,000_g_ for 40 min. The supernatant was collected and centrifuged at 80,000_g_ for 60 min at 4 °C to pellet the CPV virions. The pellet was then directly re-suspended in 10mM PBS (pH 7.4) and immediately flash-frozen for cryo-electron microscopy imaging. We eliminated the subsequent sucrose gradient centrifugation steps used in previous protocols4,13.
Cryo-electron microscopy imaging
The CPV virions were embedded in a thin layer of vitreous ice suspended across the holes of holey carbon films for cryo-electron microscopy imaging. The images were collected at liquid-nitrogen temperature in a 300-kV FEI Polara cryo-electron microscope with a dosage of ~20 e−per Å 2 at a magnification of 154,380 and a step size of 0.97Å per pixel. Focal pairs of micrographs were recorded on a 16-megapixel CCD camera (TVIPS) using the EMMENU software. The first close-to-focus micrographs in the focal pairs were aimed at 0.5–1.0 µm underfocus. The second micrographs were taken at 1.5 µm farther underfocus and were used only for the determination of preliminary orientation parameters. The defocus values were estimated from the positions of the CTF rings in the incoherently averaged Fourier transforms of particle images. As compared to conventional imaging on photographic films, imaging on CCD exposes a much smaller sample area, thus reducing possible beam-induced specimen movement/charging and the effect of defocus variation within each image. The latter allows a more accurate defocus value determination.
Three-dimensional reconstruction and visualization
Determination of particle orientation, centre parameters and subsequent three-dimensional reconstruction were carried out using the IMIRS package29, which uses integrated approaches of common lines32, projection matching30, Fourier–Bessel synthesis 31,32 and spherical harmonics reconstruction33 methods. Before merging of particles for three-dimensional reconstruction, the Fourier transform values of individual images were corrected for the CTF with 15% amplitude contrast and a decay factor of 35Å 2. Reconstructions were performed independently from images recorded in individual cryo-electron microscopy imaging sessions and were evaluated based on resolvability of high-resolution structural features. Many data sets were discarded owing to difficulties in pushing beyond 5Å resolution, and the final reconstruction was only from images selected from two best cryo-electron microscopy sessions. The images from these two extended sessions were independently processed to 5Å and combined for further refinement. The final refined data set was split into two halves for Fourier shell correlation calculation.
The three-dimensional reconstruction was visualized using Chimera34. The density maps were displayed by contouring at 1.5σ (standard deviation) above the average density values unless otherwise specified. Each molecular component was manually segmented out using Chimera. Molecular boundaries were well delineated by interactive three-dimensional examination of the continuity of mass densities by varying the contouring levels (1.5σ–3.0 σ).
Atomic model building and visualization
The sequences of CPV proteins are highly diversified, as no significant sequence homologies have been detected (<8% sequence identity). So far, we have not been able to perform homology modelling for CSP, LPP and TP, despite extensive efforts using a variety of bioinformatics tools. Interpretation of the final experimental density map and model building into the densities were performed using the software O (ref. 20). To facilitate polypeptide chain tracing, the density map was skeletonized in O. For CSP-A and CSP-B, Cα models were built in O according to the skeleton and following the overall topology of orthoreovirus λ1 proteins8. For the GTase domain of the TP, tentative Cα tracing was based on the density map only. Subsequently, a poly-Ala model was built from the Cα model using the lego_auto_MC command in O. Fitting of the model to the map and adjusting of clashing atoms were done manually in O. Local stereochemistry was improved using the real-space refinement (the refine_zone command) in O. Tentative registration of amino acids was based on the density map, the secondary structure profiles predicted from the primary amino-acid sequences and the overall topology. The current model contains residues 1–26, 48–275 and 291–391 of the 1,057 amino acid residues for TP; residues 174–255, 261–720, 889–1095, 1098– 1214, 1220–1255 and 1265–1287 of the 1,333 residues for CSP-A; and residues 174–255, 261–720, 889–1214, 1222–1255 and 1269–1285 of the 1,333 residues for CSP-B. This accounts for a total of 2,195 amino acids for the icosahedral asymmetric unit.
Secondary structures were clearly recognizable for the small protrusion domain of CSP, and the two methylase domains of TP, even though their backbones could not be traced from the map, probably due to their inherent flexibility. For LPP, its identity and sequence are still ambiguous. One possible candidate for LPP is the VP5 protein; however, the predicted molecular weight based on the sequence of VP5 is much larger than the density for LPP, probably as a result of proteolysis35. Therefore, those domains or molecules were not included in the current model.
Figures and Supplementary Movies were prepared using Chimera34, O (ref. 20), Molscript36 and Pymol37.
Supplementary Material
Acknowledgements
This research is supported in part by grants from NIH and the Welch Foundation. We are grateful to W. Chiu for his advice and support of this project. We thank Y. Liang, J. Jakana, M. Baker and W. Chiu for their participation at the preliminary stage of this project; J.-Q. Zhang for providing the CPV-containing polyhedra sample; I. Atanasov for assistance during cryo-electron microscopy imaging; X. Zhang for graphics illustration; and P. Lo for reading our manuscript.
Footnotes
Author Information The cryo-electron microscopy density map has been deposited to the EM Data Bank with accession codes EMD-1508. Coordinates for CSP-A, CSP-B and the N-terminal two domains of TP have been deposited in the Protein Data Bank with accession code 3CNF. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Mertens PPC, Attoui H, Duncan R, Dermody TS. In: Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. London: Elsevier/Academic Press; 2005. pp. 447–454. [Google Scholar]
- 2.Zhou ZH. In: Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology. Patton JT, editor. Norfolk: Caister Academic Press; 2008. pp. 27–43. [Google Scholar]
- 3.Hill CL, et al. The structure of a cypovirus and the functional organization of dsRNA viruses. Nature Struct. Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, et al. Visualization of protein-RNA interactions in cytoplasmic polyhedrosis virus. J. Virol. 1999;73:1624–1629. doi: 10.1128/jvi.73.2.1624-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeda K, et al. Immobilization of diverse foreign proteins in viral polyhedra and potential application for protein microarrays. Proteomics. 2006;6:54–66. doi: 10.1002/pmic.200500022. [DOI] [PubMed] [Google Scholar]
- 6.Coulibaly F, et al. The molecular organization of cypovirus polyhedra. Nature. 2007;446:97–101. doi: 10.1038/nature05628. [DOI] [PubMed] [Google Scholar]
- 7.Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 8.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa A, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Gouet P, et al. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 11.Diprose JM, et al. Translocation portals for the substrates and products of a viral transcription complex: the bluetongue virus core. EMBO J. 2001;20:7229–7239. doi: 10.1093/emboj/20.24.7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu P, Miller SE, Joklik WK. Generation of reovirus core-like particles in cells infected with hybrid vaccinia viruses that express genome segments L1, L2, L3,and S2. Virology. 1993;197:726–731. doi: 10.1006/viro.1993.1648. [DOI] [PubMed] [Google Scholar]
- 13.Zhou ZH, Zhang H, Jakana J, Lu X-Y, Zhang J-Q. Cytoplasmic polyhedrosis virus structure at 8Å by electron cryomicroscopy: structural basis of capsid stability and mRNA processing regulation. Structure. 2003;11:651–663. doi: 10.1016/s0969-2126(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 14.Hagiwara K, Naitow H. Assembly into single-shelled virus-like particles by major capsid protein VP1 encoded by genome segment S1 of Bombyx mori cypovirus 1. J. Gen. Virol. 2003;84:2439–2441. doi: 10.1099/vir.0.19216-0. [DOI] [PubMed] [Google Scholar]
- 15.Di X, Sun Y-k, McCrae MA, Rossmann MG. X-ray powder pattern analysis of cytoplasmic polyhedrosis virus inclusion bodies. Virology. 1991;180:153–158. doi: 10.1016/0042-6822(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 16.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 17.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou ZH, Chiu W. Determination of icosahedral virus structures by electron cryomicroscopy at subnanometer resolution. Adv. Protein Chem. 2003;64:93–124. doi: 10.1016/s0065-3233(03)01003-9. [DOI] [PubMed] [Google Scholar]
- 19.Inaba K, et al. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 20.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Xia Q, Jakana J, Zhang JQ, Zhou ZH. Structural comparisons of empty and full cytoplasmic polyhedrosis virus: protein-RNA interactions and implications for endogenous RNA transcription mechanism. J. Biol. Chem. 2003;278:1094–1100. doi: 10.1074/jbc.M205964200. [DOI] [PubMed] [Google Scholar]
- 22.Lawton JA, Estes MK, Prasad BV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nature Struct. Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 23.Hakansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda K, et al. Molecular characterization of Bombyx mori cytoplasmic polyhedrosis virus genome segment 4. J. Virol. 2001;75:988–995. doi: 10.1128/JVI.75.2.988-995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway JF, et al. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl Acad. Sci. USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang W, et al. Backbone structure of the infectious ε15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 28.Ludtke SJ, et al. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution3Dreconstruction package integrated with a relational image database. J. Struct. Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 30.Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 31.Crowther RA, Amos LA, Finch JT, DeRosier DJ, Klug A. Three dimensional reconstructions of spherical viruses by Fourier synthesis from electron micrographs. Nature. 1970;226:421–425. doi: 10.1038/226421a0. [DOI] [PubMed] [Google Scholar]
- 32.Crowther RA. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. R. Soc. Lond. B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, et al. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J. Struct. Biol. 2008;161:64–73. doi: 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Hagiwara K, Matsumoto T. Nucleotide sequences of genome segments 6 and 7 of Bombyx mori cypovirus 1, encoding the viral structural proteins V4 and V5,respectively. J. Gen. Virol. 2000;81:1143–1147. doi: 10.1099/0022-1317-81-4-1143. [DOI] [PubMed] [Google Scholar]
- 36.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 37.DeLano WL. The PyMOL User’s Manual. Palo Alto, California: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.