FACS-Assisted Microarray Profiling Implicates Novel Genes and Pathways in Zebrafish Gastrointestinal Tract Development (original) (raw)
. Author manuscript; available in PMC: 2010 Oct 1.
Published in final edited form as: Gastroenterology. 2009 Jun 27;137(4):1321–1332. doi: 10.1053/j.gastro.2009.06.050
Abstract
Background and Aims
The zebrafish Danio rerio is an excellent model system for mammalian gastrointestinal development. To identify differentially regulated genes important in gastrointestinal organogenesis, we profiled the transcriptome of the zebrafish developing gastrointestinal tract.
Methods
Embryos from a transgenic zebrafish line expressing GFP in the developing intestine, liver, and pancreas were dissociated at four developmental timepoints, their cells sorted based on GFP expression using fluorescence—activated cell sorting (FACS), and analyzed with microarrays. To improve our analysis, we annotated the Affymetrix Zebrafish GeneChip with human orthologs.
Results
Transcriptional profiling showed significant differences between GFP+ and GFP− cells. Upregulated genes and pathways were consistent with mammalian gastrointestinal development, such as HNF gene networks, and cancer. We implicate the PI-3 Kinase pathway and show that inhibition with LY294002 causes gastrointestinal defects in zebrafish. We identified novel genes, such as the microRNAs miR-217 and miR-122, the tight junction protein claudin c, the gene fam136a, and a zebrafish tetraspanin. Novel pathways include genes containing a putative transcription factor binding sequence, GGAANCGGAANY, and a nucleolar gene network. The zebrafish microarrays also identify a set of 32 genes that may mediate the effects of gain of chromosome arm 8q in human colon, liver, and pancreatic cancers.
Conclusions
We successfully combine fluorescence—activated cell sorting and microarray profiling to follow organogenesis throughout development. These experiments identify novel genes and pathways that likely play a role in mammalian gastrointestinal development and are potential targets for therapeutic intervention in the management of gastrointestinal disease and cancer.
Introduction
Among vertebrates, the zebrafish is particularly well suited to study organogenesis of the gastrointestinal (GI) tract, because this vertebrate can be easily manipulated using both forward and reverse genetics and because the zebrafish organs have been described in physiological detail and are highly similar to their mammalian counterparts.1-8 To better understand molecular aspects of zebrafish and mammalian GI development, we transcriptionally profiled the developing zebrafish gastrointestinal tract, using the gutGFP transgenic line, expressing GFP in the developing liver, gut, and pancreas.2-5
In gutGFP zebrafish, a rod of endodermal cells can be seen in the mid-section of the embryo at 1 day post fertilization (dpf).5 From this rod, the liver and the pancreas begin to successively bud, grow, and differentiate into mature organs,2, 3 but alternative models are possible.7, 8 Specification of hepatocytes requires integration of FGF, BMP, and Wnt signals.9-11 By 4dpf, much of liver and pancreatic development appears to be complete.2, 3 Intestinal development begins after 1dpf, when the endodermal rod itself thickens anteriorly, followed by lumen formation, intestinal cell differentiation, epithelial folding, and onset of gut motility by 5dpf.4, 6, 7, 12 These processes are controlled by several transcription factors and the gene networks they control, including hhex, hnf4a, and hnf6/onecut3 in liver development, and which are conserved from zebrafish to human.13-17 Due to this conservation, lessons from zebrafish likely also apply to mammalian GI development.
We selected zebrafish embryos at four critical developmental time points and separated them by FACS into GFP+ and GFP− cell populations, representing GI tissues and the remainder of the embryo. We then profiled the transcriptome by microarray and analyzed our data for known and novel genes expressed in the GI tract during organogenesis and for known and novel pathways and cellular components that may be important in GI development and function. We find evidence for some aspects of GI organ function as early as 2dpf, although substantial functionality is likely not attained until 4dpf. Further evidence suggests a role for PI3-kinase signaling in GI development; treatment with a PI3-kinase inhibitor results in liver defects. We also identify a novel DNA sequence pattern that is significantly enriched in the promoters of GI-specific genes that may be bound by a potentially critical, unknown transcription factor. Our results also support a regulatory role for microRNAs in GI development. Finally, we identify candidate genes located on human chromosome arm 8q that may mediate the effects of early gain of 8q in the development of liver, pancreas, and colon cancers. This manuscript presents proof that combining spatially and temporally restricted GFP expression with flow sorting and microarray analysis is instructive for understanding organogenesis and human disease processes.
Methods
Fish husbandry
All fish husbandry was carried out in accordance with local IACUC protocols. We selected a line of homozygous Tg(XlEef1a1:GFP)s854 transgenic fish (gutGFP) with the strongest, non-variegating GFP expression throughout the GI tract. We grew embryos in standard embryo media, supplemented with 0.003% 1-phenyl-2-thiourea (PTU) to reduce autofluorescence, and harvested about 700 embryos at 2, 3, 4, and 6dpf on ice at the same daily timepoint.
Isolation of gastrointestinal cells by FACS and RNA purification
Embryos were washed in PBST three times, resuspended in Hank’s Balanced Salt Solution with 0.25% Trypsin and 0.1% EDTA (CellGro) supplemented with 40 g/ml Proteinase K and 10 μg/ml Collagenase, and dissociated in a Wheaton tapered tissue grinder. Cells were collected by centrifugation and resuspended in TMI.18 After straining the slurry through a 40 μm filter, cell suspensions were separated into GFP+ and GFP− fractions on a flow cytometer (UPCI Flow Cytometry Facility, Pittsburgh; Supplemental Table 1 and Supplemental Figure 1). RNA was isolated by column purification (Stratagene) and assayed on an Agilent BioAnalyzer. Only samples with intact, distinct ribosomal peaks were chosen for further analysis.
Hybridization of Affymetrix GeneChips
For each time point, we selected three sets of GFP+ and GFP− RNA from a single flow sort, amplified 50 ng of each sample with Ovation (NuGEN), and hybridized them to Affymetrix Zebrafish GeneChips (UPCI Clinical Genomics Facility, Pittsburgh), as per manufacturer’s instructions.
We extracted expression values using robust multi-array averaging (http://rmaexpress.bmbolstad.com/). Expression values were log transformed, centered by median subtraction, and scaled by dividing with the standard deviation. Microarrays were not individually weighted. All primary microarray data and a table of all expression values after normalization are available (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12189). A linear model was fitted and genes identified as differentially expressed at a threshold of p < 0.05 after false-positive correction using the Benjamini-Hochberg algorithm.19
Annotation of Affymetrix Zebrafish GeneChip using a BLAST strategy
To use mammalian gene interaction databases, we improved the available annotation of the Affymetrix Zebrafish GeneChip. We linked as many probesets as possible with zebrafish RefSeq records using information provided by Affymetrix and Ensembl or through nucleotide BLAST for unassigned probesets (GenBank nt Database from 04/23/2007, Ensembl Zebrafish Genome Build Zv6 from 04/27/2007). We extracted zebrafish annotation information. To define protein domains and function, we screened corresponding peptide sequences for PFAM protein domains using HMMer (Pfam_ls and Pfam_fs databases v.21 from 10/13/2006; http://pfam.janelia.org and http://hmmer.janelia.org)20 and collated GO annotation from GenBank, ZFIN, and Pfam. Finally, we screened human protein sequences with zebrafish sequences using TBLASTX (Supplemental Table 2).
Whole mount in situ hybridization of zebrafish embryos
See Supplemental Table 3 for a list of genes, accession numbers, ZFIN IDs, primer sequences, and antibodies for Figures 1, 3, 5 and Supplemental Figures 2, 8. In situs and immunohistochemistry were carried out as described.11, 21
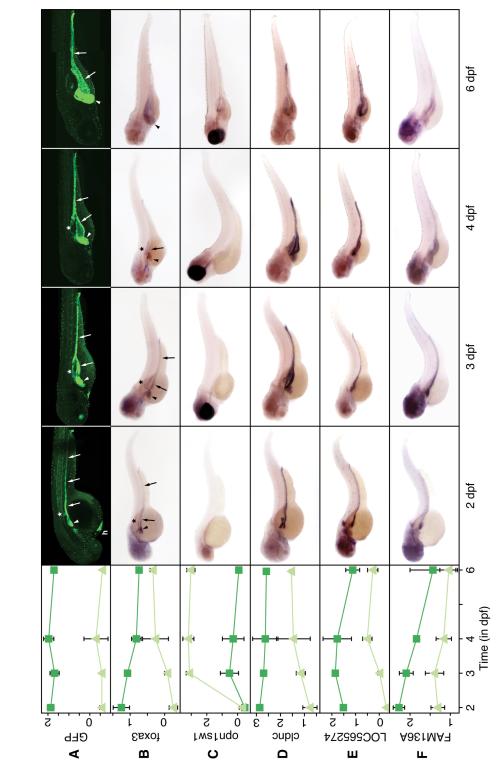
Figure 1.
Microarray profiles and in-situ hybridization of selected genes.
Shown in the first column is the microarray expression profile. Dark green lines with squares show expression in GFP+ cells, encompassing the intestine, liver, and pancreas; light green lines with triangles show expression in GFP− cells. Confocal images (A) and in-situ hybridization of gutGFP embryos fixed at different timepoints (B-F). For probesets and primers, see Supplemental Table 3. Arrowhead: liver; arrow: gut; asterisk: pancreas; h: hatching gland.
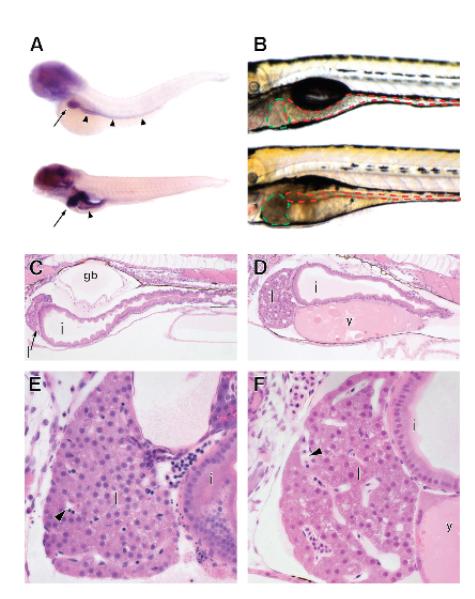
Figure 3.
Phosphatidylinositol signaling is required for normal GI development.
In-situ hybridization for expression of zebrafish akt2l (A) at 4 (top) and 6dpf (bottom). Arrows: liver; arrowheads: intestine. Embryos treated with LY294002 show GI defects (B-F). Brightfield images of treated (bottom) and untreated 6dpf zebrafish (top, B). Liver outlined in green, intestine in red. 4 μM thick sections of untreated (C, E) and treated (D, F) 6dpf embryos. Liver: l; intestine: i; gasbladder: gb; y: yolk; arrowheads: sinusoids.
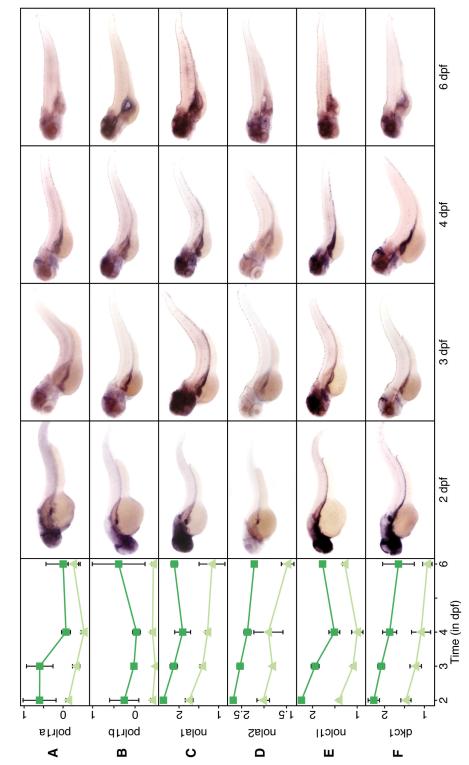
Figure 5.
Nucleolar genes are preferentially expressed in the developing GI tract. Same format as Figure 1.
PI3-Kinase inhibitor treatment
DMSO was added to 0.05% either alone or with 10 μM LY294002 (Sigma) from 2dpf to 5dpf. Embryos were analyzed at 6dpf by brightfield microscopy or embedding in JB4 plastic (Polysciences), sectioning, and standard H&E staining.
Results
GFP transgenic fish, RNA processing, and initial microarray analysis
As basis of our study, we selected the transgenic fish line _Tg(XlEef1a1:GFP)_s854, gutGFP.2-5 After early, ubiquitous expression, transgenic GFP expression was restricted to the developing gut, liver, and pancreas at 2dpf, likely by the foxa3 promoter (our own results corroborate 4), which controls a similar expression pattern (Figure 1A-B). By 3dpf, GFP expression became restricted to the liver, exocrine pancreas, and intestine and continued in GI tissue until at least 6dpf. From 4dpf onward, GFP expression in the gut and exocrine pancreas became variable, but remained strong in the liver (Figure 1B). Although variability of GFP expression can be a limitation in this kind of study, our data argue that cells from all three organs were retained since we identified hepatic, intestinal, and pancreatic genes and pathways.
We dissociated homozygous gutGFP embryos at four time points, 2, 3, 4, and 6dpf, and sorted them based on GFP expression, obtaining average purities of 92% for GFP+ cells and 98% for GFP− cells (Supplemental Figure 1, Supplemental Table 1). For each timepoint, RNA from paired sets of GFP+ and GFP− cells of three independent sorts was amplified and hybridized to Affymetrix Zebrafish GeneChips. After normalization of expression values, we identified probe sets differentially expressed between GFP+ and GFP− cells at each time point (p < 0.05; Table 1A). Because the GFP+ cells represented three very different organs that carry out specialized functions, we find a larger number of differentially expressed probesets (peaking at 3,600 at 6dpf) than might be expected. To avoid confounding our analysis by changes in gene expression in GFP− cells, we also compared only the profiles of GFP+ cells between different timepoints to identify genes changing in expression within GI cells irrespective of expression in GFP− tissue (p < 0.05) and found the largest change in gene expression between 3 and 4dpf (Table 1B). To better analyze this critical transition point between 3 and 4dpf, we also compared expression at 2 and 3dpf to 4 and 6dpf and identified over 4,000 differentially regulated genes (p < 0.05; Table 1B). Lists of differentially expressed probesets with the annotation reported in this paper are available in Supplemental Table 4.
Table 1.
Probesets differentially expressed during GI development
Table showing the numbers of differentially expressed genes at each time point in a comparison between GFP+ and GFP− cells, p < 0.05 (A). Comparison of expression profile in GFP+ cells between different timepoints regardless of expression in GFP− cells, p < 0.05 (B).
| A) | ||||
|---|---|---|---|---|
| 2dpf | 3dpf | 4dpf | 6dpf | |
| Upregulated | 1,498 | 2,246 | 2,971 | 3,641 |
| Downregulated | 1,141 | 1,863 | 1,755 | 2,316 |
| Total | 2,639 | 4,109 | 4,726 | 5,957 |
| B) | ||||
|---|---|---|---|---|
| 2dpf vs. 3dpf | 3dpf vs. 4dpf | 4dpf vs. 6dpf | 2, 3dpf vs. 4, 6dpf | |
| More activeearly than late | 45 | 475 | 466 | 2,159 |
| More activelate than early | 82 | 592 | 243 | 2,095 |
| Total | 127 | 1,067 | 709 | 4,254 |
Validation of microarray data
We carried out whole mount in situ hybridization to compare the predicted expression profile with the observed pattern. In 83 of 89 genes tested (93%), we found good correlation between expression profile and in situ pattern (data not shown). If expression in GI tissue was predicted by profiling (78 genes), we generally observed GI staining by in-situ hybridization. The most common in-situ pattern was expression in the liver and intestine, often with expression in the head (39 genes; Figures 1, 5 and unpublished data). We also found genes whose GI expression was restricted to gut, liver, or pancreas (24, 3, and 1 gene(s), respectively; Figure 1, Supplemental Figure 2, and data not shown). Twelve genes were expressed in all GI tissues (Figure 1 and data not shown). The small relative size of the pancreas may have minimized the statistical significance of the observed expression differences and contributed to the comparative paucity of pancreatic genes. Figure 1 shows microarray expression profiles compared with whole mount in-situ hybridization patterns for a few representative genes: foxa3, putatively controlling GFP expression in the gutGFP zebrafish; short-wave sensitive opsin 1 (opn1sw1) as a negative control; claudin c (cldnc), a tight junction protein; LOC565274, a zebrafish ortholog of the tetraspanin family; and the novel gene fam136a. For a few genes (gcga, hnf4a, and prox1), we also determined protein distribution by immunohistochemistry (Supplemental Figure 2). The in-situ hybridization and antibody staining patterns were consistent with the microarray expression profile.
Annotation of the Affymetrix Zebrafish GeneChip
We wanted to identify molecular pathways that were differentially regulated in the developing gastrointestinal tract. To use existing databases, we added human orthologs to the annotation of the Affymetrix Zebrafish GeneChip. We first replaced EST sequences for as many probesets as possible with full-length, GenBank-annotated zebrafish RefSeqs (12,362 of 15,617; 79%) and extracted from the RefSeq sequences and remaining ESTs all GeneIDs (81%), UnigeneIDs (93%), and ZFIN IDs (57%). We were able to functionally annotate 56% and 66% with GO terms or PFAM families, respectively. Using the zebrafish nucleotide sequences, we identified human orthologs for 12,038 probesets (77%) by TBLASTX, representing 7,761 different human genes (Supplemental Table 2). With these orthologs and our expression data, we used Ingenuity’s Pathway Analysis (IPA, http://www.ingenuity.com) and Gene Set Enrichment Analysis (GSEA, http://www.broad.mit.edu/gsea/).22
Upregulation of known mammalian GI functions and networks in zebrafish
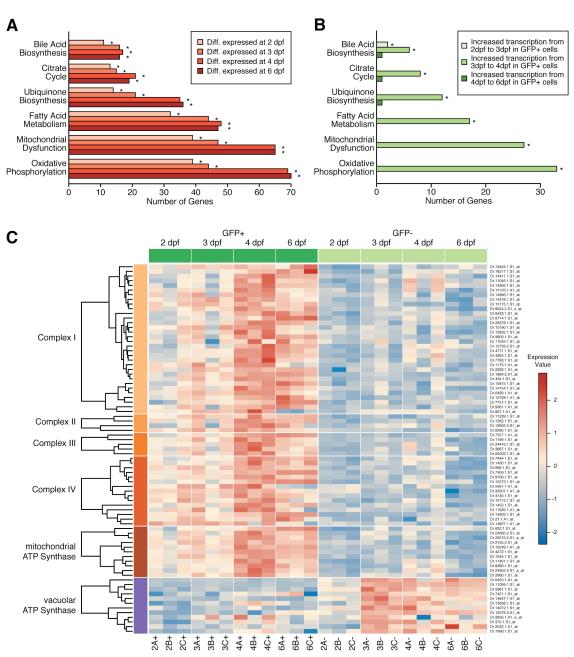
GSEA identified mammalian GI organ-specific gene signatures enriched in GFP+ cells as early as 2dpf (Supplemental Figures 3, 4, and Table 6). Analyzing expression changes at different timepoints solely within GFP+ cells, we observed significant increases in liver and pancreas function from 3 to 4dpf (Supplemental Figures 4E-F, 5-6), concomitant with the onset of organ function.2, 3 Similarly, IPA detected several pathways indicative of GI function as early as 2 dpf, and a significant increase from 3 to 4dpf (Figures 2A-B). Several of the top upregulated functions implicated the electron transport chain and oxidative phosphorylation, including the mitochondrial ATP synthase. In contrast, the vacuolar ATP synthase, which is important in kidney and osteoclasts,23 was active primarily in GFP− cells from 3dpf forward. A heatmap of genes in these pathways showed preferential activation in GFP+ cells, with a strong increase in gene activity from 3 to 4dpf (Figure 2C). These results are consistent with the substantial energy requirements of the GI tract, especially of liver.24
Figure 2.
GI specific functions are upregulated in GFP+ cells.
IPA showing enrichment for metabolic pathways in GFP+ cells within differentially expressed genes at different timepoints (A) and increases in transcriptional activity of genes in GI pathways when comparing GFP+ cells between different timepoints (B). Bars with an asterisk are statistically significant (p < 0.05). Heatmap of probesets involved in the electron transport chain (Complex I — Complex IV), the mitochondrial ATP Synthase, and vacuolar ATP Synthase (C).
Since the hepatic nuclear factor (HNF) family of genes mediates critical GI developmental functions, we asked whether known HNF target genes in zebrafish14 or mammals (Ingenuity), or genes shown to be bound by HNF transcription factors in liver and pancreas tissue explants25 were also upregulated in GFP+ cells. Already at 2dpf, a large percentage of genes controlled by HNF factors were differentially expressed, and their percentage further increased with developmental time, but seemed to plateau at 4dpf, consistent with our earlier results (Table 2, Supplemental Figure 7). Because we can identify both genes and pathways known to be involved in GI development and disease, we are confident that novel genes and pathways contained within this dataset will be relevant to GI organogenesis and disease.
Table 2.
Differential expression of target genes of HNF transcription factor network
Target Probesets show the number of HNF target genes from Ingenuity or from the literature (Cheng et al.14 and Odom et al.25) present on the Affymetrix Zebrafish GeneChip. At each time point, differentially expressed probesets are shown both as number and percentage of target probesets (top line). Downregulated genes are shown below upregulated genes.
| TargetProbesets | 2dpf | 3dpf | 4dpf | 6dpf | |
|---|---|---|---|---|---|
| Ingenuity | 85 | 41 (48%)(37/4) | 52 (61%)(49/3) | 63 (74%)(60/3) | 63 (74%)(60/3) |
| Cheng et al. | 295 | 132 (44%)(124/8) | 189 (64%)(179/10) | 242 (82%)(239/3) | 261 (88%)(259/2) |
| Odom et al. | 806 | 211 (26%)(173/38) | 309 (38%)(247/62) | 376 (47%)(306/70) | 425 (53%)(320/105) |
| Combined | 1,056 | 329 (31%)(277/52) | 462 (44%)(383/79) | 568 (54%)(487/81) | 632 (60%)(515/117) |
Novel genes, gene networks, and pathways implicated in GI development
During our initial analysis, we found that expression levels of several genes in the PI-3 signaling and inositol metabolism pathway were significantly higher in GI tissue or changed throughout development. Akt2l, a key mediator of PI-3 signaling, was notably upregulated in the GI tract by microarray and in-situ hybridization (Supplemental Figure 8A-D; Figure 3A). Using GSEA, we showed that genes upregulated in glioblastoma cells in response to LY294002, a small molecule inhibitor of most PI-3 kinases which prevents activation of akt2,26, 27 were active in early zebrafish GI tissue (Supplemental Figure 8E). Consequently, we tested the hypothesis that PI3K signaling and akt2l activation are required for normal GI development. We treated embryos with LY294002 and found that they had a globular liver, decreased intestinal volume, unconsumed yolk, and partially inflated gasbladders. Liver histology revealed a spongy appearance, enlarged sinusoids, and reduced cell number (Figure 3B-F) consistent with a critical role of PI3K signaling in GI development.
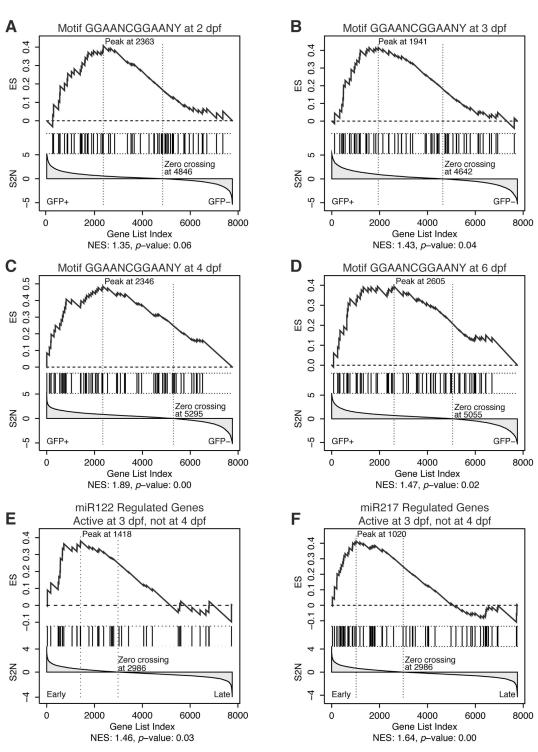
Using GSEA, we then looked for evidence of novel transcription factors in GFP+ cells and found a significant enrichment for 33 human genes containing the sequence element GGAANCGGAANY within 2 kb of the transcriptional start site (Figure 4A-D, Supplemental Table 6).28 Among these genes were subunits of the mitochondrial ribosome, consistent with the important role mitochondria play in GI function. IPA identified a network surrounding β-estradiol and containing 12 genes from our set alongside MYC, HGF, MAPK1, and others (Supplemental Figure 9A). Top functions in this network included organ morphology and cellular movement, suggesting that it may integrate hormone signaling with organogenesis. Interestingly, estrogen has been shown to play a role in hepatocellular carcinomas (HCC)29 and estrogen dependent responses have been observed in zebrafish by 4dpf.30
Figure 4.
GSEA enrichment plots for GGAANCGGAANY-containing genes and miR122 and miR217.
Each panel shows the running enrichment score (ES) in the top and the signal-to-noise ratio (S2N) used for gene ranking in the bottom. Vertical lines in the middle of the panel indicate the position of gene set members within the sorted microarray data. Normalized ES (NES) and nominal _p_-value are listed below each graph.22, 58 Enrichment for genes containing GGAANCGGAANY within 2 kb of the start codon at 2, 3, 4, and 6dpf (A-D). Enrichment for genes regulated by miR122 (E) and miR217(F) within genes that are active in GFP+ cells at 3dpf, but not at 4dpf.
GSEA also identified a significant enrichment for targets of microRNAs miR-12228 and miR-21728 among genes active in GFP+ cells at 3, but not at 4dpf (Figure 4E-F; Supplemental Table 6), suggesting that they may function in controlling GI organogenesis in zebrafish. The zebrafish ortholog of miR-122 is expressed in liver and pancreas at 3dpf; mir-217 is expressed ubiquitously at 2dpf and becomes highly enriched in the pancreas at 5dpf.31 MiR-122 and miR-217 are significantly downregulated in GI cancers.32, 33
Nucleolar genes are differentially expressed in the developing GI tract
IPA identified a network of nucleolar genes (Supplemental Figure 9B), which were unexpectedly overexpressed in GFP+ cells. This network consisted of three subunits of RNA Polymerase I (polr1a and polr1b, Figure 5A-B; polr1c, data not shown), two small nucleolar ribonucleoproteins (nola1 and nola2; Figure 5C-D), an ion channel protein (nolc1l, Figure 5E), and another nucleolar protein (diskerin, dkc1, Figure 5F). We confirmed the predicted profiles by in-situ hybridization, noting a GI specific enrichment of genes generally thought to be ubiquitously expressed (Figure 5). These data support other studies suggesting that the nucleolus is actively regulated and is critical to the proper development of the zebrafish GI tract.34
Correlation between Danio rerio GI development and human GI cancer progression
Since gene signatures of embryonic GI development can inform our understanding of cancer,35-37 we correlated our zebrafish microarray data with data from human GI cancer. Lam et al.38 identified 126 genes, expressed in both zebrafish adult liver cancer and human liver cancer. Using GSEA, we show that this gene set is significantly enriched at the early time points of 2 and 3dpf, but not at the later time points, suggesting a similarity of gene expression between HCCs and earlier, less differentiated hepatocytes (Supplemental Figure 10, Supplemental Table 6). At late timepoints, we also find significant downregulation of genes that are highly expressed in HCCs with poor prognosis. In contrast, genes highly expressed in HCCs with good prognosis, are upregulated at all timepoints (Supplemental Figure 11, Supplemental Table 6). Generally, our data are most consistent with a model positing that cancer cells are normally more like early, less differentiated cells.
Colorectal cancer develops from normal colonic epithelium through a series of well-described stages with associated chromosomal aberrations and may form liver metastases in some patients.39-41 We asked if there was an enrichment of developmentally active zebrafish genes in GFP+ cells with orthologs localizing to areas of the human genome frequently altered in colorectal cancer. We defined active probesets for this comparison as a) having a human ortholog, b) being expressed at levels significantly higher than other probesets (_p_-value < 0.01, Wilcoxon-Rank Sum Test), and c) being expressed uniformly across all three replicates (its standard deviation less than twice the mean standard deviation). Applying these criteria, we identified 1446, 1355, 1265, and 1465 active probesets at each timepoint and ranked them by their expression levels (Supplemental Table 7). We queried whether gene sets representing chromosome regions frequently gained or lost during cancer progression were overrepresented among these genes.39-41
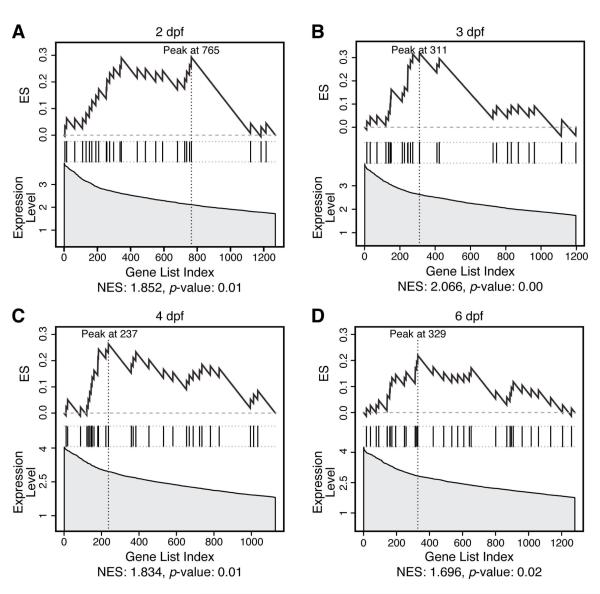
At all four timepoints, we established a significant correlation between genes on 8q and our lists of active genes (Figure 6 A-D, Supplemental Table 6). The enriched genes at each time point are similar, suggesting that only a small number of genes mediate the effect of gain of 8q and contain ribosomal subunits (RPL7, RPL8, and RPL30) and translation initiation factors (EIF3S6), again showing that ‘housekeeping’ genes might function in tissue- and organ-specific development and disease. We also find genes with putative DNA binding domains (CHD7, ZF16 and ZF706), a cadherin specific to liver and intestine (CDH17), and genes implicated in epithelial differentiation (NDRG1) or colorectal neoplasia (CA2). Additionally, gain of 8q was also frequent in liver and pancreatic cancers.42, 43
Figure 6.
GSEA plots show enrichment of human orthologs on 8q among genes active in zebrafish GI development.
GSEA enrichment plots for genes with orthologs on human chr8q among genes active in zebrafish GI tissue ranked by expression level (A—D). Graphs are similar to Figure 3, except the ranking statistic is based on expression level, not signal-to-noise ratio.
Discussion
Our experiments help establish a widely applicable technique to define the developmental transcriptome of any organ over time that should not be limited to zebrafish.44-47 We were able to exploit the strong conservation between zebrafish and mammalian organ physiology and gene regulatory networks to find conserved gene networks active throughout vertebrate development. We expect that the genes and pathways identified here will lead to novel zebrafish GI development and cancer models, which should increase our knowledge of mammalian GI development and disease.
Using our microarray data, we were able to identify individual genes that may play an important role in GI development, some for the first time. However, a major strength of microarray analysis is that the thousands of genes assayed allow analysis of pathways and complexes that may play an important role in GI development. Already at 2dpf, we show beginning upregulation of GI-specific pathways which appears complete by 4dpf, suggesting that the organs have attained much of their functionality at this point. Nevertheless, cells may exert additional control over protein activity through non-transcriptional mechanisms that would go undetected in our study and thus further upregulate GI-specific pathways.
Using GSEA, we identified a novel network of genes with GGAANCGGAANY in their promoter region. With IPA, we found a gene network centered around β-estradiol, which was previously shown to affect gene transcription in zebrafish by 4dpf.30 Evaluating this response in the context of the novel network of GGAANCGGAANY—containing genes may yield clues to the role of β-estradiol in normal liver development and cancer.29
We present evidence that components of the basal cell machinery, particularly the nucleolus, play a specific, developmentally regulated role in organogenesis. Our microarray data confirm previous results from an insertional mutagenesis screen in zebrafish which found that mutations in RNA polymerases I and III, nucleolar proteins, and mitochondrial ATP synthase genes, including some genes described in this paper, caused a small liver and intestine phenotype (e.g., mutants hi4050 — nolc1l, hi1159 — polr1a, hi1125, hi1942, hi2578, or hi3619).48 Independently isolated zebrafish mutations in PolIII49 and the nucleolar protein RBM1950 also have specific GI defects. Despite a requirement for nucleolar function in every cell, mutations in individual nucleolar proteins appear to cause human diseases with cell-type-specific defects, such as Diamond-Blackfan anemia or dyskeratosis congenita.34 Our results predict that defects in nucleolar pathway function may similarly contribute to the development of specific GI diseases.
One pathway previously associated with metastasis formation posits the formation of a protein complex containing claudin-7, the tetraspanin CO-029 and others in membrane microdomains.51 We show here that both the claudin gene cldnc and the tetraspanin LOC565274 are preferentially expressed in the zebrafish GI tract, raising the possibility that they interact to form a similar complex in the developing zebrafish. Since claudins play a role in epithelial tight junctions and tetraspanins in transmembrane signaling,52, 53 we hypothesize that such a complex might relay signals during stratification of the intestinal epithelium and its dysregulation in adults might aid metastasis development. Indeed, human orthologs of cldnc, Claudin 3 and Claudin 4, and the orthologs of LOC565274, TM4SF1 and TM4SF4, are frequently upregulated in human GI cancers.54-56
On a genomic scale, similarities in gene expression patterns have been observed between developmental processes and cancers of the colon and liver35-38 and were also present in our data. By comparing genes active during zebrafish GI development with human orthologs on chromosome arms often gained or lost during GI tumorigenesis, we identified a set of genes located on 8q that might mediate tumorigenesis. Since gain of 8q is one of the earliest hallmarks of colorectal cancer,39 occurs in most colon carcinoma cell lines,58 HCC,42 and pancreatic cancer,43 these genes located on 8q are specific candidates for possible contributors to the colorectal, liver and pancreatic cancer phenotypes; a hypothesis that will be tested in subsequent experiments.
Multiple lines of evidence support the utility of our data for identifying critical mediators of GI development and disease. Available zebrafish mutants for nolc1l and polr1a show defects in GI development.48 We predict a function of miR122 in GI development and disease and experiments on murine miR122 show a role as a hepatic tumor suppressor.33 The HNF4α network is highly enriched statistically in our dataset and GI-organ specific HNF4α knockouts have defects in their respective organs.25 While beyond the scope of this paper, data from morpholino knockdown and overexpression analyses of genes differentially expressed in this study showed specific GI phenotypes (authors’ unpublished data). Finally, we have demonstrated a developmental role for the PI3K/Akt pathway in normal GI development and PI3K signaling is currently evaluated as a chemotherapeutic target.57
Genomic assays on a defined subpopulation of cells yielded detailed insight into the transcriptional profile of developing GI cells in zebrafish. We showed that known pathways were active in developing GI cells as expected and also derived new hypotheses about formation of a functional GI tract and GI cancer. We are certain that future analysis of our dataset with forthcoming data will reveal additional trends and pathways, and vice versa. As methods for RNA amplification and microarray analysis continue to improve and new transgenic lines become available, combining FACS-sorting with microarray profiling will become a useful method to describe the transcriptome of a specific, well-defined population of cells of interest.
Supplementary Material
01 - suppl figure 1
10 - suppl table 6
11 - suppl table 7
12 - suppl figure 5
13 - suppl table 8
14 - suppl figure 6
15 - suppl figure 7
16 - suppl figure 8
17 - suppl figure 9
18 - suppl figure 10
19 - suppl figure 11
02 - suppl figure 2
03 - suppl figure 3
04 - suppl figure 4
05 - suppl table 1
06 - suppl table 2
07 - suppl table 3
08 - suppl table 4
09 - suppl table 5
Acknowledgements
We thank Sarah Berman, Erin McClelland (UPCI Flow Cytometry Facility), Christin Sciulli (UPCI Clinical Genomics Facility), Drs. Paul Carlson, Duc Dong, Gerard Nau, and Martin Schmidt for excellent technical assistance and suggestions on data analysis and bioinformatics. We are grateful to Zi-Qing Sun for critically reading the manuscript. This research was supported by grants from the March of Dimes (FY04-109) and the NIH (R21DK073177).
Grant Support: March of Dimes FY04-109 NIH R21DK073177
Abbreviations Used in Manuscript
dpf
days post fertilization
FACS
fluorescence-activated cell sorting
GFP
green fluorescent protein
GI
gastrointestinal
Footnotes
Financial Disclosure: The authors declare that they do not have competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Stuckenholz C, Ulanch PE, Bahary N. From guts to brains: using zebrafish genetics to understand the innards of organogenesis. Curr Top Dev Biol. 2005;65:47–82. doi: 10.1016/S0070-2153(04)65002-2. [DOI] [PubMed] [Google Scholar]
- 2.Field HA, Dong PD, Beis D, et al. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- 3.Field HA, Ober EA, Roeser T, et al. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–90. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 4.Ng AN, de Jong-Curtain TA, Mawdsley DJ, et al. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286:114–35. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Ober EA, Field HA, Stainier DY. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev. 2003;120:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 6.Wallace KN, Akhter S, Smith EM, et al. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–73. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- 8.Yee NS, Lorent K, Pack M. Exocrine pancreas development in zebrafish. Dev Biol. 2005;284:84–101. doi: 10.1016/j.ydbio.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 9.Ober EA, Verkade H, Field HA, et al. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–91. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 10.Shin D, Shin CH, Tucker J, et al. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–50. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 11.Dong PD, Munson CA, Norton W, et al. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- 12.Bagnat M, Cheung ID, Mostov KE, et al. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9:954–60. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- 13.Zorn AM, Wells JM. Molecular basis of vertebrate endoderm development. Int Rev Cytol. 2007;259:49–111. doi: 10.1016/S0074-7696(06)59002-3. [DOI] [PubMed] [Google Scholar]
- 14.Cheng W, Guo L, Zhang Z, et al. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev Biol. 2006;294:482–96. doi: 10.1016/j.ydbio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–31. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- 16.Matthews RP, Lorent K, Russo P, et al. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–59. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Wallace KN, Yusuff S, Sonntag JM, et al. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis. 2001;30:141–3. doi: 10.1002/gene.1050. [DOI] [PubMed] [Google Scholar]
- 18.Kaur S, Castellone MD, Bedell VM, et al. Robo4 signaling in endothelial cells implies attraction guidance mechanisms. J Biol Chem. 2006;281:11347–56. doi: 10.1074/jbc.M508853200. [DOI] [PubMed] [Google Scholar]
- 19.Smyth GK. Limma: linear models for microarray data. In: Gentleman RC, Carey DJ, Dudoit S, Irizarry RA, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 20.Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahary N, Goishi K, Stuckenholz C, et al. Duplicate VegfA genes and orthologues of the KDR receptor tyrosine kinase family mediate vascular development in the zebrafish. Blood. 2007;110:3627–36. doi: 10.1182/blood-2006-04-016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–29. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 24.Sato N. Central role of mitochondria in metabolic regulation of liver pathophysiology. J Gastroenterol Hepatol. 2007;22(Suppl 1):S1–6. doi: 10.1111/j.1440-1746.2007.04963.x. [DOI] [PubMed] [Google Scholar]
- 25.Odom DT, Zizlsperger N, Gordon DB, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–81. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okano J, Gaslightwala I, Birnbaum MJ, et al. Akt/protein kinase B isoforms are differentially regulated by epidermal growth factor stimulation. J Biol Chem. 2000;275:30934–42. doi: 10.1074/jbc.M004112200. [DOI] [PubMed] [Google Scholar]
- 27.Terragni J, Graham JR, Adams KW, et al. Phosphatidylinositol 3-kinase signaling in proliferating cells maintains an anti-apoptotic transcriptional program mediated by inhibition of FOXO and non-canonical activation of NFkappaB transcription factors. BMC Cell Biol. 2008;9:6. doi: 10.1186/1471-2121-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannitrapani L, Soresi M, La Spada E, et al. Sex hormones and risk of liver tumor. Ann N Y Acad Sci. 2006;1089:228–36. doi: 10.1196/annals.1386.044. [DOI] [PubMed] [Google Scholar]
- 30.McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 32.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 33.Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–82. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 34.Boisvert FM, van Koningsbruggen S, Navascués J, et al. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–85. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser S, Park YK, Franklin JL, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam SH, Gong Z. Modeling liver cancer using zebrafish: a comparative oncogenomics approach. Cell Cycle. 2006;5:573–7. doi: 10.4161/cc.5.6.2550. [DOI] [PubMed] [Google Scholar]
- 37.Lepourcelet M, Tou L, Cai L, et al. Insights into developmental mechanisms and cancers in the mammalian intestine derived from serial analysis of gene expression and study of the hepatoma-derived growth factor (HDGF) Development. 2005;132:415–27. doi: 10.1242/dev.01579. [DOI] [PubMed] [Google Scholar]
- 38.Lam SH, Wu YL, Vega VB, et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24:73–5. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 39.Diep CB, Kleivi K, Ribeiro FR, et al. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer. 2006;45:31–41. doi: 10.1002/gcc.20261. [DOI] [PubMed] [Google Scholar]
- 40.Hermsen M, Postma C, Baak J, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–19. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- 41.Postma C, Hermsen MA, Coffa J, et al. Chromosomal instability in flat adenomas and carcinomas of the colon. J Pathol. 2005;205:514–21. doi: 10.1002/path.1733. [DOI] [PubMed] [Google Scholar]
- 42.Poon TC, Wong N, Lai PB, et al. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006;131:1262–70. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 43.Kitoh H, Ryozawa S, Harada T, et al. Comparative genomic hybridization analysis for pancreatic cancer specimens obtained by endoscopic ultrasonography-guided fine-needle aspiration. J Gastroenterol. 2005;40:511–7. doi: 10.1007/s00535-005-1577-0. [DOI] [PubMed] [Google Scholar]
- 44.Bryant Z, Subrahmanyan L, Tworoger M, et al. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc Natl Acad Sci U S A. 1999;96:5559–64. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bezencon C, Furholz A, Raymond F, et al. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J Comp Neurol. 2008;509:514–25. doi: 10.1002/cne.21768. [DOI] [PubMed] [Google Scholar]
- 46.von Stetina SE, Watson JD, Fox RM, et al. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 2007;8:R135. doi: 10.1186/gb-2007-8-7-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Covassin L, Amigo JD, Suzuki K, et al. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol. 2006;299:551–62. doi: 10.1016/j.ydbio.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amsterdam A, Nissen RM, Sun Z, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yee NS, Gong W, Huang Y, et al. Mutation of RNA Pol III subunit rpc2/polr3b Leads to Deficiency of Subunit Rpc11 and disrupts zebrafish digestive development. PLoS Biol. 2007;5:e312. doi: 10.1371/journal.pbio.0050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenzen JA, Bonacci BB, Palmer RE, et al. Rbm19 is a nucleolar protein expressed in crypt/progenitor cells of the intestinal epithelium. Gene Expr Patterns. 2005;6:45–56. doi: 10.1016/j.modgep.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Kuhn S, Koch M, Nubel T, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–67. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- 52.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–29. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 53.Zoller M. Gastrointestinal tumors: metastasis and tetraspanins. Z Gastroenterol. 2006;44:573–86. doi: 10.1055/s-2006-926795. [DOI] [PubMed] [Google Scholar]
- 54.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–6. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 56.Nishino R, Honda M, Yamashita T, et al. Identification of novel candidate tumour marker genes for intrahepatic cholangiocarcinoma. J Hepatol. 2008 doi: 10.1016/j.jhep.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 57.Pang RW, Poon RT. From molecular biology to targeted therapies for hepatocellular carcinoma: the future is now. Oncology. 2007;72(Suppl 1):30–44. doi: 10.1159/000111705. [DOI] [PubMed] [Google Scholar]
- 58.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01 - suppl figure 1
10 - suppl table 6
11 - suppl table 7
12 - suppl figure 5
13 - suppl table 8
14 - suppl figure 6
15 - suppl figure 7
16 - suppl figure 8
17 - suppl figure 9
18 - suppl figure 10
19 - suppl figure 11
02 - suppl figure 2
03 - suppl figure 3
04 - suppl figure 4
05 - suppl table 1
06 - suppl table 2
07 - suppl table 3
08 - suppl table 4
09 - suppl table 5