Thiol-Based Redox Switches in Eukaryotic Proteins (original) (raw)
Abstract
For many years, oxidative thiol modifications in cytosolic proteins were largely disregarded as in vitro artifacts, and considered unlikely to play significant roles within the reducing environment of the cell. Recent developments in in vivo thiol trapping technology combined with mass spectrometric analysis have now provided convincing evidence that thiol-based redox switches are used as molecular tools in many proteins to regulate their activity in response to reactive oxygen and nitrogen species. Reversible oxidative thiol modifications have been found to modulate the function of proteins involved in many different pathways, starting from gene transcription, translation and protein folding, to metabolism, signal transduction, and ultimately apoptosis. This review will focus on three well-characterized eukaryotic proteins that use thiol-based redox switches to influence gene transcription, metabolism, and signal transduction. The transcription factor Yap1p is a good illustration of how oxidative modifications affect the function of a protein without changing its activity. We use glyeraldehyde-3-phosphate dehydrogenase to demonstrate how thiol modification of an active site cysteine re-routes metabolic pathways and converts a metabolic enzyme into a pro-apoptotic factor. Finally, we introduce the redox-sensitive protein tyrosine phosphatase PTP1B to illustrate that reversibility is one of the fundamental aspects of redox-regulation. Antioxid. Redox Signal. 11, 997–1014.
Reactive Oxygen Species and Oxidative Stress
Reactive oxygen species (ROS) include a broad variety of different chemical oxidants, including hydrogen peroxide (H2O2), superoxide anion (•O2−), singlet oxygen (1O2), and hydroxyl radicals (•OH). Inside the cells, reactive oxygen species are constantly generated as side products of neutrophil-mediated phagocytosis (46), cellular respiration (20), and various NADPH oxidases (131). This ROS production appears not to be just a harmful byproduct of aerobic metabolism, but seems to involve a tightly regulated process (41). It allows low levels of ROS to serve as important signaling molecules in processes such as gene transcription, protein tyrosine de- and phosphorylation, stress protection, apoptosis, and metabolism (29, 41, 118, 128). Similarly, reactive nitrogen oxide species (RNS), which include the free radical nitric oxide (•NO), peroxynitrite (ONOO–), and nitrogen dioxide (NO2), also play significant roles as redox active molecules (89). In the presence of oxygen, endogenously produced •NO can rapidly lead to _S_-nitrosation of protein thiols (SNO) and glutathione (GSNO), presumably via the formation of other reactive nitrogen oxide species, such as N2O3, NO2, and thiyl radicals (116). Over 200 different proteins have been found to be targeted by _S_-nitrosation, including metabolic enzymes, phosphatases, transcription factors, and others (48).
To maintain redox homeostasis under conditions where ROS and RNS concentrations begin to increase (see below), cells can draw on a wide arsenal of enzymatic and nonenzymatic defense and repair strategies. For rapid ROS detoxification and scavenging, most cells use a combination of antioxidant enzymes with very high catalytic activity, such as superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (Gpx), or of high cellular abundance, such as peroxiredoxins (>0.5% of total cell protein) (107). The original oxidation status of cytosolic protein thiols is maintained and rapidly restored by the action of two redox- balancing systems, thioredoxin (Trx) and glutaredoxin (Grx), which draw their reducing power ultimately from NADPH (49). The overall reducing environment of the cytosol of −230 to −260 mV, which represents the cellular standard redox potential, is preserved by the equilibrium between the small tripeptide glutathione (GSH) and its oxidized form (GSSG) (58, 63). The GSH/GSSG couple constitutes the redox buffer of most eukaryotic and prokaryotic cells. The low reactivity of the GSH thiol at physiological conditions (pKa of 9.4) (133) is counterbalanced by its high intracellular concentration (∼1–10 m_M_) and rapid enzymatic regeneration by glutathione reductase and NADPH (15).
Once ROS and RNS concentrations exceed the antioxidant capacity of the cell, oxidative damage to DNA, lipids, and proteins can occur. This puts organisms into a state defined as oxidative stress (125). It triggers a highly conserved response in both prokaryotic and eukaryotic cells (42). The response includes increased expression of oxidant scavengers, chaperones, proteases, and DNA repair enzymes to mitigate and reverse oxidative damage on proteins and DNA. Severe oxidative damage, that will eventually lead to apoptosis and cell death, has been shown to accompany many pathological conditions, such as cancer, neurodegenerative diseases, and diabetes (25, 80). Moreover, it is the damaging effect of increasing oxidant concentrations that is thought to be the underlying culprit of eukaryotic aging (41).
Cysteine Thiols–Central Components of Redox-Sensitive Nanoswitches
Cysteine residues play important roles in the biochemistry of many proteins due to the unique chemical properties of their side chain. The high reactivity, redox properties, and ability of thiol groups to coordinate metal ions designate cysteines as the amino acids of choice to form key catalytic components of enzymes (10). Over the past 10 years, however, a new role for cysteine thiols emerged. They form the central building block of thiol-based redox switches in redox-regulated proteins. Redox-sensitive cysteines undergo reversible thiol modifications in response to reactive oxygen or nitrogen oxide species, thereby modulating protein function, activity, or localization. They play important roles in many signal transduction cascades (e.g., PTEN and PTP1B) (74, 150), gene transcription (e.g., p53) (102), as well as in the immediate defense against oxidative stress conditions (e.g., Hsp33, Hsf-1) (2, 52).
What makes cysteine residues particularly redox-sensitive and proteins potentially redox-regulated? Regulatory thiol modifications involve one or more cysteines, whose reactivity is largely determined by the cysteine's structural environment and its pKa value. Most cytoplasmic protein thiols have pKa values of greater than 8.0, which render the thiol groups predominantly protonated and largely nonreactive at intracellular pH (43). Thiol groups of redox-sensitive cysteines, on the other hand, have characteristically much lower pKa values, ranging from as low as ∼3.5 in thiol transferase to ∼5.1–5.6 in protein tyrosine phosphatases. Under physiological pH conditions, these thiols are therefore present as deprotonated, highly reactive thiolate anions (RS-) (78, 81). The low pKa values of redox-sensitive cysteines arise primarily from stabilizing charge–charge interactions between the thiolate anion and neighboring positively charged or aromatic side chains (106, 152).
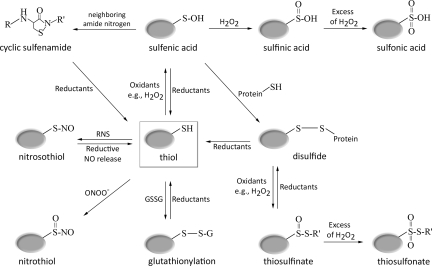
Thiolate anions, in contrast to their protonated counterparts, are highly susceptible to oxidation by ROS and RNS and can undergo a diverse spectrum of oxidative modifications (141). These include sulfenic (SOH), sulfinic (SO2H), and sulfonic (SO3H) acids, disulfide bonds (PrSSPr), or nitrosothiols (SNO) (27, 128, 129) (Fig. 1). Cysteine sulfenic acids and their deprotonated cysteine-sulfenates are remarkably reactive and versatile oxidation products, which are frequently formed (Fig. 1) first upon reaction of protein thiols with H2O2 (112). Due to their high reactivity, sulfenic acids are often considered as metastable intermediates that undergo further reactions to form stable modifications, such as disulfides with other protein thiols or _S_-glutathionylation with the small redox buffer component glutathione (27, 112, 128, 129). Importantly, most oxidative thiol modifications are fully reversible in vivo and utilize dedicated oxidoreductases, such as the thioredoxin or glutaredoxin system, to quickly restore the original redox state upon the cell's return to nonstress conditions (26, 30, 119) (Fig. 1). It appears that it is the reaction rate with these dedicated oxidoreductases that often determines the lifespan of oxidized proteins and supports their accumulation even in an overall reducing environment (7, 77).
FIG. 1.
Oxidative thiol modifications. Oxidation of cysteine thiol groups by H2O2 leads to sulfenic acid (R-SOH) formation. Sulfenic acids are either stabilized by nearby charges or react with neighboring thiols or proximal nitrogen to form disulfide bonds (R′-S-S-R″) or sulfenamide bonds (R′-S-NH-R′), respectively. In the presence of high H2O2 concentrations, overoxidation to sulfinic (R-SO2H) or sulfonic acid (R-SO3H) occurs. Although a few protein-specific sulfinic acid reductases have been identified, overoxidation is still considered to be largely irreversible in vivo. Alternatively, reaction of thiolate anions (RS-) with oxidized cysteines of other proteins or low molecular weight thiols such as glutathione (GSSG) leads to mixed disulfide bond formation (R′-S-S-R″) or _S_-glutathionylation (R-S-SG), respectively. Overoxidation of disulfide bonds in the presence of strong oxidants can cause thiosulfinate (R′-SO-S-R″) or irreversible thiosulfonate (R′-SO2-S-R″) formation. Most oxidative thiol modifications are reduced by members of the glutaredoxin (Grx) system and thioredoxin (Trx) system (reductants), which draw their reducing power from cellular NADPH. Exposure of thiolate anions to reactive nitrogen oxide species causes _S_-nitrosothiol formation, whereas treatment with peroxynitrite yields _S_-nitrothiol formation. The exact mechanism by which individual RNS cause oxidative thiol modifications in vivo is still under investigation.
Extended and/or extensive exposure of proteins to oxidants can lead to the “overoxidation” of cysteine residues to form sulfinic and sulfonic acids (82). These oxidative modifications are considered largely irreversible in vivo, although a small number of protein-specific sulfinic acid reductases (e.g., sulfiredoxin, sestrin) have recently been identified (16, 142). Moreover, strong oxidative stress conditions often result in excessive disulfide bonding, protein misfolding, aggregation, and degradation, and will eventually lead to cell death (28, 127).
The type and extent of oxidative modifications in redox-regulated proteins depends on the specific type of oxidant (75), its absolute level and persistency, the subcellular location of oxidant production and its distance to the target protein (29, 41). Even small changes in the basal level of intracellular ROS and RNS can cause oxidative modifications in proteins that are specifically sensitive to these oxidants. Oxidative thiol modifications most often lead to changes in the structure of proteins and cause either their activation (e.g., OxyR, Hsp33) (52, 99) or inactivation (e.g., PTEN, GapDH) (120, 150). With the help of innovative in vivo and in vitro thiol trapping techniques (reviewed in refs. 72 and 149) and global redox proteomic methods (reviewed in ref. 76), the number of redox-regulated proteins that have been identified is steadily increasing (3, 27, 59). Redox-regulated proteins turn out to be involved in nearly every physiological process, including metabolism, signaling, cell growth, gene expression, transcription factor activation, differentiation, senescence, and apoptosis (41, 118, 128, 129). Interestingly, more often than not, redox-regulated proteins appear to be central components of these pathways, suggesting that protein redox regulation is the cell's fundamental strategy to fine-tune cellular processes in response to ROS and RNS. Here, we focus on a subset of proteins with thiol-based redox switches, whose redox regulation influences distinct processes in eukaryotic cells: transcription, metabolism, and signal transduction.
Changing Gene Transcription to Protect Cells Against Oxidative Stress
The ability to rapidly adapt gene expression to environmental changes is crucial for the growth and survival of every organism. Over the past few years, an increasing number of transcription factors have been shown to be directly regulated by accumulating ROS (59). These include the activator protein-1 (AP-1), NF-κB, Myb, p53, USF (upstream stimulatory factor), NF-1 (nuclear factor 1), and others (9, 87, 101). Redox regulation of these proteins is mediated by the modification of one or more cysteine residues, which causes either the activation (e.g., AP-1, NF-κB) or inactivation (e.g., USF) of the respective transcription factor (87, 101).
We focus here primarily on AP-1-like transcription factors, which are well- characterized leucine-zipper (bZIP) DNA-binding proteins, that are implicated in the regulation of multiple cellular processes, including proliferation, differentiation, stress response, and apoptosis (55, 61,134). In Saccharomyces cerevisiae, the AP-1-like transcription factor Yap1p plays an important role in the cellular response to oxidative stress and various xenobiotics (8, 68, 93). The Yap1p regulon includes as many as 70 genes encoding most of yeast's antioxidant enzymes and components of the cellular thiol-reducing pathway, including Trx, Grx, GPx, SOD, and CAT (22, 54, 68).
Yap1—Changing the Redox State Means Switching Homes
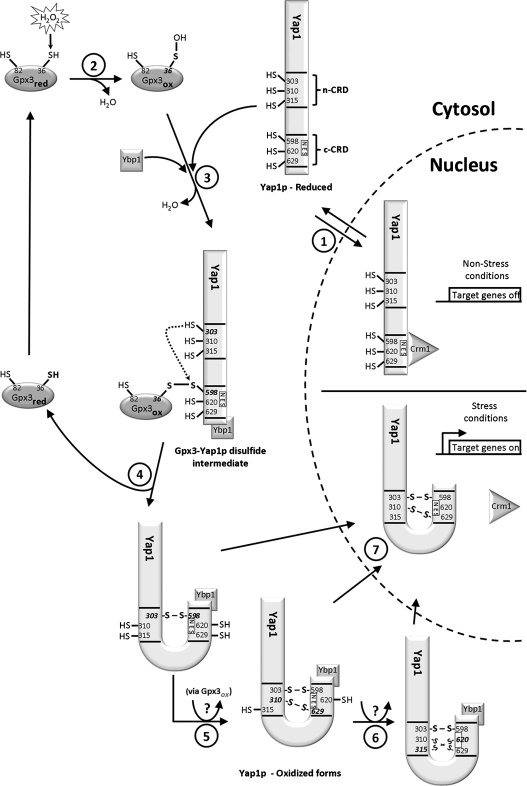
The activation of Yap1p upon peroxide stress (i.e., H2O2) is triggered by highly conserved cysteine residues, which are reduced under nonstress conditions and rapidly form intramolecular disulfide bonds upon exposure to oxidative stress conditions (31, 143). The redox-active cysteines in Yap1p are clustered in two cysteine-rich domains (CRD) (Fig. 2). Each CRD contains three cysteines (Cys303, Cys310, and Cys315) positioned in the n-CRD at the center of the protein, and three cysteines (Cys598, Cys620, and Cys629) located in the c-CRD at the C-terminus of Yap1p. Embedded in the c-CRD is also a leucine-rich nuclear export signal (NES), which is directly responsible for Yap1p's subcellular distribution and indirectly responsible for its functional regulation (69, 147).
FIG. 2.
Model of the Yap1p-Gpx3 redox relay. Under nonstress conditions, the cysteines of Yap1, which are clustered in two cysteine-rich domains (n-CRD, c-CRD), are reduced. (1) The nuclear export sequence (NES) of Yap1p is accessible for interaction with Crm1, and Yap1p shuttles between the cytosol and the nucleus. It prevents Yap1p's nuclear accumulation and guarantees low constitutive activity under nonstress conditions. (2) Upon exposure of yeast cells to oxidative stress conditions (i.e., H2O2) the active site cysteine (Cys36) of glutathione peroxidase Gpx3 attacks the O–O bond of H2O2, thereby forming a sulfenic acid at the active site cysteine. At the same time, Ybp1 binds to Yap1p. (3) In the next step, the active site sulfenic acid of Gpx reacts with Cys598 of Yap1p, causing the formation of a Gpx3-Yap1p disulfide intermediate. (4) A thiolate anion formed at Cys303 of Yap1p attacks the intermolecular disulfide bond, thereby causing the formation of the inter-domain Cys303–Cys598 disulfide bond and the recovery of reduced Gpx3. (5) Additional inter-domain disulfide bonds form in a process likely also involving Gpx3. While formation of the Cys310–Cys629 appears to increase transcriptional activation, (6) formation of Cys315–Cys620 might prolong the H2O2-mediated signal. The exact role of Ybp1 has yet to be determined, but it appears that Ybp1 mediates the redox signal from Gpx3 to Yap1p. (7) Disulfide bond formation appears to lead to conformational changes in Yap1p, which mask the NES and prevent nuclear export. Yap1p accumulates in the nucleus and activates the oxidative stress response. For reasons of simplicity, only one oxidized form of Yap1p is shown in the nucleus.
Yap1p's function as a transcription factor is controlled by redox-regulated changes in the accessibility of the NES (69, 70). In nonstressed yeast cells, the NES is exposed and Yap1p shuttles between the nucleus and the cytosol. Upon migration into the nucleus, Yap1p is rapidly exported in a process that is mediated by binding to the nuclear export receptor Crm1 (also called Xpo1) (70, 147). The outcome is a relatively low nuclear level of Yap1p and therefore a low constitutive activity in unstressed yeast cells. Exposure of yeast cells to H2O2, however, leads to rapid redox-mediated conformational changes in Yap1p, that disrupt the interaction with Crm1. This, in turn, leads to the nuclear accumulation of Yap1p and results in significantly increased antioxidant gene transcription (31, 69).
Yap1p—Member of a Two-Component Redox Relay
Over the past few years it became evident that Yap1p is not directly oxidized by H2O2 but functions in a two-component system with its partner protein glutathione peroxidase Gpx3 (also named Orp1), a protein originally identified as a hydroperoxide scavenger (54). Upon exposure to H2O2, the catalytic site cysteine Cys36 of Gpx3 attacks the O–O bond of the substrate, thereby forming a sulfenic acid (Cys36-SOH) at its active site cysteine (81) (Fig. 2). Cys36-SOH subsequently reacts with Cys598 of Yap1p's c-CRD to form a mixed Yap1p-Gpx3 disulfide intermediate. Rearrangement of this disulfide by intramolecular thiol-disulfide exchange with Cys303 of Yap1p then leads to the formation of the first inter-domain disulfide bond between Cys303-Cys598 in Yap1 and the regeneration of reduced Gpx3 (32, 81). Triggered by this inter-domain disulfide bond formation, Yap1p undergoes further conformational changes that apparently mask the NES and disrupt the Yap1p–Crm1 interaction (31, 96) (Fig. 2) Subsequently, additional disulfide bonds between cysteines of n-CRD and c-CRD of Yap1p form (see below) presumably in a similar Gpx3-mediated process (29). Upon return to nonstress conditions, the disulfide bonds are reduced by the thioredoxin-system, leading to the reversal of the structural rearrangements and to the re-exposure of NES (31). Thus, Crm1-mediated nuclear export of Yap1p can resume and Yap1's transcriptional activity returns to its low pre-stress levels (143).
A similar relay system has also been described for the redox-regulation of the mammalian AP-1 transcription factor that is formed upon dimerization of c-Fos and c-Jun. In contrast to Yap1p, however, which requires the sulfenic acid in Gpx3 for its oxidative activation, mammalian AP-1 uses the reduced cysteines of the redox factor Ref-1 for its reductive activation (139, 145). In the c-Fos/c-Jun AP-1 transcription factor, a single conserved cysteine residue in the DNA-binding domains of each of the two individual proteins appears to form the molecular redox switch (1). In vitro treatment of AP-1 with alkylating agents, such as _N_-ethylmaleimide (NEM) has been shown to cause a decrease in transcriptional activity while exposure to reductants, such as DTT, or mutation of the cysteines results in enhanced DNA binding (1, 97). Once oxidized and inactivated, AP-1 cannot be directly reduced and activated by thioredoxin (Trx) (1). Activation of AP-1 requires Ref-1, which appears to draw its electrons from thioredoxin and ultimately from NADPH (146).
Redox-Mediated Fine-Tuning of Yap1p's Functional Activity
In vitro studies demonstrated that Yap1p's activation by H2O2 involves a multistep formation of three inter-domain disulfide bonds. This finding led to a model suggesting that all six cysteine residues in Yap1p are important for optimal H2O2 sensing and signal transduction (96). Transcriptional activity of Yap1p appears to be directly determined by the balance between Gpx3-mediated disulfide bond formation in Yap1p, which is regulated by intracellular H2O2 levels, and thioredoxin-mediated disulfide bond reduction, which is regulated by intracellular NADPH levels (32), suggesting that the cellular ratio of H2O2 to NADPH ultimately determines the number of disulfide bonds that form and the level of Yap1 activity (96).
In vitro activation generates at least three oxidation forms of Yap1p, which differ in the number of disulfide bonds (96). As mentioned above, formation of the Cys303–Cys598 disulfide bond seems to initiate the activation of Yap1p by directly causing conformational changes that bury the NES. Formation of the second inter-domain disulfide bond between Cys310 and Cys629 appears to further increase Yap1p's transcriptional activity. Both inter-domain disulfide bonds seem to be required for the recruitment of Rox3p to the TRX2 promoter, a prerequisite for maximum Yap1p-mediated TRX2 expression (45). Moreover, Yap1p mutant strains carrying substitutions at either Cys310 or Cys629 show a mixture of oxidized and reduced Yap1p species upon H2O2 stress in vivo. This result suggests that the formation of the Cys310–Cys629 bond stabilizes the Cys303–Cys598 disulfide and prevents its premature reduction (143). The arrangement of the third disulfide bond between Cys315 and Cys620 appears to be important for maintaining Yap1p in the nucleus and upholding its activity in the later stages of the H2O2 response (96).
Noteworthy at this point is the observation that a third protein appears to be involved in peroxide-induced Yap1p oxidation. In vivo studies showed that the protein Ybp1 forms an H2O2-induced complex with the c-CRD-containing region of Yap1p, thereby stimulating nuclear accumulation and activity of the transcription factor (137). Although Ybp1 has several cysteine residues, this interaction appears to be based on a nonredox function of Ybp1 (29). Because Δybp1/Δgpx3 double mutants show very similar effects on the regulation of Yap1p's activity as the individual single mutants, a model was proposed in which Ybp1 and Gpx3 act in the same pathway (137) (Fig. 2). The H2O2 stress-induced stimulation of the Yap1p–Ybp1 complex formation, together with the cytoplasmic localization of Ybp1, suggests that the molecular function of Ybp1 might be important for disulfide bond formation or signal transduction between Gpx3 and Yap1p (95, 137). Alternatively, Ybp1 might function as mediator in the formation of the Gpx3–Yap1p disulfide intermediate or in preventing Gpx's sulfenic acid intermediate from reacting with its own second cysteine, which would short-cut the oxidation relay.
Activation of Yap1p—More Than One Way to Get the Response
In contrast to the activation of Yap1p in response to ROS, such as peroxide, which depends on Gpx3 and/or Ybp1, activation of Yap1p in response to diamide, electrophiles, and divalent heavy metal cations appears to function independently of Gpx3 and does not involve any inter-domain disulfide bonds (8, 31). In the presence of diamide, thiol modification of the three cysteines in c-CRD seems sufficient to disrupt interaction between Yap1p and Crm1 and to promote the nuclear accumulation of Yap1p (67). Diamide-mediated thiol modifications might either lead to the formation of an intra-domain disulfide bond between two of the three cysteines in c-CRD (67) or cause individual cysteine modification, such as the formation of sulfenylhydrazine (8, 31, 67). Activation of Yap1p without disulfide bond formation was also observed in response to the superoxide generator and alkylating agent menadione and _N_-ethylmaleimide under anaerobic conditions as well as in a Δgpx3 strain (8). Based on these findings, it is likely that other non-ROS inducers of Yap1p, such as cadmium or selenate, activate Yap1p by either binding to or chemically modifying individual C-terminal cysteines (8, 31). This type of activation is similar to the diethylmaleate-mediated activation of the Yap1 homologue Pap1 in Saccharomyces pombe, which is suggested to operate through covalent cysteine adduct formation (23).
Yap1p—A Prototype for Emerging Concepts in Redox Regulation?
Yap1p's mechanism of activation combines a number of features that have become increasingly common among redox-regulated proteins. One concept that emerges is that redox-regulated disulfide bond formation causes extensive conformational changes in the affected proteins, which either lead to the folding of previously unfolded regions (e.g., Yap1p, Pap1) (23, 143) or to the unfolding of previously folded regions (e.g., Hsp33, RsrA) (52, 98). Whether disulfide bond formation mediates folding or unfolding of the protein depends largely on the individual protein, its function, and the mechanism of regulation. Interestingly, it appears not to correlate with the final activity of the oxidized protein. Disulfide bond formation in Yap1p, for instance, converts a poorly folded region into a compactly folded protein domain in a process that effectively buries the NES and activates the transcriptional response (143). In contrast, ROS-mediated disulfide bond formation in the redox-regulated chaperone Hsp33 causes large portions of the protein to adopt a natively unfolded protein conformation (52). This unfolding is crucial for the exposure of a substrate-binding site in Hsp33, which is capable of binding proteins that undergo stress-induced unfolding and protects bacteria against oxidative damage (52). In the anti-sigma factor RsrA, on the other hand, disulfide-mediated unfolding abolishes its binding to the sigma factor, which is released and induces antioxidant gene transcription in Streptomyces coelicolor (98). Extensive biophysical studies on other redox-regulated proteins will reveal how many redox-regulated proteins use this rapid and fully reversible mechanism to translate changes in their redox state into functional changes.
A second mechanistic aspect that might turn into a new paradigm is that thiol- based regulatory switches such as found in Yap1p or the related Pap1p do not function as primary ROS sensors. Instead, both redox-regulated transcription factors use peroxidases as ROS sensors, which relay the redox signal from their active site cysteine to the acceptor protein and trigger disulfide bond formation (138). It is conceivable that other thiol peroxidases serve as central relay stations for yet to be identified redox-regulated proteins in vivo.
Redox Theme With Variations—The Nrf2-Keap1 Connection
In multicellular organisms, protection against ROS, nitric oxide, heavy metals, and electrophilic compounds is primarily mediated by the redox-sensitive Nrf2-Keap1 signaling pathway (35, 65). Nrf2, the nuclear factor erythroid-2 related factor, functions as master transcription factor. It forms heterodimers with other transcription factors such as c-Jun, Jun B, Jun D, ATF3, and ATF4, thereby regulating the transcription of >200 eukaryotic genes. The gene products are involved in cellular protection against oxidative stress, in detoxification of electrophiles, and in proteasomal activity (65).
Under nonstress conditions, Nrf2 forms a tight complex with the redox-sensitive stress receptor protein Keap1 (Kelch-like ECH-associated protein-1). Keap1 functions as a negative regulator of Nrf2 by retaining the transcription factor in the cytoplasm and targeting it for ubiquitin-mediated proteasomal degradation (64). Keap1 facilitates Nrf2's proteasomal degradation by acting as a substrate adaptor protein for Cul-containing E3 ubiquitin ligases, which catalyze the ubiquitination and subsequent proteasomal degradation of Nrf2 (reviewed in ref. 151). Exposure of cells to ROS or other inducers, however, promotes the escape of Nrf2 and its subsequent translocation into the nucleus, where it activates the antioxidant response element (ARE) (94).
Stress sensing is mediated by the cysteine-rich protein Keap1. At least three (Cys151, Cys273, Cys288) of the 25 to 27 cysteines present in mammalian Keap1 appear to play critical roles. Interestingly, any type of thiol modifications at these three cysteine residues seem to be sufficient to induce conformational rearrangements in Keap1, which abrogate its binding to Nrf2 and activate the oxidative stress response (65). Recently, Keap1 was identified as a metal-binding protein, which binds zinc in a 1:1 stochiometry (35). Mutant studies suggested that two of the three redox-sensitive cysteines (Cys273 and Cys288) are directly involved in zinc coordination under reducing nonstress conditions (35). Exposure to ROS was found to induce disulfide bond formation and lead to zinc release and conformational rearrangements in Keap1, suggesting that Keap1 senses ROS via a redox-sensitive zinc center. This model is very reminiscent of the redox-regulated anti-sigma factor RsrA (98) and the bacterial chaperone Hsp33 (52). Both proteins use redox-sensitive zinc-centers as ROS-sensors and undergo dramatic conformational changes upon disulfide bond formation and zinc release. The additional layers of regulation that are involved in the Nrf2-Keap1 signaling pathway have been expertly reviewed in the past (151), and their discussion goes beyond the scope of this review.
GapDH—Putting Cellular Metabolism Under Redox-Control
A variety of key metabolic enzymes in eukaryotic cells have been identified to contain redox sensitive active site cysteine residues, which become oxidatively modified upon exposure to ROS and/or RNS. These redox-regulated metabolic enzymes include among others carbonic anhydrase, creatine kinase (CK ), and glyeraldehyde-3-phosphate dehydrogenase (GapDH) (62, 88, 105). Here, we focus on GapDH, which has a central function in glycolysis in both prokaryotic and eukaryotic cells. Glycolysis is the almost universal central pathway of glucose catabolism and represents the main source of metabolic energy for many cells and organisms. GapDH catalyzes the reversible oxidative phosphorylation of glyeraldehyde-3-phosphate (GAP) to 1,3-bisphosphoglycerate in the presence of nicotinamide adenine dinucleotide and inorganic phosphate. It uses a highly conserved and reactive active site cysteine (Cys149 or Cys150, respectively) for the initial nucleophilic attack on the aldehyde of GAP (18). This active site cysteine has been shown to be highly sensitive to oxidative modifications by a variety of different reactive oxygen and nitrogen oxide species. In addition, GapDH appears to play an important role in a plethora of other processes that are unrelated to its glycolytic function (126). These include transcription (153), regulation of Ca2+-homeostasis (100), membrane fusion, microtubule bundling, phosphotransferase activity, nuclear RNA export, and DNA replication and repair (24, 126). Recent studies have furthermore shown an active role of GapDH in apoptosis (47) and an involvement in Huntington's (117), Parkinson's (135), and Alzheimer's disease (83). The proapoptotic role of GapDH in these diseases seems to be dependent upon its nuclear accumulation and its formation of insoluble aggregates (28, 114).
GapDH functions as a homotetramer. Each subunit contains one active site cysteine and a second, somewhat less conserved cysteine three amino acids apart. Upon exposure to oxidative stress conditions, the active-site thiolate in GapDH was shown to rapidly become the target of both reversible and irreversible thiol modifications, whose nature largely depends on the type and concentration of oxidants. In the presence of H2O2, for instance, GapDH's active site cysteine was found to be oxidized to either sulfenic, sulfinic, sulfonic acid, or engaged in an intramolecular disulfide bond with the neighboring Cys154 (75). Incubation with HNO (nitroxyl anion) or the NO-donor DEA/NO (diethylamine NONOate), on the other hand, was shown to cause _S_-nitrosation (47, 88), _S_-glutathionylation (26, 88, 120), carbonylation (21), and ADP-ribosylation (34). All these modifications cause the reversible or irreversible inhibition of GapDH enzyme activity and lead to the attenuation of glycolysis in vivo.
Re-Routing the Metabolic Flux to Protect Cells Against Oxidative Damage
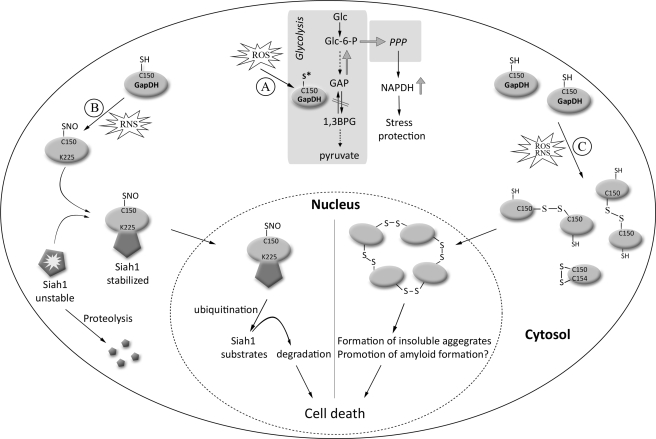
Oxidative stress-induced inhibition of GapDH as a central component of glycolysis leads to the rapid re-routing of glucose-6-phosphate into the pentose phosphate pathway (PPP). This serves as immediate response to protect against oxidative insults (44, 104) (Fig. 3). The PPP is one of the key routes for nicotinamide adenine dinucleotide phosphate (NADPH) production in the eukaryotic cytoplasm. The product NADPH serves as electron donor for both glutathione reductase (Grx) and thioredoxin reductase (TrxR), and is therefore essential to restore and maintain the reducing environment of the cytosol (6). Oxidative inactivation of triose phosphate isomerase (TPI) causes a similar re-direction of the metabolic flux from glycolysis to PPP. TPI, which directly precedes GapDH in glycolysis, catalyzes the isomerization of dihydroxyacetone phosphate to GAP. Yeast cells with reduced TPI activity are highly resistant to treatment with the oxidant diamide (103), supporting the model that re-routing the glucose flux maintains the cytoplasmic NADPH/NADP+ equilibrium and counteracts oxidative stress. When cells return to nonstress conditions, reduced glutathione and thioredoxin (77) rapidly restore GapDH activity and therefore the fine-tuned metabolic flux of glucose.
FIG. 3.
Oxidative thiol modifications of GapDH—From cytoprotection to cytotoxicity. (A) ROS-mediated inhibition of GapDH re-routes metabolic flux as immediate defense against oxidative insult. Oxidative stress-induced inhibition of GapDH as a central component of glycolysis leads to the rapid re-routing of glucose-6-phosphate into the pentose phosphate pathway (PPP), one of the key routes for NADPH production in the eukaryotic cytoplasm. NADPH serves as electron donor for glutathione reductase and thioredoxin reductase, and is therefore essential to restore and maintain the reducing environment of the cytosol. (B) Model of GapDH/Siah1-signaling cascade-dependent cell death. Exposure of GapDH to RNS causes _S_-nitrosation of Cys150 in GapDH. _S_-nitrosation stimulates the interaction between the E3-ubiquitin ligase Siah1 and GapDH. Siah1, which possesses a nuclear localization signal (NLS), escorts GapDH into the nucleus where the association between GapDH and Siah1 appears to stabilize the E3-ligase. This stabilization, in turn, increases the rate of degradation for a set of different target proteins, and ultimately triggers cell death. (C) Model of apoptosis triggered by ROS-mediated GapDH aggregation. Exposure of GapDH to excess ROS leads to the initial disulfide bond formation between either Cys150 and Cys154 (intramolecular disulfide bond) or between two active site cysteines Cys150. The latter leads to extensive conformational changes and exposes additional cysteines (e.g., Cys281) that undergo further intermolecular disulfide bond formation. This results in the accumulation of insoluble aggregates composed of multiple GapDH subunits in the cytosol and the nucleus. The aggregates are either intrinsically cytotoxic or interact with other aggregation-prone proteins (e.g., Aß, tau, α-synuclein) to cause cytotoxicity. For reasons of simplicity, only one subunit of the GapDH tetramer is shown.
These findings show that ROS-mediated changes in the activity of glycolytic enzymes trigger important and immediate response mechanisms that control the redox state of the cell and protect cells against increased oxidative damage. These observations also form the basis for discussions that inhibitors of glycolysis such as 2-deoxy-D-glucose (71) might serve as potential therapeutics for oxidative stress-related neuronal disorders such as Alzheimer's disease and Parkinson's disease (104).
ROS-Mediated GapDH Aggregation and Its Role in Apoptosis
Both nitrosative and oxidative stress conditions have been shown to lead to intermolecular disulfide bond formation between GapDH subunits, causing extensive GapDH oligomerization both in vitro and in vivo (28, 92) (Fig. 3). Interestingly, these aggregates were very similar in their characteristic features (e.g., Congo red staining) to amyloid-like fibrils previously reported for α-synuclein (66) and ß-amyloid (14). The ability of thiol reductants such as dithiothreitol (DTT) to dissolve these aggregates was dependent on the extent of oxidation, supporting the model that intermolecular disulfide bonds are involved in aggregate formation. Two cysteines appear to play the major role in this oxidative stress-induced aggregation process, the active site cysteine (Cys149 in rabbit GapDH, Cys150 in yeast GapDH) and, to a lesser extent, Cys281 (28, 92). The precise mechanism by which these large disulfide-linked GapDH oligomers form has yet to be determined. Given that the active site cysteine is located at the bottom of GapDH's catalytic site, the distance between these cysteines on the individual subunits is too far to allow inter-subunit disulfide bond formation to occur (92). It is conceivable, however, that initial thiol modification of Cys150 causes extensive conformational changes that lead to the exposure of additional cysteines such as Cys281 and allow them to undergo intermolecular disulfide bond formation (92).
Importantly, in vivo studies demonstrated that the degree of GapDH aggregation correlates well with the rate of oxidative stress-induced cell death (92). These results suggested that insoluble GapDH aggregates are either cytotoxic per se, or promote apoptosis indirectly by interacting with other intracellular aggregate-prone proteins, such as tau or α-synuclein (91, 110) (Fig. 3). This mechanism appears to contradict an earlier model proposed by Hara et al. in which NO-induced alterations of the GapDH/Siah1 signaling cascade were thought to cause the GapDH-dependent cell death under nitrosative stress conditions (47) (Fig. 3). According to this model, _S_-nitrosation of Cys150 in GapDH of HEK293 cells stimulates the interaction between the ATP-dependent E3-ubiquitin ligase Siah1, and Lys225 in GapDH (47, 50). Siah1, which possesses a nuclear localization signal (NLS), escorts GapDH into the nucleus where the association between GapDH and Siah1 appears to stabilize the E3-ligase (47). Stabilization of Siah1, in turn, increases the rate of degradation for a set of different target proteins and ultimately triggers apoptosis (109) (Fig. 3). As so often in biology, it is more than likely that both mechanisms are linked to oxidative stress-induced cell death. More research is required to identify whether the two mechanisms work synergistically or independently, and to elucidate whether oxidant specificity of GapDH's active site cysteine determines the apoptotic route that cells take (47). In either case, these studies demonstrate that GapDH, a protein best known for its central role in glycolysis, probably plays an equally important role as a redox-regulated proapoptotic protein (47, 92).
Role of GapDH's Redox-Regulation in Signaling
Nuclear translocation of GapDH mediated by oxidative modifications of its active site cysteine appears to correlate with the initiation of cell death and the physiology of a number of neurodegenerative diseases (28, 47, 114, 135). In addition, GapDH translocation might also play a role in intracellular signaling (24, 47). GapDH has long been shown to associate with DNA (108), and translocation into the nucleus might further stimulate this interaction. It is conceivable that oxidative stress-mediated nuclear translocation of GapDH serves as mediator for relaying extranuclear stress signals into the nucleus, which would translate changes in the cellular redox state directly into changes in transcriptional activity (122). This is in line with data that show the direct involvement of metabolic enzymes or homologs such as GapDH or CtBP in transcription (121, 153). P38, a nuclear form of GapDH, was found to be a key component of OCA-S, a multicomponent Oct-1 co- activator complex, which is essential for S phase-dependent histone H2B transcription during DNA replication (153). GapDH's direct interaction with the promoter-bound Oct-1 supports the binding of the OCA-S complex to Oct-1. Noteworthy, both the interaction of GapDH with Oct-1 as well as the transcriptional activity of OCA-S are altered by the NAD+/NADH redox status (153). This NAD+/NADH influence might link the metabolic and/or redox state of the cell to histone transcription and DNA replication. However, the structural basis for NAD+/NADH modulation and the mechanism by which GapDH supports gene transcription have yet to be determined (153).
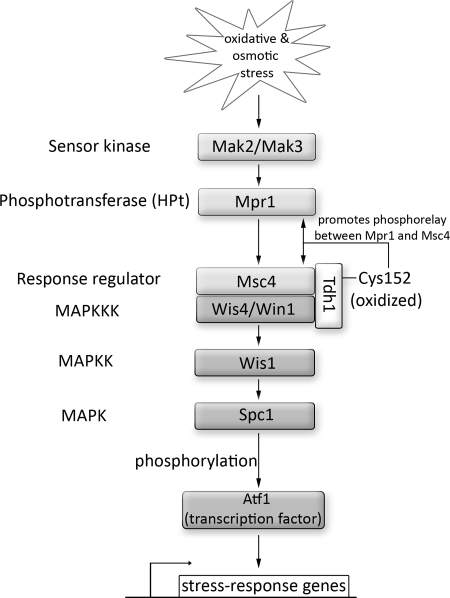
Very recently, the involvement of GapDH's redox regulation in a multistep phosphorelay signaling in Schizosaccharomyces pombe was reported (90). In phosphorelays, which are variations of the bacterial two-component signaling system, a histidine-containing phosphotransfer (HPt) protein mediates the phosphotransfer from a sensor kinase to a response regulator, which in turn activates a kinase cascade and eventually causes induction of the corresponding response genes (111). In Schizosaccharomyces pombe, osmotic and oxidative stress conditions activate the mitogen-activated protein kinase (MAPK ) Spc1 (124) (Fig. 4). Activated Spc1 phosphorylates the Atf1 transcription factor, which induces several stress response genes, including ctt1+ encoding for cytoplasmic catalase (140). The transmission of the stress signals to Spc1 in S. pombe occurs via a multistep phosphorelay system consisting of the phosphotransfer protein Mpr1 and the response regulator Mcs4, which forms a complex with the stress-responsive Wis4/Win1 MAP kinase kinase kinase (MAPKKK ) (5). Morigasaki et al. showed that the Msc4-MAPKKK complex contains Tdh1, an isoform of GapDH (90). Oxidative modification of Tdh1's active site cysteine Cys152 was found to result in enhanced binding between Tdh1 and Msc4, which appears to be essential for the interaction between Msc4 and Mpr1 and promotes the transmission of H2O2 stimuli to the downstream components of the MAP-kinase cascade (i.e., Wis1, Spc1) (5) (Fig. 4). Mutation of the active site cysteine (C152S) or deletion of the tdh1 gene was demonstrated to cause a decrease in Msc4 and Mpr1 interaction, resulting in a defect in H2O2-signaling through the phosphorelay. These results reveal that peroxide-induced oxidative modification of the redox-sensitive cysteine in GapDH plays a key role in transmitting H2O2 stimuli to a response regulator, which is part of a multistep phosphorelay cascade, thereby modulating cellular signaling (90). As GapDH is conserved in multiple eukaryotic organisms, GapDH might play an important role in other signaling relay systems in different species.
FIG. 4.
Role of redox regulation by GapDH in stress signaling. In the fission yeast Schizosaccharomyces pombe, a multistep phosphorelay composed of the sensor kinase Mak2/Mak3, the histidine-containing phosphotransferase (HPt) Mpr1 and the Msc4 response regulator transmit stress signals, such as H2O2, to the Spc1 MAPK cascade. This cascade is composed of the Wis4/Win1 MAPKKKs, the MAPKK Wis1, and Spc1 MAPK as final receptor. Wis4/Win1 and Msc4 form a complex with the GapDH isoform Tdh1. H2O2-mediated oxidative modification of Tdh1's active site cysteine Cys152 enhances the interaction between Tdh1 and the Msc4 response regulator, which, in turn, promotes the interaction and phosphorelay signaling between the response regulator Msc4 and the HPt protein Mpr1. This interaction is required for the transmission of the H2O2 stress signal to Spc1.
GapDH—One of Many Redox-Regulated Metabolic Enzymes
GapDH is only one of many metabolic enzymes whose activity is regulated by oxidative thiol modifications (155). Creatine kinase, for instance, which plays a central role in controlling cellular energy homeostasis in energy demanding tissues such as muscle and brain, contains one highly oxidation-sensitive active site cysteine Cys283 (105). ROS and RNS-mediated thiol modification of Cys283 leads to the reversible inactivation of creatine kinase, which appears to affect muscle performance and Ca2+ homeostasis (130). This inactivation seems to be of particular importance during oxidative stress conditions encountered in ischemia, cardiomyopathy, and several neurodegenerative disorders (reviewed in ref. 115). Similarly, both glycogen synthase and protein phosphatase-I are readily inactivated upon treatment with oxidized glutathione (38, 123). This inactivation might explain the high glycogenolytic activity that has been observed in liver tissue treated with thiol oxidants (115). Carbonic anhydrase 3, which maintains pH homeostasis in mammals, has been found to be sensitive to oxidative modifications of Cys186 and Cys181 (62). Although earlier results suggested already a role of carbonic anhydrase 3 in the cellular response to oxidative stress (19), it remains to be determined whether these observed oxidative thiol modifications are involved. Interestingly, for many of these enzymes, oxidative modifications such as _S_-glutathionylation have been considered to be a protective measure rather than a redox-regulatory feature, with the purpose to protect active site cysteines against irreversible overoxidation and inactivation (30, 32). However, because loss of enzyme activity is inevitably connected with these oxidative thiol modifications and rapid reactivation is usually achieved upon return to reducing nonstress conditions, this argument might be purely semantic.
Redox Regulation of Eukaryotic Signal Transduction Cascades
Reversible protein tyrosine phosphorylation and dephosphorylation are important posttranslational protein modifications involved in a variety of cellular signal transduction cascades that control metabolism, motility, cell growth, proliferation, differentiation, and survival (4, 11). Whereas phosphorylation reactions are carried out by specific tyrosine kinases, dephosphorylation reactions are catalyzed by members of a large superfamily of protein tyrosine phosphatases (PTPs). In the human genome, 107 PTP encoding genes have been identified (4). Even though members of the PTP superfamily show only low sequence similarities and possess different topologies (12, 39), they are all characterized by an active site cysteine located within a highly conserved 11-residue motif [I/V]HCXXGXXR[S/T]. This active site cysteine exhibits a very low pKa value [∼5.6 in PTP1B, ∼4.7 in YOP phosphatase (78, 152)]. It is present as a thiolate anion at physiological pH, which renders the catalytic site cysteine highly reactive and promotes its nucleophilic attack at a phosphotyrosine (pTyr) substrate (11, 40). This reaction results in the formation of a covalent phospho-cysteine intermediate, which is subsequently hydrolyzed by an activated water molecule (33). The very same properties that make PTP's active site cysteine such an excellent nucleophile, also makes it highly susceptible to oxidants such as H2O2 and superoxide, and make the proteins highly sensitive to oxidative stress-mediated inactivation (33). By regulating tyrosine phosphorylation- dependent signaling events using redox-mediated posttranslational modification of PTP activity, ROS can actively function as second messenger (33, 106).
PTP1B—Using Cyclic Sulfenamide Formation as Redox Switch
PTP1B, a prototypic member of the PTP family, is widely expressed in multiple cell types and tissues including brain, liver, and skeletal muscle (36). It is considered to be the physiological regulator of glucose homeostasis via its control of leptin sensitivity and its function as a major negative regulator of the insulin pathway (36, 37). Furthermore, PTP1B appears to have oncogenic properties in various cancer types such as in colon cancer, where it activates Src by catalyzing the dephosphorylation of the negative regulator residue Tyr530 (154).
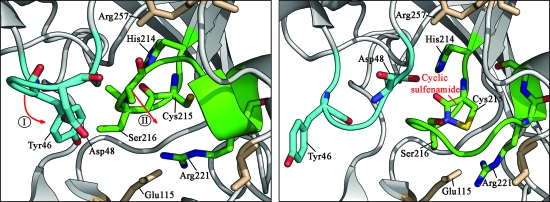
In active PTP1B, the catalytic cysteine Cys215 is reduced and located at the base of a cleft that lies within the pTyr substrate binding pocket (11, 12) (Fig. 5). Oxidation of Cys215 in the presence of mild oxidative conditions, such as 100 μ_M_ H2O2, causes the reversible formation of a cyclic sulfenamide species (Fig. 6). This oxidation is accompanied by major changes in the structure of the active site and leads to the inhibition of enzyme activity (112, 136). The generation of a cyclic sulfenamide bond in PTP1B probably occurs through the initial oxidation of Cys215 to a sulfenic acid intermediate. This step is followed by a nucleophilic attack of the backbone nitrogen atom of Ser216 on the sulfenated Cys215 forming cyclic sulfenamide and concomitantly releasing water (112, 136). Amino acids in the active site of PTP1B, such as the highly conserved His214, facilitate the cyclization by providing steric and/or electrostatic effects (112) (Fig. 5).
FIG. 5.
Redox-mediated conformational changes in the catalytic site of PTP1B. Oxidation and sulfenamide bond formation of PTP1B's active site cysteine causes significant conformational changes in the catalytic site of PTP1B. Large rearrangements of both pTyr loop (blue) and PTP loop (green) inhibit substrate binding and apparently protect the active site Cys215 against irreversible overoxidation. Figures were made with PyMOL using the coordinates of reduced human PTP1B protein (2BGE) and the sulfenamide species (1OEM) deposited in the Protein Data Bank. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
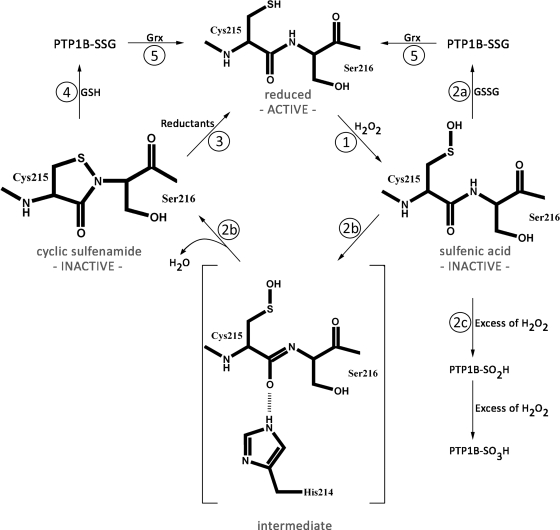
FIG. 6.
PTP1B—Redox regulation by cyclic sulfenamide formation. Under nonstress conditions, the active site cysteine (Cys215) of tyrosine phosphatase PTP1B is reduced and PTP1B is active. (1) Upon exposure to oxidative stress conditions (e.g., H2O2), Cys215 becomes oxidized to sulfenic acid, which inactivates the enzyme. (2a) Reaction of the active site sulfenic acid with oxidized glutathione leads to _S_-glutathionylation (PTP1B-SSG). (2b) Alternatively, a nucleophilic attack of the backbone nitrogen of Ser216 on the Sγ-atom of Cys215 occurs, which results in the formation of a cyclic sulfenamide and the release of water. Sulfenamide formation is promoted by the environment of the catalytic site, with His214 playing a prominent role in polarizing the amide bond of Ser216. (2c) In the presence of excess H2O2, PTP1B's sulfenic acid is overoxidized to either sulfinic or sulfonic acid. (3) Active PTP1B can be either directly regenerated by reducing agents such as DTT in vitro or (4) indirectly via the reaction of cyclic sulfenamide with GSH and the formation of PTP1B-SSG. (5) Reduced active PTP1B is then formed by the subsequent reaction of PTP1B-SSG with the glutaredoxin (Grx) system.
Upon formation of the active site sulfenamide, PTP1B undergoes dramatic tertiary structural rearrangements at its catalytic site (Fig. 5). These involve conformational rearrangements of both PTP and pTyr loops, which are central to catalysis and substrate recognition. In addition, converting the PTP loop into an open conformation exposes the Sγ atom of Cys215, which might enhance the accessibility of the catalytic cysteine for reducing agents once nonstress conditions are restored (112). Reducing agents such as GSH or DTT have been shown to reduce the active site sulfenamide, reverse the conformational changes, and fully restore the phosphatase activity (112, 136). In contrast, however, in the presence of excess H2O2, overoxidation of the active site cysteine to sulfinic and sulfonic acid occurs. This process is irreversible and causes permanent inactivation of the phosphatase. Because cyclic sulfenamide formation has been found to be stable for >5 h in 50 μ_M_ H2O2 (112), it is thought that this oxidative thiol modification functions as a protective mechanism to prevent PTP1B against irreversible overoxidation (112, 136). A similar protection against overoxidation via sulfenamide bond formation was also shown for OhrR in Bacillus subtilis (73). In addition to sulfenamide bond formation, glutathionylation of the catalytic cysteine in PTP1B might also protect the enzyme against overoxidation (13). So far, what different roles glutathionylation and sulfenamide bond formation might play in the protection of the catalytic cysteine remains unclear. Salmeen et al. (113) suggest that the formation of a sulfenamide bond may be the first step in protection, followed by glutathionylation, which can occur as the enzyme is being reduced (Fig. 6).
Overoxidation of PTP1B—More Than a Dead-End Product?
A recently applied MS-based approach revealed the extent of oxidative thiol modification of PTP1B both in vitro and in vivo (79). This approach verified that only the active site cysteine of PTP1B is the target of thiol oxidative modifications. Surprisingly, however, quantitative analysis of the in vivo oxidation state of PTP1B in cancer cell lines that continuously produce ROS revealed that ∼40% of PTP1B molecules were irreversibly overoxidized. Between 25% and 50% of all PTP1B molecules were found to be reversibly oxidized to either sulfenic acid or sulfonamide, whereas only 10%–15 % of cellular PTP1B was found in its reduced and active form (79). These results indicated that the level of ROS in these cell lines is sufficient to cause terminal oxidation of a significant amount of PTP1B protein, despite the presence of high levels of glutathione (51). Interestingly, overoxidation of the active site cysteine in PTP1B appears to fully reverse the conformational changes that accompany ROS-mediated sulfenamide formation (112, 136). It is therefore not surprising that overoxidized PTP1B does not trigger the cellular machinery that is usually responsible for the rapid degradation of oxidatively damaged proteins (79). It will now be interesting to determine whether the overoxidized form of PTP1B is indeed an end product that is destined for degradation or whether it fulfills a yet to be determined alternative function in the cell. The latter would be reminiscent of 2-Cys peroxiredoxins, which are also highly susceptible to overoxidation under high H2O2 concentration (144). Sulfinic acid formation in 2-Cys peroxiredoxins leads to the inactivation of their peroxidase activity, but at the same time, appears to convert 2-Cys peroxiredoxins into proteins with molecular chaperone activity (57). This observation then raises the question as to whether PTP1B-specific sulfinic acid reductases exist, which, similar to sulfiredoxin in the case of overoxidized 2-Cys peroxiredoxins (16), return PTP1B into its original redox and functional state.
Other Redox-Regulated Signal Transduction Cascades in Eukaryotes
Many physiological ligands such as hormones, growth factors, and cytokines trigger intrinsic ROS production. ROS-mediated regulation of protein tyrosine phosphatases (PTPs) appears therefore to represent a general mechanism by which ligands control PTP activity and therefore signal transduction cascades. ROS- induced active site thiol modifications were demonstrated for a variety of PTPs in different cell types and included formation of disulfide bonds (e.g., LMW-PTPs, Cdc25, and PTEN), cyclic sulfenamide (PTP1B and RPTPa) (112, 148), and a potentially stabilized sulfenic acid (e.g., VHR and LAR) (60, 85, 118). The active site cysteine of the tumor suppressor PTEN (phosphatase with sequence homology to tensin), for instance, which regulates the overall activity of the PI3 kinase signaling pathway (132), is reversibly oxidized by either RNS or H2O2. Oxidative inactivation of PTEN leads to increased PtdIns(3,4,5)P3 (phosphatidylinositol (3,4,5)-trisphosphate) levels, increased Akt–phosphorylation and ultimately in cell growth (132). Reversible inactivation of MAP kinase phosphatase 3 (MKP3) upon H2O2 treatment, on the other hand, promotes TNFα-induced cell death (60). Interestingly, MKP3 does not form a disulfide with a single neighboring cysteine but can use multiple cysteines distributed in both the N-terminal and C-terminal domain to trap the sulfenic acid of the active site cysteine (Cys293) as a disulfide (118). The availability of multiple redox active cysteines, which has also been reported for other redox-sensitive proteins (86), might represent a general mechanism to prevent irreversible overoxidation of the active site sulfenic acids.
It is interesting to note at this point that ROS generated by distinct physiological stimuli often affect only a particular subset of redox-sensitive PTPs. This specificity is well illustrated by the PDGF-induced specific oxidation of SHP-2 (85) or the insulin-induced specific oxidation of PTP1B and TC-PTP (84). Furthermore, in-gel phosphatase assays revealed that ROS generated by cancer cells selectively affect a subset of cellular PTPs (79) and do not, as previously assumed, oxidize all redox-sensitive PTPs that are expressed. As of now, the reason behind this specificity is unknown. One explanation might be a more localized generation of ROS upon endogenous stimuli, which promotes oxidation of only those PTPs that are close to the source of ROS production. Alternatively, however, metabolites and/or proteins specific for individual signaling pathways might modulate the redox sensitivity of a subset of phosphatases, thereby making them more susceptible to ROS-mediated modification than others. Similar observations have been made with protein kinase C, whose redox regulation appears to be modulated by different retinoids (53).
Concluding Remarks
Over the past 10 years, a vastly increasing number of proteins have been identified that use thiol-based redox switches to quickly regulate their function, activity, or structure in response to changes in the redox homeostasis of the cell (3, 44, 48). As many of these redox-regulated proteins are central players of metabolic and signal transduction pathways, this strategy ensures that most cellular processes are under tight redox control. Here, we reviewed current knowledge about three eukaryotic proteins with thiol-based redox switches, Yap1p, GapDH, and PTP1B, and provided a short overview of other redox-regulated proteins similarly involved in gene expression, metabolic pathways or signal transduction. We used these model proteins to illustrate the variety of mechanisms by which cysteine thiols sense and respond to reactive oxygen and nitrogen oxide species. However, as the field expands, so does the knowledge that no singly unifying concept can be used to characterize redox-regulated proteins. While some redox-regulated proteins harbor only one oxidative stress-sensitive cysteine (e.g., GapDH, PTP1B) (13, 112, 120), others make use of cysteine clusters (e.g., Yap1b) (31, 69) or even redox-sensitive zinc centers (e.g., Hsp33, RsrA) (52, 56). The nature of the oxidative thiol modifications depends largely on the type of oxidant as well as on the environment of the cysteine within the protein. Moreover, because most oxidative modifications affect cysteines in structurally or functionally important regions, variations in the type of oxidative modifications often create distinct conformational changes, which can result in different functional states. Finally, whereas only a few proteins harbor generally oxidative stress-sensitive cysteines (e.g., GapDH), most proteins have been found to be highly specific for distinct oxidative or nitrosative stressors (17); the factors that are important for this specificity have yet to be determined.
Oxidative and nitrosative stress is involved in many disease conditions, and numerous redox-regulated proteins have been identified that use their redox sensitivity to protect cells and organisms against these stress conditions. Despite the tremendous increase in the number of redox-regulated proteins identified, it is still unclear what makes cysteines particularly sensitive to specific oxidants in some proteins and not in others. Our challenge for the future will be to obtain a comprehensive view about what makes individual proteins redox-sensitive. Can we predict, based on sequence and structure similarities, the redox sensitivity of individual cysteines, and more importantly, a protein's potential of being redox- regulated? This capability would greatly facilitate efforts to identify proteins that are potentially involved in the oxidative stress protection of the cell. A number of global proteomic techniques have been developed that allow the identification of redox-regulated proteins in cells and organisms (76, 77, 149). With the use of highly quantitative techniques such as the recently developed OxICAT method (75), the thiol oxidation status of hundreds of different proteins can be quantified in a single experiment. These techniques, in combination with extensive biochemical and structural studies, will have the potential to reveal novel redox-regulated proteins. These studies will deepen our understanding of redox regulation and further confirm the importance of oxidative thiol modifications as one of the key posttranslational modifications in pro- and eukaryotic organisms.
Abbreviations
AP-1, activator protein-1; ARE, antioxidant response element; ATF, activating transcription factor; bZIP, basic leucine zipper; CAT, catalase; CRD, cysteine-rich domain; Crm1, nuclear export receptor; GAP, glyeraldehyde-3-phosphate; GapDH, glyceraldehyde-3-phosphate dehydrogenase; Gpx, glutathione peroxidase; Grx, glutaredoxin; GSH, glutathione (reduced); GSNO, _S_-nitrosoglutathione; GSSG, glutathione disulfide; Hsf-1, heat shock transcription factor 1; Hsp33, heat shock protein 33; Keap1, Kelch-like ECH-associated protein-1; MAPK, mitogen-activated protein kinase; MKP3, MAP kinase phosphatase 3; NES, nuclear export signal; NF1, nuclear factor 1; NF-κB, nuclear factor kappa B; NLS, nuclear localization signal; Nrf2, NF-E2-related factor-2; PDGF, platelet-derived growth factor; PI3 kinase, phosphoinositide 3-kinase; PTEN, phosphatase and tensin homolog; PTP, protein tyrosine phosphatase; pTyr, phosphotyrosine; SOD, superoxide dismutase; Spc1, suppressor of phosphatase 2C; TC-PTP, T-Cell protein tyrosine phosphatase Tdh1, glyceraldehyde-3-phosphate dehydrogenase, isozyme 1; TPI, triose phosphate isomerase; Trx, thioredoxin; TrxR, thioredoxin reductase; USF, upstream stimulatory factor; Yap1p, yeast AP-1; Ybp1, Yap1p binding protein.
References
- 1.Abate C. Patel L. Rauscher FJ., 3rd Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 2.Ahn SG. Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17:516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen RG. Tresini M. Oxidative stress and gene regulation. Free Radic Biol Med. 2000;28:463–499. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 4.Alonso A. Sasin J. Bottini N. Friedberg I. Friedberg I. Osterman A. Godzik A. Hunter T. Dixon J. Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Aoyama K. Mitsubayashi Y. Aiba H. Mizuno T. Spy1, a histidine-containing phosphotransfer signaling protein, regulates the fission yeast cell cycle through the Mcs4 response regulator. J Bacteriol. 2000;182:4868–4874. doi: 10.1128/jb.182.17.4868-4874.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arner ES. Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 7.Aslund F. Zheng M. Beckwith J. Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Nat Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azevedo D. Tacnet F. Delaunay A. Rodrigues–Pousada C. Toledano MB. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic Biol Med. 2003;35:889–900. doi: 10.1016/s0891-5849(03)00434-9. [DOI] [PubMed] [Google Scholar]
- 9.Bandyopadhyay S. Gronostajski RM. Identification of a conserved oxidation-sensitive cysteine residue in the NFI family of DNA-binding proteins. J Biol Chem. 1994;269:29949–29955. [PubMed] [Google Scholar]
- 10.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Barford D. Das AK. Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 12.Barford D. Flint AJ. Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 13.Barrett WC. DeGnore JP. Konig S. Fales HM. Keng YF. Zhang ZY. Yim MB. Chock PB. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry. 1999;38:6699–6705. doi: 10.1021/bi990240v. [DOI] [PubMed] [Google Scholar]
- 14.Benzinger TL. Gregory DM. Burkoth TS. Miller–Auer H. Lynn DG. Botto RE. Meredith SC. Two-dimensional structure of beta-amyloid(10-35) fibrils. Biochemistry. 2000;39:3491–3499. doi: 10.1021/bi991527v. [DOI] [PubMed] [Google Scholar]
- 15.Berndt C. Lillig CH. Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am J Physiol Heart Circ Physiol. 2007;292:H1227–1236. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 16.Biteau B. Labarre J. Toledano MB. ATP-dependent reduction of cysteinesulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 17.Brandes N. Rinck A. Leichert LI. Jakob U. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol Microbiol. 2007;66:901–914. doi: 10.1111/j.1365-2958.2007.05964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodie AE. Reed DJ. Cellular recovery of glyceraldehyde-3-phosphate dehydrogenase activity and thiol status after exposure to hydroperoxides. Arch Biochem Biophys. 1990;276:212–218. doi: 10.1016/0003-9861(90)90028-w. [DOI] [PubMed] [Google Scholar]
- 19.Cabiscol E. Levine RL. Carbonic anhydrase III. Oxidative modification in vivo and loss of phosphatase activity during aging. J Biol Chem. 1995;270:14742–14747. doi: 10.1074/jbc.270.24.14742. [DOI] [PubMed] [Google Scholar]
- 20.Cadenas E. Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 21.Cahuana GM. Tejedo JR. Jimenez J. Ramirez R. Sobrino F. Bedoya FJ. Nitric oxide-induced carbonylation of Bcl-2, GAPDH and ANT precedes apoptotic events in insulin-secreting RINm5F cells. Exp Cell Res. 2004;293:22–30. doi: 10.1016/j.yexcr.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Carmel–Harel O. Stearman R. Gasch AP. Botstein D. Brown PO. Storz G. Role of thioredoxin reductase in the Yap1p-dependent response to oxidative stress in Saccharomyces cerevisiae. Mol Microbiol. 2001;39:595–605. doi: 10.1046/j.1365-2958.2001.02255.x. [DOI] [PubMed] [Google Scholar]
- 23.Castillo EA. Ayte J. Chiva C. Moldon A. Carrascal M. Abian J. Jones N. Hidalgo E. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol Microbiol. 2002;45:243–254. doi: 10.1046/j.1365-2958.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 24.Chuang DM. Hough C. Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- 25.Cook JA. Gius D. Wink DA. Krishna MC. Russo A. Mitchell JB. Oxidative stress, redox, and the tumor microenvironment. Semin Radiat Oncol. 2004;14:259–266. doi: 10.1016/j.semradonc.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Cotgreave IA. Gerdes R. Schuppe–Koistinen I. Lind C. S-glutathionylation of glyceraldehyde-3-phosphate dehydrogenase: Role of thiol oxidation and catalysis by glutaredoxin. Methods Enzymol. 2002;348:175–182. doi: 10.1016/s0076-6879(02)48636-3. [DOI] [PubMed] [Google Scholar]
- 27.Cumming RC. Andon NL. Haynes PA. Park M. Fischer WH. Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 28.Cumming RC. Schubert D. Amyloid-beta induces disulfide bonding and aggregation of GAPDH in Alzheimer's disease. FASEB J. 2005;19:2060–2062. doi: 10.1096/fj.05-4195fje. [DOI] [PubMed] [Google Scholar]
- 29.D'Autreaux B. Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 30.Dalle–Donne I. Rossi R. Giustarini D. Colombo R. Milzani A. S-glutathionylation in protein redox regulation. Free Radic Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Delaunay A. Isnard AD. Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaunay A. Pflieger D. Barrault MB. Vinh J. Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 33.Denu JM. Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 34.Dimmeler S. Lottspeich F. Brune B. Nitric oxide causes ADP-ribosylation and inhibition of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1992;267:16771–16774. [PubMed] [Google Scholar]
- 35.Dinkova–Kostova AT. Holtzclaw WD. Kensler TW. The role of Keap1 in cellular protective responses. Chem Res Toxicol. 2005;18:1779–1791. doi: 10.1021/tx050217c. [DOI] [PubMed] [Google Scholar]
- 36.Dube N. Tremblay ML. Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: From diabetes, obesity to cell cycle, and cancer. Biochim Biophys Acta. 2005;1754:108–117. doi: 10.1016/j.bbapap.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 37.Elchebly M. Payette P. Michaliszyn E. Cromlish W. Collins S. Loy AL. Normandin D. Cheng A. Himms–Hagen J. Chan CC. Ramachandran C. Gresser MJ. Tremblay ML. Kennedy BP. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 38.Ernest MJ. Kim KH. Regulation of rat liver glycogen synthetase D. Role of glucose 6-phosphate and enzyme sulfhydryl groups in activity and glycogen binding. J Biol Chem. 1974;249:5011–5018. [PubMed] [Google Scholar]
- 39.Fauman EB. Saper MA. Structure and function of the protein tyrosine phosphatases. Trends Biochem Sci. 1996;21:413–417. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- 40.Finkel T. Oxygen radicals and signaling. Curr Opin Cell Biol. 1998;10:248–253. doi: 10.1016/s0955-0674(98)80147-6. [DOI] [PubMed] [Google Scholar]
- 41.Finkel T. Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 42.Gasch AP. Spellman PT. Kao CM. Carmel–Harel O. Eisen MB. Storz G. Botstein D. Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giles GI. Tasker KM. Jacob C. Hypothesis: The role of reactive sulfur species in oxidative stress. Free Radic Biol Med. 2001;31:1279–1283. doi: 10.1016/s0891-5849(01)00710-9. [DOI] [PubMed] [Google Scholar]
- 44.Godon C. Lagniel G. Lee J. Buhler JM. Kieffer S. Perrot M. Boucherie H. Toledano MB. Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 45.Gulshan K. Rovinsky SA. Coleman ST. Moye–Rowley WS. Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J Biol Chem. 2005;280:40524–40533. doi: 10.1074/jbc.M504716200. [DOI] [PubMed] [Google Scholar]
- 46.Hampton MB. Kettle AJ. Winterbourn CC. Inside the neutrophil phagosome: Oxidants, myeloperoxidase, and bacterial killing. Blood. 1998;92:3007–3017. [PubMed] [Google Scholar]
- 47.Hara MR. Agrawal N. Kim SF. Cascio MB. Fujimuro M. Ozeki Y. Takahashi M. Cheah JH. Tankou SK. Hester LD. Ferris CD. Hayward SD. Snyder SH. Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 48.Hess DT. Matsumoto A. Kim SO. Marshall HE. Stamler JS. Protein Snitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 49.Holmgren A. Johansson C. Berndt C. Lonn ME. Hudemann C. Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 50.House CM. Frew IJ. Huang HL. Wiche G. Traficante N. Nice E. Catimel B. Bowtell DD. A binding motif for Siah ubiquitin ligase. Proc Nat Acad Sci USA. 2003;100:3101–3106. doi: 10.1073/pnas.0534783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang ZZ. Chen C. Zeng Z. Yang H. Oh J. Chen L. Lu SC. Mechanism and significance of increased glutathione level in human hepatocellular carcinoma and liver regeneration. FASEB J. 2001;15:19–21. doi: 10.1096/fj.00-0445fje. [DOI] [PubMed] [Google Scholar]
- 52.Ilbert M. Horst J. Ahrens S. Winter J. Graf PC. Lilie H. Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imam A. Hoyos B. Swenson C. Levi E. Chua R. Viriya E. Hammerling U. Retinoids as ligands and coactivators of protein kinase C alpha. FASEB J. 2001;15:28–30. doi: 10.1096/fj.00-0329fje. [DOI] [PubMed] [Google Scholar]
- 54.Inoue Y. Matsuda T. Sugiyama K. Izawa S. Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J Biol Chem. 1999;274:27002–27009. doi: 10.1074/jbc.274.38.27002. [DOI] [PubMed] [Google Scholar]
- 55.Izawa S. Maeda K. Sugiyama K. Mano J. Inoue Y. Kimura A. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J Biol Chem. 1999;274:28459–28465. doi: 10.1074/jbc.274.40.28459. [DOI] [PubMed] [Google Scholar]
- 56.Jakob U. Muse W. Eser M. Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 57.Jang HH. Lee KO. Chi YH. Jung BG. Park SK. Park JH. Lee JR. Lee SS. Moon JC. Yun JW. Choi YO. Kim WY. Kang JS. Cheong GW. Yun DJ. Rhee SG. Cho MJ. Lee SY. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Jones DP. Redox potential of GSH/GSSG couple: assay and biological significance. Methods Enzymol. 2002;348:93–112. doi: 10.1016/s0076-6879(02)48630-2. [DOI] [PubMed] [Google Scholar]
- 59.Kamata H. Hirata H. Redox regulation of cellular signalling. Cell Signal. 1999;11:114. doi: 10.1016/s0898-6568(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 60.Kamata H. Honda S. Maeda S. Chang L. Hirata H. Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 61.Karin M. Liu Z. Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 62.Kim G. Levine RL. Molecular determinants of S-glutathionylation of carbonic anhydrase 3. Antioxid Redox Signal. 2005;7:849–854. doi: 10.1089/ars.2005.7.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirlin WG. Cai J. Thompson SA. Diaz D. Kavanagh TJ. Jones DP. Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic Biol Med. 1999;27:1208–1218. doi: 10.1016/s0891-5849(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi A. Kang MI. Okawa H. Ohtsuji M. Zenke Y. Chiba T. Igarashi K. Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobayashi M. Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 66.Krishnan S. Chi EY. Wood SJ. Kendrick BS. Li C. Garzon–Rodriguez W. Wypych J. Randolph TW. Narhi LO. Biere AL. Citron M. Carpenter JF. Oxidative dimer formation is the critical rate-limiting step for Parkinson's disease alpha-synuclein fibrillogenesis. Biochemistry. 2003;42:829–837. doi: 10.1021/bi026528t. [DOI] [PubMed] [Google Scholar]
- 67.Kuge S. Arita M. Murayama A. Maeta K. Izawa S. Inoue Y. Nomoto A. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal- induced reversible disulfide bond formation. Mol Cell Biol. 2001;21:6139–6150. doi: 10.1128/MCB.21.18.6139-6150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuge S. Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuge S. Jones N. Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuge S. Toda T. Iizuka N. Nomoto A. Crm1 (XpoI) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells. 1998;3:521–532. doi: 10.1046/j.1365-2443.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- 71.Kuo SC. Lampen JO. Inhibition by 2-deoxy-D-glucose of synthesis of glycoprotein enzymes by protoplasts of Saccharomyces: relation to inhibition of sugar uptake and metabolism. J Bacteriol. 1972;111:419–429. doi: 10.1128/jb.111.2.419-429.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landino LM. Protein thiol modification by peroxynitrite anion and nitric oxide donors. Methods Enzymol. 2008;440:95–109. doi: 10.1016/S0076-6879(07)00805-1. [DOI] [PubMed] [Google Scholar]
- 73.Lee JW. Soonsanga S. Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Nat Acad Sci USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SR. Kwon KS. Kim SR. Rhee SG. Reversible inactivation of protein- tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 75.Leichert LI. Gehrke F. Gudiseva HV. Blackwell T. Ilbert M. Walker AK. Strahler JR. Andrews PC. Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Nat Acad Sci USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leichert LI. Jakob U. Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signal. 2006;8:763–772. doi: 10.1089/ars.2006.8.763. [DOI] [PubMed] [Google Scholar]
- 77.Leichert LI. Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lohse DL. Denu JM. Santoro N. Dixon JE. Roles of aspartic acid-181 and serine-222 in intermediate formation and hydrolysis of the mammalian proteintyrosine-phosphatase PTP1. Biochemistry. 1997;36:4568–4575. doi: 10.1021/bi963094r. [DOI] [PubMed] [Google Scholar]
- 79.Lou YW. Chen YY. Hsu SF. Chen RK. Lee CL. Khoo KH. Tonks NK. Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 80.Lowell BB. Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 81.Ma LH. Takanishi CL. Wood MJ. Molecular mechanism of oxidative stress perception by the Orp1 protein. J Biol Chem. 2007;282:31429–31436. doi: 10.1074/jbc.M705953200. [DOI] [PubMed] [Google Scholar]
- 82.Mallis RJ. Hamann MJ. Zhao W. Zhang T. Hendrich S. Thomas JA. Irreversible thiol oxidation in carbonic anhydrase III: Protection by S-glutathiolation and detection in aging rats. Biol Chem. 2002;383:649–662. doi: 10.1515/BC.2002.067. [DOI] [PubMed] [Google Scholar]
- 83.Mazzola JL. Sirover MA. Subcellular alteration of glyceraldehyde-3phosphate dehydrogenase in Alzheimer's disease fibroblasts. J Neurosci Res. 2003;71:279–285. doi: 10.1002/jnr.10484. [DOI] [PubMed] [Google Scholar]
- 84.Meng TC. Buckley DA. Galic S. Tiganis T. Tonks NK. Regulation of insulin signaling through reversible oxidation of the protein-tyrosine phosphatases TC45 and PTP1B. J Biol Chem. 2004;279:37716–37725. doi: 10.1074/jbc.M404606200. [DOI] [PubMed] [Google Scholar]
- 85.Meng TC. Fukada T. Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 86.Messens J. Hayburn G. Desmyter A. Laus G. Wyns L. The essential catalytic redox couple in arsenate reductase from Staphylococcus aureus. Biochemistry. 1999;38:16857–16865. doi: 10.1021/bi9911841. [DOI] [PubMed] [Google Scholar]
- 87.Meyer M. Schreck R. Baeuerle PA. H2O2 and antioxidants have opposite effects on activation of NF-kappa B and AP-1 in intact cells: AP-1 as secondary antioxidant-responsive factor. EMBO J. 1993;12:2005–2015. doi: 10.1002/j.1460-2075.1993.tb05850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohr S. Hallak H. de Boitte A. Lapetina EG. Brune B. Nitric oxide-induced Sglutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 89.Moncada S. Palmer RM. Higgs EA. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 90.Morigasaki S. Shimada K. Ikner A. Yanagida M. Shiozaki K. Glycolytic enzyme GAPDH promotes peroxide stress signaling through multistep phosphorelay to a MAPK cascade. Mol Cell. 2008;30:108–113. doi: 10.1016/j.molcel.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nagai Y. Inui T. Popiel HA. Fujikake N. Hasegawa K. Urade Y. Goto Y. Naiki H. Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 92.Nakajima H. Amano W. Fujita A. Fukuhara A. Azuma YT. Hata F. Inui T. Takeuchi T. The active site cysteine of the proapoptotic protein glyceraldehyde-3 phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J Biol Chem. 2007;282:26562–26574. doi: 10.1074/jbc.M704199200. [DOI] [PubMed] [Google Scholar]
- 93.Nguyen DT. Alarco AM. Raymond M. Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J Biol Chem. 2001;276:1138–1145. doi: 10.1074/jbc.M008377200. [DOI] [PubMed] [Google Scholar]
- 94.Nguyen T. Sherratt PJ. Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 95.Okazaki S. Naganuma A. Kuge S. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid Redox Signal. 2005;7:327–334. doi: 10.1089/ars.2005.7.327. [DOI] [PubMed] [Google Scholar]
- 96.Okazaki S. Tachibana T. Naganuma A. Mano N. Kuge S. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol Cell. 2007;27:675–688. doi: 10.1016/j.molcel.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 97.Okuno H. Akahori A. Sato H. Xanthoudakis S. Curran T. Iba H. Escape from redox regulation enhances the transforming activity of Fos. Oncogene. 1993;8:695–701. [PubMed] [Google Scholar]
- 98.Paget MS. Bae JB. Hahn MY. Li W. Kleanthous C. Roe JH. Buttner MJ. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol Microbiol. 2001;39:1036–1047. doi: 10.1046/j.1365-2958.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- 99.Paget MS. Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- 100.Patterson RL. van Rossum DB. Kaplin AI. Barrow RK. Snyder SH. Inositol 1,4,5-trisphosphate receptor/GAPDH complex augments Ca2+ release via locally derived NADH. Proc Nat Acad Sci USA. 2005;102:1357–1359. doi: 10.1073/pnas.0409657102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pognonec P. Kato H. Roeder RG. The helix-loop-helix/leucine repeat transcription factor USF can be functionally regulated in a redox-dependent manner. J Biol Chem. 1992;267:24563–24567. [PubMed] [Google Scholar]
- 102.Rainwater R. Parks D. Anderson ME. Tegtmeyer P. Mann K. Role of cysteine residues in regulation of p53 function. Mol Cell Biol. 1995;15:3892–3903. doi: 10.1128/mcb.15.7.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ralser M. Heeren G. Breitenbach M. Lehrach H. Krobitsch S. Triose phosphate isomerase deficiency is caused by altered dimerization—not catalytic inactivity—of the mutant enzymes. PLoS ONE. 2006;1:e30. doi: 10.1371/journal.pone.0000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ralser M. Wamelink MM. Kowald A. Gerisch B. Heeren G. Struys EA. Klipp E. Jakobs C. Breitenbach M. Lehrach H. Krobitsch S. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reddy S. Jones AD. Cross CE. Wong PS. Van Der Vliet A. Inactivation of creatine kinase by S-glutathionylation of the active-site cysteine residue. Biochem J 347 Pt. 2000;3:821–827. [PMC free article] [PubMed] [Google Scholar]
- 106.Rhee SG. Bae YS. Lee SR. Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 107.Rhee SG. Chae HZ. Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 108.Ronai Z. Glycolytic enzymes as DNA binding proteins. Int J Biochem. 1993;25:1073–1076. doi: 10.1016/0020-711x(93)90123-v. [DOI] [PubMed] [Google Scholar]
- 109.Roperch JP. Lethrone F. Prieur S. Piouffre L. Israeli D. Tuynder M. Nemani M. Pasturaud P. Gendron MC. Dausset J. Oren M. Amson RB. Telerman A. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21 (Waf1) Proc Nat Acad Sci USA. 1999;96:8070–8073. doi: 10.1073/pnas.96.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ross CA. Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 111.Saito H. Histidine phosphorylation and two-component signaling in eukaryotic cells. Chem Rev. 2001;101:2497–2509. doi: 10.1021/cr000243+. [DOI] [PubMed] [Google Scholar]
- 112.Salmeen A. Andersen JN. Myers MP. Meng TC. Hinks JA. Tonks NK. Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 113.Salmeen A. Barford D. Functions and Mechanisms of Redox Regulation of Cysteine-Based Phosphatases. 2005:560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 114.Sawa A. Khan AA. Hester LD. Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: Nuclear translocation participates in neuronal and nonneuronal cell death. Proc Nat Acad Sci USA. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schlattner U. Tokarska–Schlattner M. Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochim Biophys Acta. 2006;1762:164–180. doi: 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 116.Schrammel A. Gorren AC. Schmidt K. Pfeiffer S. Mayer B. S-nitrosation of glutathione by nitric oxide, peroxynitrite, and (*)NO/O(2)(*-) Free Radic Biol Med. 2003;34:1078–1088. doi: 10.1016/s0891-5849(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 117.Senatorov VV. Charles V. Reddy PH. Tagle DA. Chuang DM. Overexpression and nuclear accumulation of glyceraldehyde-3-phosphate dehydrogenase in a transgenic mouse model of Huntington's disease. Mol Cell Neurosci. 2003;22:285–297. doi: 10.1016/s1044-7431(02)00013-1. [DOI] [PubMed] [Google Scholar]
- 118.Seth D. Rudolph J. Redox regulation of MAP kinase phosphatase 3. Biochemistry. 2006;45:8476–8487. doi: 10.1021/bi060157p. [DOI] [PubMed] [Google Scholar]
- 119.Shelton MD. Chock PB. Mieyal JJ. Glutaredoxin: role in reversible proteins-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 120.Shenton D. Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shi Y. Sawada J. Sui G. Affar el B. Whetstine JR. Lan F. Ogawa H. Luke MP. Nakatani Y. Shi Y. Coordinated histone modifications mediated by a CtBP corepressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 122.Shi Y. Shi Y. Metabolic enzymes and coenzymes in transcription–a direct link between metabolism and transcription? Trends Genet. 2004;20:445–452. doi: 10.1016/j.tig.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 123.Shimazu T. Tokutake S. Usami M. Inactivation of phosphorylase phosphatase by a factor from rabbit liver and its chemical characterization as glutathione disulfide. J Biol Chem. 1978;253:7376–7382. [PubMed] [Google Scholar]
- 124.Shiozaki K. Shiozaki M. Russell P. Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol Biol Cell. 1998;9:1339–1349. doi: 10.1091/mbc.9.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sies H. Role of reactive oxygen species in biological processes. Klin Wochenschr. 1991;69:965–968. doi: 10.1007/BF01645140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 127.Sitia R. Molteni SN. Stress, protein (mis)folding, and signaling: the redox connection. Sci STKE. 2004;2004:pe27. doi: 10.1126/stke.2392004pe27. [DOI] [PubMed] [Google Scholar]
- 128.Stadtman ER. Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab Rev. 1998;30:225–243. doi: 10.3109/03602539808996310. [DOI] [PubMed] [Google Scholar]
- 129.Stamler JS. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 130.Steeghs K. Benders A. Oerlemans F. de Haan A. Heerschap A. Ruitenbeek W. Jost C. van Deursen J. Perryman B. Pette D. Bruckwilder M. Koudijs J. Jap P. Veerkamp J. Wieringa B. Altered Ca2+ responses in muscles with combined mitochondrial and cytosolic creatine kinase deficiencies. Cell. 1997;89:93–103. doi: 10.1016/s0092-8674(00)80186-5. [DOI] [PubMed] [Google Scholar]
- 131.Suh YA. Arnold RS. Lassegue B. Shi J. Xu X. Sorescu D. Chung AB. Griendling KK. Lambeth JD. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 132.Sun H. Lesche R. Li DM. Liliental J. Zhang H. Gao J. Gavrilova N. Mueller B. Liu X. Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Nat Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tajc SG. Tolbert BS. Basavappa R. Miller BL. Direct determination of thiol pKa by isothermal titration microcalorimetry. J Am Chem Soc. 2004;126:10508–10509. doi: 10.1021/ja047929u. [DOI] [PubMed] [Google Scholar]
- 134.Toone WM. Morgan BA. Jones N. Redox control of AP-1-like factors in yeast and beyond. Oncogene. 2001;20:2336–2346. doi: 10.1038/sj.onc.1204384. [DOI] [PubMed] [Google Scholar]
- 135.Tsuchiya K. Tajima H. Kuwae T. Takeshima T. Nakano T. Tanaka M. Sunaga K. Fukuhara Y. Nakashima K. Ohama E. Mochizuki H. Mizuno Y. Katsube N. Ishitani R. Pro-apoptotic protein glyceraldehyde-3-phosphate dehydrogenase promotes the formation of Lewy body-like inclusions. Eur J Neurosci. 2005;21:317–326. doi: 10.1111/j.1460-9568.2005.03870.x. [DOI] [PubMed] [Google Scholar]
- 136.van Montfort RL. Congreve M. Tisi D. Carr R. Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 137.Veal EA. Ross SJ. Malakasi P. Peacock E. Morgan BA. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J Biol Chem. 2003;278:30896–30904. doi: 10.1074/jbc.M303542200. [DOI] [PubMed] [Google Scholar]
- 138.Vivancos AP. Castillo EA. Biteau B. Nicot C. Ayte J. Toledano MB. Hidalgo E. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc Nat Acad Sci USA. 2005;102:8875–8880. doi: 10.1073/pnas.0503251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walker LJ. Robson CN. Black E. Gillespie D. Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wilkinson MG. Samuels M. Takeda T. Toone WM. Shieh JC. Toda T. Millar JB. Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- 141.Winterbourn CC. Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic Biol Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 142.Woo HA. Chae HZ. Hwang SC. Yang KS. Kang SW. Kim K. Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 143.Wood MJ. Storz G. Tjandra N. Structural basis for redox regulation of Yap1 transcription factor localization. Nature. 2004;430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- 144.Wood ZA. Poole LB. Hantgan RR. Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 145.Xanthoudakis S. Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xanthoudakis S. Miao GG. Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Nat Acad Sci USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yan C. Lee LH. Davis LI. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang J. Groen A. Lemeer S. Jans A. Slijper M. Roe SM. den Hertog J. Barford D. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–719. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- 149.Ying J. Clavreul N. Sethuraman M. Adachi T. Cohen RA. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic Biol Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yu CX. Li S. Whorton AR. Redox regulation of PTEN by S-nitrosothiols. Mol Pharmacol. 2005;68:847–854. doi: 10.1124/mol.104.010504. [DOI] [PubMed] [Google Scholar]
- 151.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 152.Zhang ZY. Dixon JE. Active site labeling of the Yersinia protein tyrosine phosphatase: The determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry. 1993;32:9340–9345. doi: 10.1021/bi00087a012. [DOI] [PubMed] [Google Scholar]
- 153.Zheng L. Roeder RG. Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 154.Zhu S. Bjorge JD. Fujita DJ. PTP1B contributes to the oncogenic properties of colon cancer cells through Src activation. Cancer Res. 2007;67:10129–10137. doi: 10.1158/0008-5472.CAN-06-4338. [DOI] [PubMed] [Google Scholar]
- 155.Ziegler DM. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu Rev Biochem. 1985;54:305–329. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]