Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy (original) (raw)
Abstract
Proximal spinal muscular atrophy (SMA), one of the most common genetic causes of infant death, results from the selective loss of motor neurons in the spinal cord. SMA is a consequence of low levels of survival motor neuron (SMN) protein. In humans, the SMN gene is duplicated; SMA results from the loss of SMN1 but SMN2 remains intact. SMA severity is related to the copy number of SMN2. Compounds which increase the expression of SMN2 could, therefore, be potential therapeutics for SMA. Ultrahigh-throughput screening recently identified substituted quinazolines as potent SMN2 inducers. A series of C5-quinazoline derivatives were tested for their ability to increase SMN expression in vivo. Oral administration of three compounds (D152344, D153249 and D156844) to neonatal mice resulted in a dose-dependent increase in Smn promoter activity in the central nervous system. We then examined the effect of these compounds on the progression of disease in SMN lacking exon 7 (SMNΔ7) SMA mice. Oral administration of D156844 significantly increased the mean lifespan of SMNΔ7 SMA mice by ∼21–30% when given prior to motor neuron loss. In summary, the C5-quinazoline derivative D156844 increases SMN expression in neonatal mouse neural tissues, delays motor neuron loss at PND11 and ameliorates the motor phenotype of SMNΔ7 SMA mice.
INTRODUCTION
Proximal spinal muscular atrophy (SMA) is an autosomal recessive neurodegenerative disease characterized by selective loss of α motor neurons in the anterior horn of the spinal cord (1). This leads to atrophy of limb and trunk muscles. In humans, the survival motor neuron (SMN) gene is duplicated and the two SMN genes (SMN1 and SMN2) differ by a single nucleotide (C → T) within an exon splice enhancer of exon 7 (2,3). SMN1 transcripts produce full-length SMN (FL-SMN) protein. Most of the transcripts from SMN2 lacking exon 7 (SMNΔ7) and therefore produce SMNΔ7 protein; however, 10–20% of SMN2 transcripts are correctly spliced and produce FL-SMN protein. SMA results from deletions or mutations of the SMN1 gene; the SMN2 gene remains intact (4). The severity of disease in SMA depends on the copy number of SMN2 and the levels of SMN protein (5–7).
Mice carry only one SMN gene (mSmn) which is orthologous to SMN1 in humans (8,9). Loss of mSmn results in embryonic lethality (10). Conditional knockdown of mSmn in neurons, muscle and hepatocytes also leads to death of those cells (11–13). Antisense morpholino-mediated reduction of SMN protein levels in zebrafish causes truncation of axons followed by excessive branching before reaching their target (14). Loss of functional SMN in the fruit fly leads to neuromuscular abnormalities and a progressive loss of motility and coordinated movement before death in the late larval stage (15–17). Deletion of the nematode orthologue of SMN (cSmn) leads to reduced locomotor activity and premature death (18). These studies suggest that SMN is essential for cell function and survival.
Transgenic insertion of SMN2 into a mSmn null background in mice rescues the embryonic lethal phenotype (19). mSmn null mice with low SMN2 copy numbers (i.e. 2) develop severe (type I-like) SMA and die at 6–8 days (19,20). mSmn null mice with higher copy numbers (i.e. 8), on the other hand, are normal when compared with non-transgenic littermates (19) demonstrating that the SMN2 gene product can correct the SMA phenotype. Enhancing the inclusion of exon 7 in SMN2 transcripts with a bifunctional U7 snRNA markedly improves the survival of severe SMA mice (SMN2 +/+;mSmn −/−) (21). Severe SMA mice that also contain an exon 7-lacking SMN (SMNΔ7) develop a less severe SMA phenotype and these mice die at 14–15 days (22). These transgenic experiments show that modulating SMN2 as well as SMNΔ7 levels can be beneficial to SMA mouse models.
Over the years, a number of groups have identified SMN-inducing compounds using cultured fibroblasts derived from SMA patients. Some of these compounds include aclarubicin (23), butyrate (24), 4-phenylbutyrate (25), valproic acid (26–28), hydroxyurea (28–30), indoprofen (31), interferons-β and -γ (32), forskolin (33), ortho-vanadate (34), cantharidin (35), tautomycin (35), aminoglycosides (36,37), resveratrol (38), suberoylanilide hydroxamic acid (39) and M344 (40). Many of these compounds would not make good therapeutic agents since high concentrations (micromolar and, in some cases, millimolar levels) are required to increase SMN expression, these compounds are rapidly metabolized in vivo or are toxic and many are unable to cross the blood-brain barrier.
A recently published study (41) described a series of compounds that increased SMN promoter activity in a reporter gene cell line. One such compound was a C5-quinazoline that increased SMN promoter activity with an EC50 = 330 ± 110 nm. This compound resulted in a 2.5-fold increase in the amount of FL-SMN mRNA, a 2–3-fold increase in SMN protein and a 5-fold increase in the number of gems—SMN-containing subnuclear foci (or bodies)—in a fibroblast cell line derived from an SMA patient. Through a focused medicinal chemistry effort, a series of modified C5-quinazolines have been designed so as to more potently induce SMN2 promoter activity, increase SMN protein levels and more easily penetrate the blood-brain barrier [Fig. 1 and Table 1; (42)]. One such compound, a piperidine 2,4-diaminoquinazoline known as D156844, was highly potent (EC50 = 4 nm) at activating SMN2 promoter activity with a maximal response of 2.3-fold (42). D156844 also increased SMN protein levels in SMA patient fibroblast cultures and increased the number of intranuclear, SMN-containing gems to levels observed in SMA carrier fibroblasts. Recently, screening a protein microarray with 125I-labeled potent C5-quinazoline derivatives resulted in the identification of mRNA decapping scavenger enzyme (DcpS) as the cellular protein target of D156844 (43).
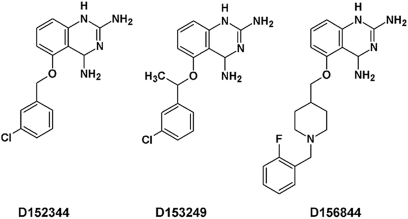
Figure 1.
The chemical structures of the three 2,4-diaminoquinazoline derivatives (D152344, D153249 and D156844) that were used in these experiments. These compounds are labeled as 5b, (S)-7a and 11a, respectively, in Ref. (42).
Table 1.
Summary of SMN2 promoter data for the 2,4-diaminoquinazoline derivatives described in this study
| Compound ID | Original ID number | SMN2 promoter EC50 (nm) | Maximum fold induction | DHFR IC50 (nm) |
|---|---|---|---|---|
| D152344 | 5b | 78 | 1.7 | 98 |
| D153249 | (S)-7a | 2 | 1.7 | 25 |
| D156844 | 11a | 4 | 2.4 | >125 000 |
In this study, we examine the efficacy of 2,4-diaminoquinazolines as SMN inducers in the central nervous system of mice and as therapeutic candidates for SMA. Three quinazoline derivatives were selected—D152344, D153249 and D156844 (Fig. 1)—based on their SMN promoter induction activity (Table 1), pharmacokinetics in adult mice and blood-brain barrier penetration. Each of the three compounds was derived from successive rounds of optimization with the last compound, D156844, being the most highly optimized. D152344 and D153249 were earlier compounds with a known major off target activity that led to a cellular liability (42). These three compounds were tested in SMNΔ7 SMA mice (SMN2 +/+;SMN_Δ_7 +/+;mSmn −/−) (22,44) for their ability to improve the SMA motor degenerative phenotype and to increase the lifespan of affected mice. One derivative, D156844, increased SMN protein levels in the spinal cord, ameliorated the SMA motor phenotype and improved the survival of SMNΔ7 SMA mice when administered prior to the onset of motor neuron loss.
RESULTS
2,4-Diaminoquinazoline derivatives cross the blood-brain barriers of neonatal mice to increase Smn expression in the central nervous system
A series of 2,4-diaminoquinazoline derivatives were recently generated and tested for their induction of SMN2 promoter activity as well as their oral bioavailability and blood-brain barrier penetration (42). Three compounds were identified as lead candidates: D152344, D153249 and D156844 (Fig. 1). Pharmacokinetic studies conducted in adult rodents indicated that all compounds were orally bioavailable and distributed into the brain. Since SMA is a childhood motor neuron disease and the model of choice for testing the effectiveness of these therapeutic candidates is the neonatal SMNΔ7 SMA mouse (22), we wanted to determine the pharmacological properties of these compounds in neonatal mice. The maximum tolerable doses (MTDs) for these three compounds were first determined in SMNΔ7 carrier (SMN2 +/+;SMN_Δ_7 +/+;mSmn +/−) mice. These mice received different oral doses of D152344 (10–100 mg/kg/day, b.i.d.), D153249 (10–100 mg/kg/day, b.i.d.), D156844 (0.1–30 mg/kg/day) or the appropriate vehicle—40% PrOHβCD for D152344 and D153249 and ddH2O for D156844—for 5 days beginning at postnatal day (PND) 04. The highest dose of D152344 (100 mg/kg/day) was lethal and mice receiving 60 mg/kg/day exhibited diminished increases in body mass and skin abnormalities. The MTD for D152344 was 30 mg/kg/day. Although there was no lethality associated with oral administration of D153249, higher doses of this compound (60–100 mg/kg/day) did result in diminished growth and hair development abnormalities. The MTD for D153249, therefore, was 30 mg/kg/day. No adverse effects were observed for D156844 at any of the doses tested. The trend in clinical signs observed the MTD analysis among these compounds correlates with the presence or absence of an identified off-target activity against dihydrofolate reductase (DHFR) (42) (Table 1).
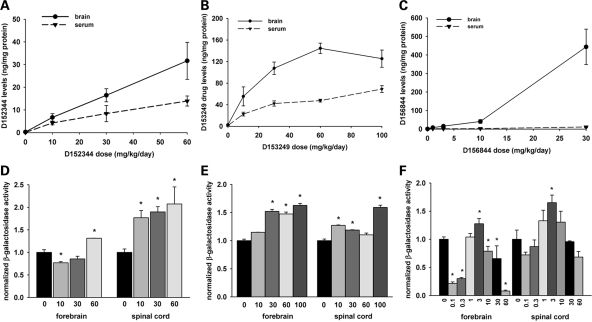
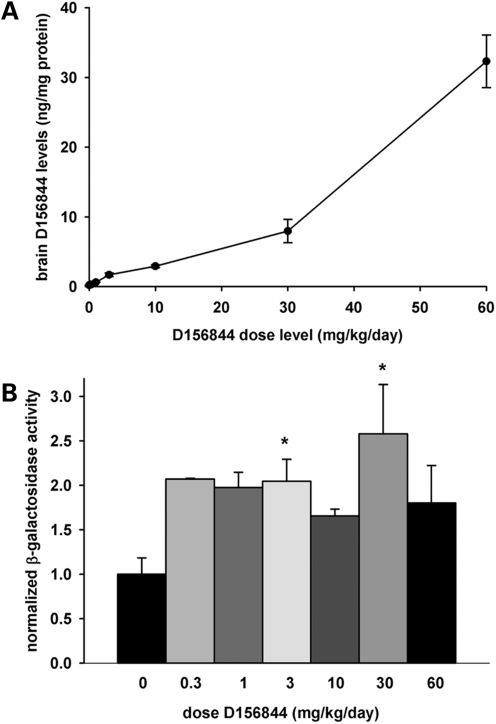
Sera and forebrain extracts from neonatal mice treated with these drugs were collected for bioanalyses using liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS). All three of the compounds tested were orally bioavailable and crossed the blood-brain barriers of neonatal mice. D152344 exhibited a linear increase in exposure with dose in both serum (dashed line) and brain (solid line in Fig. 2A) whereas serum and brain D153249 levels (Fig. 2B) exhibited a hyperbolic relationship between dose and exposure. Brain D156844 levels increased with administered dose in a linear manner at lower doses whereas serum D156844 levels were low and fairly constant (Fig. 2C). The central nervous system (CNS) clearance (ratio of brain to serum drug levels) of D156844 was much higher (∼9) than it was for either D152344 (∼1.5) or D153249 (∼2.6). On the basis of these observations, all three quinazolines were tolerable at low milligram/kilogram doses in neonates and were able to traverse the blood-brain barrier with exposure predicted to reach efficacious doses as predicted by in vitro activity (42).
Figure 2.
Drug bioavailability and mSmn promoter activities in the CNS of neonatal SMNΔ7 carrier mice treated with 2,4-diaminoquinazoline derivatives. (A–C) Dose-dependent serum (dashed line with triangles) and brain (solid line with circles) levels of D152344 (A), D153249 (B) and D156844 (C) in neonatal SMNΔ7 carrier mice. (D–F) mSmn promoter-dependent β-galactosidase activity in the forebrains and spinal cords of SMNΔ7 carrier mice receiving differing doses of D152344 (D), D153249 (E) and D156844 (F). The β-galactosidase activity at each dose was normalized to the activity of vehicle-treated (0 mg/kg/day) samples. The doses administered for each compound are listed under each bar (n = 4/treatment dose). Key: *P ≤ 0.05 when compared with 0 mg/kg/day-treated mice.
We examined the effect of D152344, D153249 and D156844 on Smn expression in vivo using neonatal mice. In the mSmn knockout cassette, a LacZ:NeoR cassette was inserted into mSmn exon 2A to nullify the production of mSmn protein (19) but also places the LacZ reporter gene under the control of the mSmn promoter. β-galactosidase activity was used to monitor SMN promoter activity since the promoter region of mSmn is very similar to those of human SMN1 and SMN2 (45–48). The data were expressed relative to β-galactosidase activity of vehicle-treated mice. Treatment of neonatal mice with D152344 (Fig. 2D) and D153249 (Fig. 2E) for 5 days resulted in a dose-dependent increase in β-galactosidase activity in forebrain and spinal cord extracts. There were no significant changes in β-galactosidase activities in non-CNS tissues at any of doses tested (data not shown). Treatment of mice with D156844 increases mSmn promoter activity in forebrain and spinal cord extracts in a dose-dependent manner to a maximum at 3 mg/kg/day; mSmn promoter activity then decreased with increasing dose of D156844 (Fig. 2F). On the basis of these observations together with the MTD analysis, the optimal doses for D152344 and D153249 in neonatal mice are 30 mg/kg/day although the optimal dose for D156844 is 3 mg/kg/day.
D156844 increases SMN protein levels in the spinal cords of SMNΔ7 SMA mice
As D156844 increases SMN protein expression in cultured fibroblasts (42), mSmn mRNA levels in NSC34 neuroblastoma cells (43) and mSmn promoter activity in vivo in the CNS (Fig. 2F), we examined SMN expression in response to D156844 treatment. In all of the experiments examining changes in SMN expression, tissue extracts from PND08 (treating mice for 5 days beginning at PND04) mice were used. This time point was chosen for monitoring changes in SMN expression because there is very little motor neuron loss at this time (22) so there is a greater likelihood that increased SMN RNA/protein levels are the result of increased SMN expression and not due to increased cell numbers.
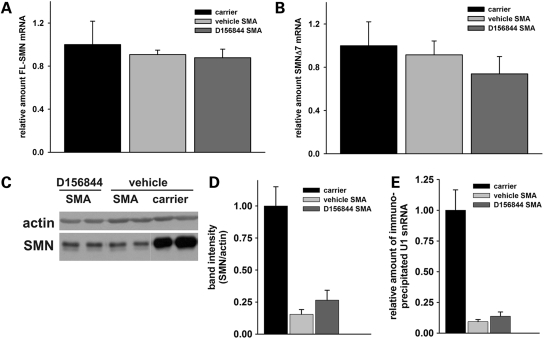
Using real-time RT–PCR, the amounts of FL-SMN and exon 7 lacking SMN (SMNΔ7) mRNAs—derived from the SMN2 and SMNΔ7 transgenes—were examined in total spinal cord RNA from SMNΔ7 SMA mice treated for 5 days with either D156844 or vehicle. The primers used for real-time RT–PCR are specific to human SMN and do not cross-hybridize with mSmn. Unexpectedly, we did not detect an increase in FL-SMN (Fig. 3A) or SMNΔ7 (Fig. 3B) mRNAs in response to D156844 treatment.
Figure 3.
Oral administration of D156844 alters SMN gene expression in the spinal cord of SMNΔ7 SMA mice. (A) Changes in human-specific, full-length SMN (FL-SMN) mRNA levels in the spinal cord of mice treated with D156844 or vehicle for 5 days (n = 3/treatment group). (B) Changes in human-specific, SMNΔ7 mRNA levels in the spinal cord of mice treated with D156844 or vehicle for 5 days (n = 3/treatment group). (C) Representative immunoblot showing SMN and β-actin protein expression in spinal cord extracts from SMNΔ7 SMA mice treated with either D156844 or vehicle for 5 days. (D) Quantitation of SMN protein levels relative to β-actin protein in D156844- or vehicle-treated spinal cord extracts (n = 3/treatment group). SMN protein levels are expressed relative to those observed in age-matched carrier mice. (E) snRNP assembly activity in the spinal cord of SMNΔ7 SMA mice treated with either D156844 or vehicle for 5 days (n = 3/treatment group). snRNP assembly activity is measured by the amount of 32P-labeled U1 snRNA precipitated by antibodies against snRNP proteins (Sm proteins) as a consequence of SMN-dependent Sm core formation taking place in vitro using spinal cord extracts (50,69). snRNP assembly activity is expressed relative to that observed in age-matched carrier mice.
The effect of D156844 treatment on SMN protein levels were also examined in the spinal cord of SMNΔ7 SMA mice. As shown in Figure 3C, treatment of SMNΔ7 SMA mice with D156844 for 5 days (n = 3/group) resulted in a reproducible but non-statistically significant 1.7-fold increase in SMN protein levels in spinal cord extracts (Fig. 3D). D156844-treated SMNΔ7 SMA mice have spinal cord SMN proteins that are ∼25% of the levels observed in SMNΔ7 carrier mice.
It has been suggested that the primary defect in SMA is the reduction of snRNP assembly (49,50). We have recently observed that snRNP assembly is reduced in severe SMA (SMN2 +/+;mSmn −/−) mice and that the magnitude of reduction in snRNP assembly correlates with phenotype severity in SMA mice (50). Treatment of SMNΔ7 SMA mice with D156844 for 5 days (from PND04 until PND08) resulted in a reproducible but non-statistically significant 1.5-fold increase in spinal cord snRNP assembly activity relative to vehicle-treated mice (Fig. 3E).
D156844 improves the survival and motor phenotype of SMNΔ7 SMA mice
Since these 2,4-diaminoquinazoline derivatives can cross the blood-brain barrier of neonatal mice to increase Smn expression in the CNS, the effects of these three compounds on the survival and phenotype of SMA mouse models were examined. The SMNΔ7 SMA mouse (SMN2 +/+;SMN_Δ_7 +/+;mSmn −/−) model was used in these experiments because their phenotype and pathology strongly resemble what is observed in SMA and these mice provide a reasonably rapid measure of therapeutic benefit since they only live on average for 13.6 ± 0.2 days (22,44). D152344, D153249 and D156844 were administered to SMNΔ7 SMA mice via oral delivery beginning at PND04. Neither D152344 (30 mg/kg/day, b.i.d.; χ2 = 1.736, P = 0.188) nor D153249 (10 mg/kg/day, b.i.d.; χ2 = 0.001, P = 0.976) affected the average lifespan of SMNΔ7 SMA mice [survival for D152344 was 14.8 ± 0.6 (n = 5) versus 13.6 ± 0.4 (n = 5) for D153249 versus 13.0 ± 0.8 (n = 7) for vehicle]. These two compounds have off-target activities towards DHFR (42).
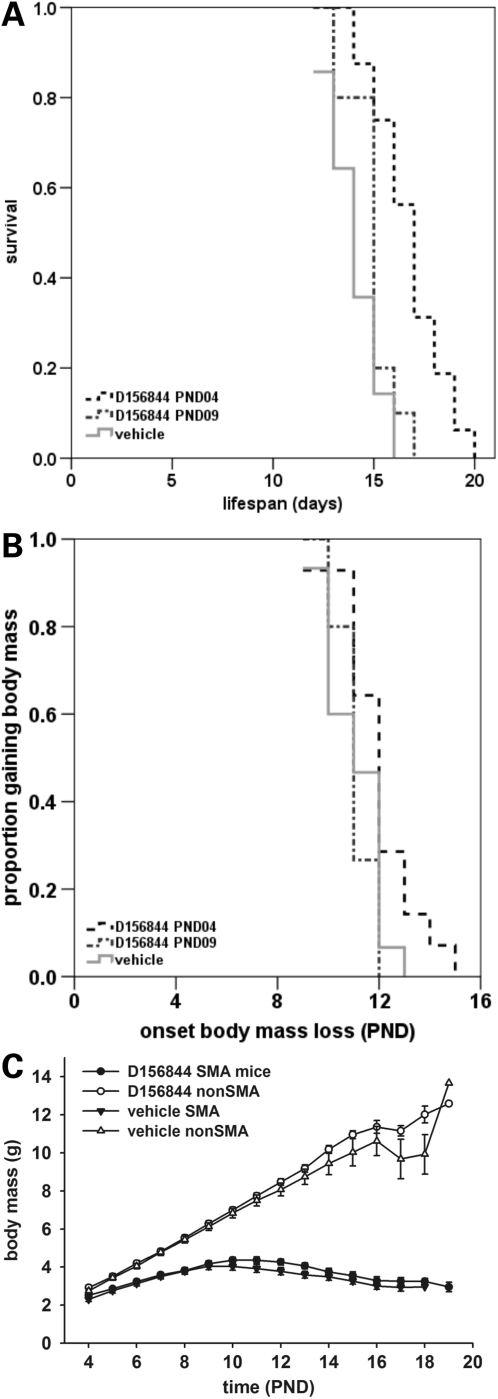
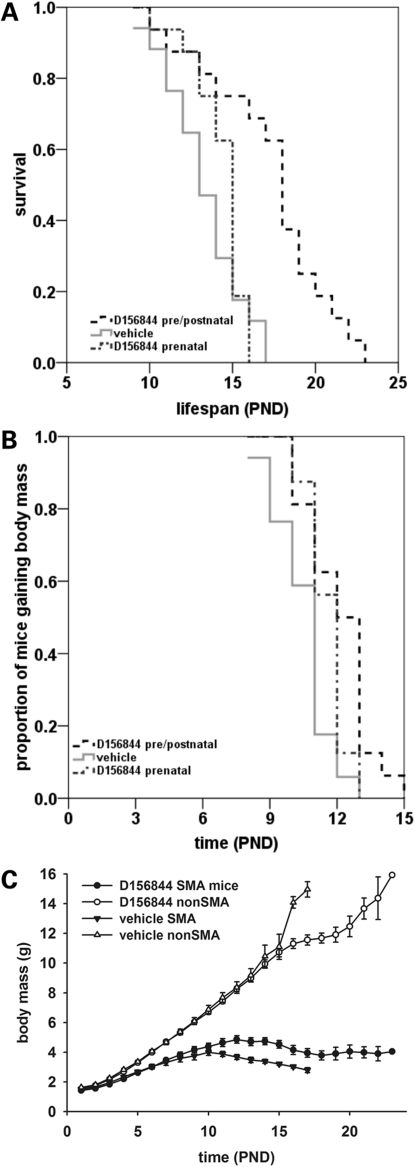
SMNΔ7 SMA mice also received D156844 (3 mg/kg/day) beginning at PND04 and continued to receive drug for the duration of their lives. Drug-treated SMNΔ7 SMA (n = 14) lived, on average, 21% longer than vehicle-treated (n = 15) SMNΔ7 SMA mice (Fig. 4A; black dashed line, 17.0 ± 0.5 versus 14.0 ± 0.4 days; χ2 = 16.783, P < 0.001). In contrast, D156844 treatment after the onset of motor neuron loss (PND09) did not improve survival of SMNΔ7 SMA mice (Fig. 4A; grey dotted line, 14.9 ± 0.4 versus 14.0 ± 0.4 days; χ2 = 2.829, P = 0.093). D156844 did delay the onset of body mass loss—an indicator for the endstage of neurodegeneration in SMNΔ7 SMA mice (44)—slightly when the treatment started at PND04 (Fig. 4B; black dashed line, 12.0 ± 0.4 versus 11.1 ± 0.3 days; χ2 = 3.247, P = 0.072). This effect was lost when SMNΔ7 SMA mice were treated starting at PND09 (Fig. 4B; grey dotted line, 14.9 ± 0.4 versus 14.0 ± 0.4 days; χ2 = 2.829, P = 0.093). Even though the onset of body mass loss was delayed in D156844-treated mice, their body mass curve (black triangles) was not different from the body mass curve of vehicle-treated (black circles) mice (Fig. 4C).
Figure 4.
Oral administration of D156844 improves the survival of and delays the onset of loss of body mass in neonatal SMNΔ7 SMA mice. (A) Kaplan–Meier survival plot for SMNΔ7 SMA mice receiving either vehicle (light grey solid line) or D156844 (3 mg/kg/day) beginning at either PND04 (black dashed line) or PND09 (grey dotted line). (B) Kaplan–Meier onset of body mass loss plot for SMNΔ7 SMA mice receiving either vehicle (light grey solid line) or D156844 (3 mg/kg/day) beginning at either PND04 (black dashed line) or PND09 (grey dotted line). (C) Body mass curves for SMNΔ7 SMA mice receiving either vehicle (triangles) or D156844 (3 mg/kg/day; circles) beginning at PND04.
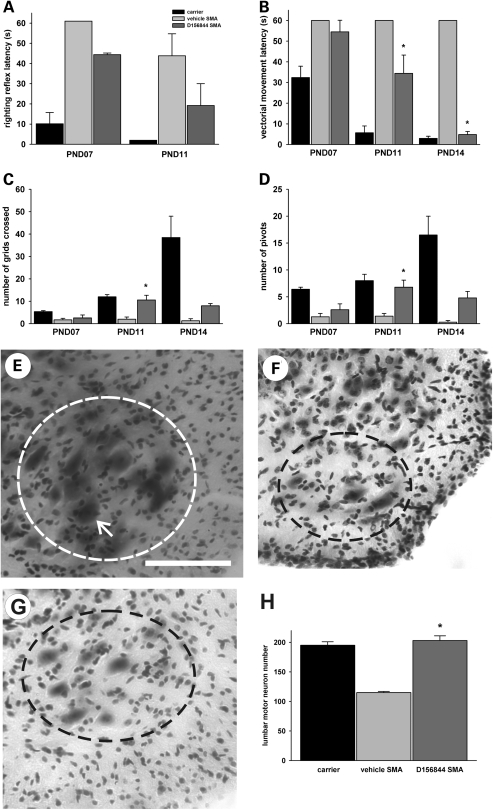
SMNΔ7 SMA mice develop a progressive impairment of neonatal motor behaviors including loss of surface righting and reduced spontaneous locomotion (44). Since D156844 improves the survival of SMNΔ7 SMA mice when administered prior to motor neuron loss, the effect of this quinazoline on the SMA motor phenotype was examined. Gross observation of D156844-treated SMNΔ7 SMA mice shows an improvement in motor activity when compared with vehicle-treated mice (Supplementary Material, Movie). Surface righting reflex responses are impaired in SMNΔ7 SMA mice; treatment with D156844 partially reduces the righting reflex latency (Fig. 5A) but this reduction is not statistically significant. When comparing the righting reflex latencies between SMNΔ7 SMA and non-SMA mice, however, the difference between SMNΔ7 SMA and non-SMA mice is no longer significant (P = 0.906) in D156844-treated mice at PND11 but is significant in vehicle-treated mice (P = 0.005). This observation suggests that D156844 does partially ameliorate surface righting reflex response impairments in SMNΔ7 SMA mice. Vectorial movement latency is the amount of time it takes a neonatal mouse to move in one direction a distance that is greater than its body length (44). Vectorial movement latency is significantly reduced in D156844-treated SMNΔ7 SMA mice relative to vehicle-treated SMNΔ7 SMA mice at PND11 and PND14 (Fig. 5B).The vectorial movement latency, however, is still longer in D156844-treated SMNΔ7 SMA mice than in carrier mice. Spontaneous locomotor activity—measured by counting the number of grids crossed in an area within 1 min—is significantly improved in D156844-treated SMNΔ7 SMA mice at PND11 when compared with vehicle-treated SMNΔ7 SMA mice (Fig. 5C). Likewise, pivoting responses, another measure of spontaneous activity, are also ameliorated in D156844-treated SMNΔ7 SMA mice (Fig. 5D). Taken together, these behavioral observations show that treatment of SMNΔ7 SMA mice with D156844 prior to motor neuron loss partially ameliorates the SMA motor phenotype.
Figure 5.
Oral administration of D156844 improves the motor phenotype of SMNΔ7 SMA mice and reduces motor neuron loss in the lumbar spinal cord. (A) Righting reflex latencies at PND07 and PND11 for SMNΔ7 SMA mice treated with D156844 (3 mg/kg/day; dark grey bar) or vehicle (light grey bar) as compared with carrier (black bar) mice. (B) Vectorial movement latencies at PND07, PND11 and PND14 for SMNΔ7 SMA mice treated with D156844 or vehicle. (C) Spontaneous locomotor activity measured as the number of grids crossed in 1 min at PND07, PND11 and PND14 for SMNΔ7 SMA mice treated with D156844 or vehicle. (D) The number of pivots (90° turns) made in 1 min at PND07, PND11 and PND14 for SMNΔ7 SMA mice treated with D156844 or vehicle. For the phenotype data shown in (A–D), the sample sizes (n) for vehicle-, D156844-treated SMA mice and carrier mice at PND07 were 7, 5 and 5, respectively, at PND11, 5, 5 and 3, respectively and at PND14, 3, 5 and 3, respectively. Key for (A–D): *P < 0.05 when comparing vehicle-treated SMNΔ7 SMA mice to D156844-treated SMNΔ7 SMA mice. (E–G) Representative images of Cresyl violet-stained lumbar spinal cord sections from carrier mice (E) and SMNΔ7 SMA mice treated with vehicle (F) or D156844 (G) from PND04 until PND11. The dashed ovals represent the regions of interest in the ventral horn and arrow in (E) points to a motor neuron. Scale, 100 µm. (H) The number of motor neurons in the lumbar (L4-5) spinal cord of SMNΔ7 SMA mice (PND11) treated with D156844 or vehicle (n = 3/group). Key for (H): * P < 0.001 when comparing vehicle-treated SMNΔ7 SMA mice to D156844-treated SMNΔ7 SMA mice.
SMNΔ7 SMA mice have a progressive loss of motor neurons in the lumbar spinal cord starting at approximately PND09 (22). The effect of D156844 administration on the number of motor neurons in the lumbar spinal cord was examined. Dosing of SMNΔ7 SMA mice began at PND04 and continued daily until PND11. Representative sections from carrier mice as well as from SMNΔ7 SMA mice treated with vehicle or D156844 are shown in Figure 5E, F and G, respectively. At PND11, vehicle-treated SMNΔ7 SMA mice have ∼41% fewer motor neurons in the lumbar spinal cord than carrier mice (Fig. 5H; 115 ± 2 versus 195 ± 6; P < 0.001). D156844-treated SMNΔ7 SMA mice, however, have motor neuron counts at PND11 similar to those observed in carrier mice (203 ± 8 versus 195 ± 6; P = 0.368). Treatment with D156844 starting at PND04, therefore, prevents the loss of motor neurons at PND11.
Prenatal administration of D156844 further improves the survival of SMNΔ7 SMA mice
Transgenic mouse studies have shown that increasing SMN levels in prenatal SMA mouse neurons (beginning at embryonic day 13) ameliorates the motor phenotype of SMA mice (51). In order to assess whether prenatal administration of D156844 will further improve the survival and motor phenotype of SMNΔ7 SMA mice, we must first determine whether this compound can cross the placental barriers to reach the CNS of fetal mice. Timed-pregnant SMNΔ7 carrier dams were treated orally with differing doses of D156844 (0–60 mg/kg/day) beginning at embryonic day 11 (ED11). After 5 days of treatment, the prenatal pups were dissected and analyzed for D156844 drug levels. As shown in Figure 6A, there is a dose-dependent increase in D156844 levels in fetal brain extracts suggesting that this compound can traverse the placental barriers to reach the fetal CNS. mSmn promoter activity, i.e. β-galactosidase expression, was also examined in these fetal brain samples. β-Galactosidase activity in fetal brain extracts was increased 2-fold in D156844-treated mice at all doses tested (Fig. 6B).
Figure 6.
CNS bioavailability and mSmn promoter activity in prenatal mice receiving D156844. (A) D156844 levels in prenatal mouse brains whose dams were dosed with differing amounts of D156844 (0–60 mg/kg/day; n = 3/group) beginning at ED11.5. (B) β-Galactosidase activity—a marker for mSmn promoter activity—in the brains of mice whose dams received different doses of D156844 (0–60 mg/kg/day; n = 3/group) beginning at ED11.5. The β-galactosidase activity at each dose was normalized to the activity of vehicle-treated (0 mg/kg/day) samples. Key: *P ≤ 0.05 when compared with 0 mg/kg/day-treated mice.
As D156844 is able to penetrate the blood-brain barrier and cross the placenta, we wanted to determine the effect of administration of D156844 on the survival of SMNΔ7 SMA mice when the drug is administered during prenatal development. Timed-pregnant SMNΔ7 carrier dams mice were treated with either D156844 (3 mg/kg/day) or vehicle beginning 11 days after coitus (ED11). Treatment continued through birth and was orally administered once the pups were born. Prenatal administration of D156844 (n = 16) increased the average lifespan of SMNΔ7 SMA mice by 30% when compared with vehicle-treated (n = 17) SMA mice (Fig. 7A; black dashed line, 17.3 ± 0.9 versus 13.3 ± 0.6 days; χ2 = 14.973, P < 0.001). If, however, D156844 was discontinued after birth, there was no significant difference in average lifespan between prenatally treated D156844 (n = 16) SMNΔ7 SMA mice and vehicle-treated mice (Fig. 7A; grey dotted line, 14.1 ± 0.6 versus 13.3 ± 0.6 days; χ2 = 0.26, P = 0.61). Although prenatal and postnatal administration of D156844 appears to increase the survival of SMNΔ7 SMA mice to a greater extent than only postnatal D156844 administration, the difference was not statistically significant (χ2 = 2.577; P = 0.108). Prenatal and postnatal administration of D156844 to SMNΔ7 SMA mice delayed the onset of body mass loss by 15% (Fig. 7B; black dashed line, 12.1 ± 0.4 versus 10.5 ± 0.3 days; χ2 = 9.016, P = 0.003). Interestingly, treating SMNΔ7 SMA mice with D156844 only during the prenatal period resulted in a small delay (10%) in the onset of body mass loss (Fig. 7B; grey dotted line, 11.6 ± 0.2 versus 10.5 ± 0.3 days; χ2 = 5.056, P = 0.025). SMNΔ7 SMA mice treated prenatally and postnatally with D156844 tended to have larger body masses (black circles in Fig. 7C) than those of vehicle-treated (black triangles in Fig. 7C) SMNΔ7 SMA mice especially after PND11.
Figure 7.
Prenatal administration of D156844 improves the survival and phenotype of SMNΔ7 SMA mice. (A) Kaplan–Meier survival plot for SMNΔ7 SMA mice receiving either vehicle (light grey solid line) or D156844 (3 mg/kg/day) beginning ED11.5 and continuing after birth (black dashed line) or ending at birth (grey dotted line). (B) Kaplan–Meier onset of body mass loss plot for SMNΔ7 SMA mice receiving either vehicle (light grey solid line) or D156844 [beginning ED11.5 and continuing after birth (black dashed line) or ending at birth (grey dotted line)]. (C) Body mass curves for SMNΔ7 SMA mice receiving either vehicle (triangles) or D156844 (3 mg/kg/day; circles) beginning at ED11.5.
DISCUSSION
These experiments suggest that increasing SMN expression in the spinal cord can improve the phenotype of mouse models of SMA and that survival is the most sensitive indicator of changes in SMN expression in the SMNΔ7 SMA mouse. A low increase in SMN protein does impact the SMA phenotype in mice. The protective effect of D156844-mediated increase in SMN expression is similar in magnitude to that observed with lentivirus-driven SMN gene replacement in SMNΔ7 SMA mice (52). Densitometric analysis revealed that D156844 increases SMN protein expression in spinal cord extracts from SMNΔ7 SMA mice by ∼1.7-fold (Fig. 3C and D). This change in SMN protein levels is similar to the fold change observed in response to trichostatin A (TSA) treatment (53,54). One difference between the TSA studies and this study is that we examined changes in SMN protein expression at PND08—prior to the onset of motor neuron loss (22)—although the TSA studies monitored changes in SMN expression after the onset of motor neuron loss [PND10 and PND14 using our nomenclature for mouse age; (53,54)]. There is significant motor neuron death in the spinal cords of these mice at the time points examined in the TSA studies. TSA treatment increases motor neuron counts in the spinal cord (53,54) so it remains uncertain whether the changes in SMN protein levels observed in response to TSA treatment are truly due to increased SMN expression in a cell (presumably a motor neuron) or due to the increased number of living cells in the spinal cord at PND10 or PND14. The advantage of measuring changes in SMN levels at PND08 is that there is very little motor neuron loss at this time (22) so there is a greater likelihood that increased SMN RNA/protein levels are the result of increased SMN expression and not due to increased cell numbers.
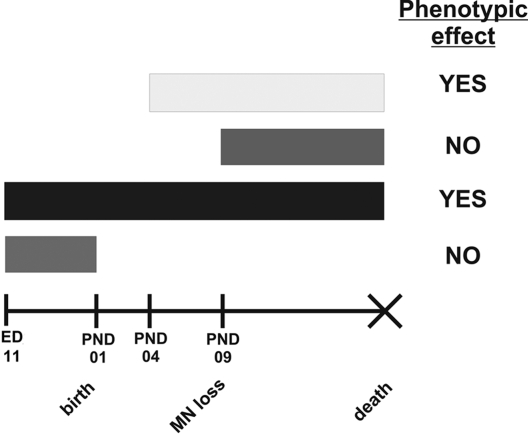
Treatment of SMA mice beginning at PND04 has a marked effect on phenotype and prenatal administration of this drug has a larger effect on survival of SMNΔ7 SMA mice. If treatment with D156844 begins after the onset of motor neuron loss in SMNΔ7 SMA mice, there is no improvement in motor phenotype or survival. Prenatal treatment alone does not result in improved survival. Severe SMA mice that also harbor a SMN transgene driven by the prion protein promoter—which activates expression in neurons during early prenatal development (ED13)—show no signs of neurodegeneration (51). Treatment of SMNΔ7 SMA mice with TSA improves the survival of these mice if drug treatment begins prior to motor neuron loss (53,54). These observations suggest that there may be an optimal window of therapeutic opportunity (Fig. 8) for SMN induction in SMA. This window of therapeutic opportunity will become very important for the design of clinical trials in SMA using SMN inducers. The identification of SMA patients as soon as possible using newborn genetic screening (55) will be the most effective means to treating these patients with an SMN inducer.
Figure 8.
Therapeutic window of opportunity for protective effects of SMN2 induction by D156844.
We have shown here that treatment of SMNΔ7 SMA with D156844 ameliorates some aspects of the motor neuron disease phenotype, prevents motor neuron loss at PND11 and significantly increases the survival of these mice. This compound increases SMN protein expression and function, i.e. snRNP assembly, in spinal cord extracts from SMNΔ7 SMA mice but these changes are not statistically significant. How can we account for the discrepancy between a statistically non-significant increase in SMN protein levels and a significant improvement in the SMA phenotype in these mice? One possible explanation for this discrepancy is a result of the limited sensitivities of the assays used to monitor changes in SMN expression in vivo. Due to the inherent limited sensitivities of these assays, large samples sizes would be required to reach statistical significance when real but small changes in SMN expression are observed.
A second possible explanation may relate the heterogeneous nature of the tissue we examined for changes in SMN expression and activity. The spinal cord is composed of many different types of cells including motor neurons, interneurons, sensory neurons, astrocytes, oligodendrocytes and microglia. D156844 may markedly increase SMN expression in motor neurons but only have a modest effect on SMN expression in other cells types. In our assays using whole tissue extracts, this putative marked increase in motor neurons may be blunted by the minimal putative changes in SMN expression in these other cell types. Future experiments can determine if D156844 differentially affects SMN expression in specific cell types.
The target for D156844 may offer a third possible explanation for the discrepancy between changes in SMN protein expression and changes in survival of SMNΔ7 SMA mice. Protein microarray scanning was recently used to identify DcpS, a scavenger mRNA decapping enzyme, as the binding target for C5-substituted 2,4-diaminoquinazolines such as D156844 (43). DcpS has a 3′ → 5′ exonuclease activity that removes the residual 5′ 7-methylguanosine cap from degrading mRNAs (56). C5-quinazolines such as D156844 are potent allosteric modulators of DcpS activity that hold the dimer in an open but catalytically inactive conformation (43). How this inhibition leads to increased SMN production is unclear. Inhibition of DcpS may increase SMN2 transcription, stabilize SMN2 mRNA, alter splicing of SMN2 pre-mRNAs or influence the translation of SMN2 mRNA transcripts through both direct and indirect mechanisms. Ongoing experiments will identify the DcpS activity critical to enhancing SMN levels and determine if inhibition of DcpS directly will ameliorate the phenotype of SMA models. Additionally, it is possible that the phenotypic improvement of SMNΔ7 SMA mice treated with D156844 may result from DcpS inhibition independent of alterations in SMN expression. Future experiments will address this question by determining if DcpS inhibition by D156844 can improve the phenotypes of other models for motor neuron disease such as the SOD1(G93A) transgenic mouse—a commonly used model for familial amyotrophic lateral sclerosis.
Treatment of SMNΔ7 SMA mice with D156844 significantly prevents motor neuron loss in the ventral horn of the spinal cord at PND11 (Fig. 5E–H). In fact, motor neuron cell body counts in the lumbar spinal cord of D156844-treated SMNΔ7 SMA mice at PND11 are the same as those observed in carrier mice. Even though there is a marked increase in motor neuron numbers at PND11, D156844 increases the average lifespan of SMNΔ7 SMA mice by 21–30% depending on when dosing starts. Treatment of D156844 may delay motor neuron loss in SMNΔ7 SMA mice but may not prevent it. A detailed examination of changes in motor neuron numbers in the lumbar spinal cord of D156844-treated SMNΔ7 SMA mice over time would address this hypothesis. Another question that arises is whether or not the motor neurons in D156844-treated SMNΔ7 SMA mice are functional. On the basis of observations where D156844-treated SMNΔ7 SMA mice have improved locomotor activity at PND11, we would suggest that these motor neurons are at least partially functional at PND11. We (22) and others (57–59) have shown that there is a progressive loss of innervation in neuromuscular junctions of SMNΔ7 SMA mice Future experiments examining changes in neuromuscular junction morphology and innervation status would help determine if the motor units from D156844-treated SMNΔ7 SMA mice exhibit any amelioration relative to vehicle-treated SMNΔ7 SMA mice.
The first two compounds tested, D152344 and D153249, did not improve the survival of SMNΔ7 SMA mice even though they were able to induce mSmn promoter activity in vivo. Profiling the compounds in assays of cellular toxicity revealed that both compounds strongly depleted cellular ATP levels. Chemical substructure searches revealed that the 2,4-diaminoquinazoline moiety of the C5-quinazolines closely overlapped with the 2,4-diamino-6-pteridinyl core of methotrexate, a potent inhibitor of DHFR (42). Methotrexate was used in cancer therapy but it can have neurotoxic effects in children (60). Subsequent testing of D152344 and D153249 revealed that they were potent DHFR inhibitors. If the DHFR inhibitory activity is reduced (as is the case with D156844), then ATP depletion activity is also reduced (42). The off-target inhibition of DHFR activity by D152344 and D153249 may explain the lack of protective effects these compounds have on SMNΔ7 SMA mice. D156844 was designed not only to be a more potent activator of SMN expression but also to remove the off-target DHFR inhibitory activity and associated toxicity. As a result, this quinazoline derivative can increase SMN protein expression in the spinal cord of neonatal mice and improve the phenotype of SMNΔ7 SMA mice. This observation underscores the fact that drug optimization needs to balance multiple compound properties simultaneously—such as enhancing target activity whereas eliminating off-target activity—in order to result in an efficacious drug.
This study is the first reported small molecule SMN inducer that was identified by an ultrahigh throughput screen (41), chemically optimized for potency, oral bioavailability and blood-brain barrier penetration using ligand-based design strategies (42) and displayed therapeutic benefit to SMA mice. This hit-to-lead-to-preclinical candidate strategy has recently been successfully applied to the development of a potent and efficacious polyglutamine aggregation inhibitor (C2-8) which is a preclinical candidate for Huntington's disease (61,62). Indoprofen (31) and aclarubicin (23) were also identified as SMN inducers using chemical library screening but neither compound can cross the blood-brain barrier. Using a similar strategy to the one used for the optimization of D156844, potent, blood-brain barrier penetrant SMN inducers can be designed so as to develop additional preclinical candidates for SMA.
Previous studies have identified sodium butyrate (BA) (24) and TSA (53) as inducers of SMN expression in vitro that have beneficial effects on mouse models of SMA. Neither of these compounds are promising clinical candidates because of poor pharmacokinetics (BA), low oral bioavailability (BA and TSA) and toxicity (TSA). Both BA and TSA act as histone deacetylase inhibitors (63). As the target for D156844 has been identified as DcpS, it is now possible to test these compounds in combination with D156844 so as to maximize SMN protein induction using different targets and to enhance the beneficial effects of these compounds in SMA. Initial data on the SMN2 promoter assay used to identify the quinazoline compounds indicates this combination therapy has an additive effect on reporter gene activity in this assay (41).
In summary, we suggest that increasing SMN protein expression in the spinal cord can ameliorate the phenotype of SMNΔ7 SMA mice if expression is induced prior to motor neuron loss. D156844 is the first reported compound that has been optimized for potent increase in SMN expression in the neonatal central nervous system, blood-brain barrier penetration and oral bioavailability. This study demonstrates the feasibility of drug discovery and optimization for the development of clinical therapeutic candidates for SMA and the validity of the use of SMA mouse models (64) for drug discovery. This study along with others (53,54) also demonstrates that survival is the most sensitive indicator of therapeutic benefit in SMNΔ7 SMA mice.
MATERIALS AND METHODS
Drug formulations
The syntheses of the 2,4-diaminoquinazoline derivatives D152344 (5-(3-chlorobenzoxyl)quinazoline-2,4-diamine), D153249 (5-(S)-[1-(3-chlorophenyl)ethoxyl]quinazoline-2,4-diamine) and D156844 ([5-(1-(2-fluorobenzyl)piperidin-4-ylmethoxy]quinazoline-2,4-diamine dihydrochloride) were completed by deCODE as described in (42). D152344 and D153249 were complexed with 40% hydroxypropyl-β-cyclodextrin (PrOHβCD) in ddH2O. D156844 was dissolved in ddH2O.
Animals
For survival and phenotype analyses, SMNΔ7 SMA mice (SMN2 +/+;SMN_Δ_7 +/+;mSmn −/−) were used (22). These mice were generated from males and females with a SMN2 +/+;SMN_Δ_7 +/+;mSmn +/− [line 4299; FVB.Cg-Tg(SMN2*delta7)4299Ahmb Tg(SMN2)89Ahmb Smn1 _tm1Msd_] genotype. These mice originated from our colony but can be obtained from the Jackson Laboratory (#005025). Breeder mice along with their pups were fed on a standard laboratory diet (Harlan-Teklad 22/5 rodent diet). Neonatal offspring were genotyped using a PCR-based assay on genomic DNA from tail biopsies as described previously (22). Only pups of the genotypes SMN2 +/+;Smn_Δ_7 +/+;mSmn +/− (carrier) and SMN2 +/+;Smn_Δ_7 +/+;mSmn −/− (SMA) were used in these experiments. All experiments were conducted in accordance with the protocols described in the National Institutes of Health ‘Guide for the Care and Use of Animals’ and were approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee.
Neonatal MTD analysis
SMNΔ7 carrier mice received differing doses of D152344 (10–100 mg/kg/day, b.i.d.), D153249 (10–100 mg/kg/day, b.i.d.), D156844 (0.1–30 mg/kg/day) or the appropriate vehicle beginning at PND04. Compounds were delivered via oral administration as described previously (65). Drug treatment lasted for 5 days for the MTD analysis at which time the pups were euthanized 1 h after final dosing. Serum samples were taken and the brains were dissected and rapidly frozen in N2(l). Tail snips were also taken for genotyping.
Neonatal drug administration
Carrier and SMA littermate mice were treated with D152344 (30 mg/kg/day, b.i.d.), D153249 (10 mg/kg/day, b.i.d.), D156844 (3 mg/kg/day) or the appropriate vehicle via oral administration beginning at PND04—with birth being defined at PND01—or PND09 as described previously (65). Mice were monitored daily for changes in body mass and for death.
Prenatal MTD analysis and drug administration
For prenatal drug administration, carrier dams received either D156844 (0.3–60 mg/kg/day for MTD and 3 mg/kg/day for survival analysis) or vehicle via oral administration (65) beginning at embryonic day 11.5 (ED11.5). Drug treatment lasted for 5 days for the MTD analysis at which time the dams were euthanized 1 h after final dosing and the brains of the fetuses were dissected and rapidly frozen in N2(l). Tail snips from the fetuses were also taken for genotyping. Additionally, the forebrains of the dams were dissected and rapidly frozen in N2(l). For survival analysis, drug dosing continued through birth and the resultant pups were treated with D156844 or vehicle via oral administration beginning at PND02 until death; dosing of the dam ceased after PND01.
Behavior analysis
A cohort of drug-treated SMNΔ7 SMA mice were assessed for changes in righting reflex success and latency, spontaneous locomotor activity and pivoting activity as described by Butchbach et al. (44). For monitoring righting reflexes, each pup was turned onto its back and the time it takes to stably place all four paws on the ground was recorded (cutoff time of 60 s). Righting reflexes were assessed on PND07 and PND11. For spontaneous locomotor activity, each pup was placed in the center of a gridded (with 28 2.5-cm2 grids) arena and the number of grids crossed in 1 min was counted as well as the latency for walking a distance greater than its body length (vectorial movement latency). For pivoting, each pup was placed in the center of a gridded arena and the number of times the pup turned 90° (pivots) during a 1-min time frame was counted. Spontaneous locomotor activity and pivoting were monitored on PND07, PND11 and PND14. To minimize the stress on the pup, the spontaneous locomotor activity and pivoting tests was conducted simultaneously.
Detection of drug levels
Serum and forebrain samples were obtained from neonatal mice treated with differing doses of each compound and processed as described in (42). Drug levels were measured using LC-MS/MS. All pharmacokinetic parameters reported were calculated using WinNoLin (version 5.0). Protein concentrations were measured for each sample after LC-MS/MS analysis; serum and forebrain drug levels were expressed as nanogram/milligram protein.
For detection of D152344, the LC-MS/MS system consisted of an Hitachi L6200 gradient pump used for regeneration and trapping, HP1100 LC analytical pump, CTC PAL sample manager and a Quattro Ultima Micromass® mass spectrometer (Waters/Micromass, Milford, MA, USA). A trap column, YMC-Pack ODS-AQ, 12 nm, S-5 µm, 20 × 2.1 mm ID (YMC Europe GmbH, Schermbeck, Germany) and a SymmetryShield RP8 (3.5 µm, 50 mm × 2.1 mm i.d.) analytical column from Waters were used for the analysis. The standards and sample extracts were trapped on the trap column with mobile phase A consisting of 15% MeOH, 85% H2O and 5 mm ammonium acetate and eluted through the analytical column with mobile phase B consisting of 95% MeOH, 5% H2O and 5 mm ammonium acetate, with a total run time of 5 min. The flow through the trap column was 1.0 ml/min and the elution flow rate was 0.30 ml/min.
For detection of D153249, the LC-MS/MS system consisted of a Waters 1525µ binary pump, Waters 2777 sample manager and a Quattro Premier Micromass® mass spectrometer (Waters/Micromass, Milford, MA). A SymmetryShield RP8 (3.5 µm, 50 mm × 2.1 mm i.d.) analytical column also from Waters was used for the analysis. The standards and sample extracts were chromatographed with a gradient mobile phase consisting of 5% MeOH, 95% H2O, 5 mm ammonium acetate as mobile phase A and 95% MeOH, 5% H2O, 5 mm ammonium acetate as mobile phase B. The steep gradient was 25% B at 0.8 min to 95% B at 1.5 min with a total run time of 4 min. The flow rate was 0.35 ml/min.
The detection of D156844 was accomplished using a LC-MS/MS system consisting of a Waters 1525µ binary pump, Waters 2777 sample manager and a Quattro Premier Micromass® mass spectrometer (Waters/Micromass, Milford, MA). A SymmetryShield RP8 (3.5 µm, 50 mm × 2.1 mm i.d.) analytical column also from Waters was used for the analysis. The standards and sample extracts were chromatographed with a gradient mobile phase consisting of 15% MeOH, 85% H2O and 5 mm ammonium acetate as solvent A and 95% MeOH, 5% H2O and 5 mm ammonium acetate as mobile phase B. The steep gradient was 5% at 0.8 min to 95% B at 1.5 min with a total run time of 5 min. The flow rate was 0.35 ml/min.
β-Galactosidase assay
β-Galactosidase activity was measured using the Galacto-Light chemiluminescent reporter gene assay (Applied Biosystems, Bedford, MA, USA) according to manufacturer's directions except that the lysis buffer contained 0.1% Triton X-100. Luminescence was monitored using a Luminoskan Ascent luminometer.
Real time RT–PCR
Total RNA was isolated from forebrains and spinal cords from SMNΔ7 SMA mice treated with either D156844 or vehicle for 5 days using RNeasy Mini columns (Qiagen) according to manufacturer's directions. Quantification of FL-SMN and SMNΔ7 mRNA transcripts was accomplished using real time RT–PCR as described previously (66) with modifications. The amounts of FL-SMN and SMNΔ7 mRNAs were quantified relative to the geometric mean of three housekeeping genes (hypoxanthine phosphorylribosyltransferase, β-actin and RNA polymerase 2A) to minimize the variability in the expression of a single housekeeping gene (67).
Immunoblot
One hundred micrograms of tissue protein extract were mixed with 0.16 volumes 6×loading dye (10.28% SDS, 36% glycerol and 0.012% bromophenol blue in 350 mm Tris–HCl, pH 6.8) containing freshly added 100 mm DTT and resolved through a 12% polyacrylamide gel containing 0.1% SDS. Samples were then transferred onto a PVDF membrane via electroblotting. The resultant blots were incubated in 1×blocking buffer [5% non-fat milk, 1% BSA in PBST (0.2% Tween-20 in PBS)] overnight at 4°C and then with a primary antibody diluted in 0.2×blocking buffer for 1 h at room temperature. For detection of SMN protein, a mixture containing equivalent amounts of the following mAbs was used: 8F7 (MANSMA2), 1F1 (MANSMA21), 5E3 (MANSMA13) and 7Q12 (MANSMA19) (68). This SMN cocktail was used at a titer of 1:500. After extensive washing with PBST (3 × 10 min), the blots were incubated with a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:1000) diluted in 0.2×blocking buffer for 1 h at room temperature, washed extensively as above and the bound antibodies were detected by chemiluminescence (ECL Western Blotting Detection Reagents, Amersham Biosciences). To confirm equal loading of protein in each lane, the blots were stripped and reprobed using a mouse anti-β-actin mAb (1:20 000; clone AC-15, Sigma-Aldrich). Band intensities were measured using ImageJ (Scion Corp.).
snRNP assembly
Preparation of mouse tissue extracts and snRNP assembly experiments were carried out essentially as previously described with minor modifications (69). The amount of immunoprecipitated U1 snRNAs was quantified using a STORM 860 Phosphorimager (Molecular Dynamics) and the ImageQuant version 4.2 software.
Histology
SMNΔ7 SMA mice and carrier littermates were treated with D156844 (3 mg/kg/day) or vehicle beginning at PND04 until PND11. Treated mice were anesthetized with 2.5% Avertin and then transcardially perfused with ice-cold PBS following by 4% paraformaldehyde in sodium phosphate buffer (100 mm Na2HPO4 and 100 mm NaH2PO4, pH 7.4). The lumbar spinal cords were postfixed with 4% paraformaldehyde in sodium phosphate buffer overnight at 4°C followed by cryoprotection with 30% sucrose in ddH2O overnight at 4°C. The lumbar spinal cords were sectioned coronally at a thickness of 25 µm using the MultiBrain® Technology by NeuroScience Associates (Knoxville, TN, USA). Every 12th section block was mounted onto a glass slide and stained with 1% Cresyl violet as described by Le et al. (22).
Statistical analysis
Data are expressed as means ± standard errors. Kaplan–Meier curves were generated from the survival data and tested using the Mantel–Cox log rank test. All statistical analyses were performed with SPSS v.17.0.
AUTHOR CONTRIBUTIONS
M.E.R.B., J.S., L.R.S., L.P., J.J., A.H.M.B and M.E.G. designed research; M.E.R.B., M. Þ., L.S., E.S., J.Z. and J.D.E. performed research; J.S., J.T., T.A. and M.E.G. contributed new reagents or analytical tools; M.E.R.B., J.S., M. Þ., L.R.S., L.P. and A.H.M.B. analyzed data and M.E.R.B., J.S., M. Þ., L.R.S., L.P., J.J., A.H.M.B. and M.E.G. wrote the article.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
The development of the quinazoline compound and the testing in the SMA mice were financially supported in its entirety by Families of SMA (to M.E.R.B., A.H.M.B. and M.E.G.). The establishment and maintenance of the mice used in this study were funded in part by the National Institutes of Health (NS38650 to A.H.M.B.) and the Miracles for Madison Fund (to A.H.M.B.). The snRNP assembly assays were funded by the SMA Foundation and the Motor Neuron Center of Columbia University (to L.P.).
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
We would like to thank Dr Glenn Morris for kindly providing the SMN monoclonal antibodies, Dr Chris Spancake for advice on vehicle preparations and bioavailability analysis and the staff at NeuroScience Associates for assistance with spinal cord sectioning.
Conflict of Interest statement. Performance of research by the deCODE authors (J.S., M. Þ., T.A., J.T., J.Z. and M.E.G.) occurred pursuant to a contract with Families of SMA; deCODE was paid, as a commercial entity, by Families of SMA to conduct such research. The Families of SMA has rights to, and resulting financial interests in, the results and intellectual property obtained through such research. A.H.M.B. served as a paid consultant for a pharmaceutical company. No other authors have any potential conflicts of interest.
REFERENCES
- 1.Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 2.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing an is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H.M., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 5.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 7.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H.M. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiDonato C.J., Chen X.N., Noya D., Korenberg J.R., Nadeau J.H., Simard L.R. Cloning, characterization and copy number of the murine survival motor neuron gene: homolog of the spinal muscular atrophy-determining gene. Genome Res. 1997;7:339–352. doi: 10.1101/gr.7.4.339. [DOI] [PubMed] [Google Scholar]
- 9.Viollet L., Bertrandy S., Beuno Brunialti A.L., Lefebvre S., Burlet P., Clermont O., Cruaud C., Guénet J.L., Munnich A., Melki J. cDNA isolation, expression and chromosomal localization of the mouse survival motor neuron gene (Smn) Genomics. 1997;40:185–188. doi: 10.1006/geno.1996.4551. [DOI] [PubMed] [Google Scholar]
- 10.Schrank B., Götz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cifuentes-Diaz C., Nicole S., Velasco M.E., Borra-Cebrian C., Panozzo C., Frugier T., Millet G., Roblot N., Joshi V., Melki J. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum. Mol. Genet. 2002;11:1439–1447. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 12.Nicole S., Desforges B., Millet G., Lesbordes J., Cifuentes-Diaz C., Vertes D., Cao M.L., De Backer F., Languille L., Roblot N., et al. Intact satellite cells lead to remarkable protection against Smn gene defect in differentiation skeletal muscle. J. Cell Biol. 2003;161:571–582. doi: 10.1083/jcb.200210117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitte J.M., Davoult B., Roblot N., Mayer M., Joshi V., Courageot S., Tronche F., Vadrot J., Moreau M.H., Kemeny F., et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am. J. Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McWhorter M.L., Monani U.R., Burghes A.H.M., Beattie C.E. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan Y.B., Miguel-Aliaga I., Franks C., Thomas N., Trülzsch B., Sattelle D.B., Davies K.E., van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- 16.Rajendra T.K., Gonsalvez G.B., Walker M.P., Shpargel K.B., Salz H.K., Matera A.G. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J. Cell Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang H.C.H., Dimlich D.N., Yokokura T., Mukherjee A., Kankel M.W., Sen A., Sridhar V., Fulga T.A., Hart A.C., Van Vactor D., et al. Modeling spinal muscular atrophy in Drosophila. PLoS ONE. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briese M., Esmaeili B., Fraboulet S., Burt E.C., Christodoulou S., Towers P.R., Davies K.E., Sattelle D.B. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet. 2009;18:97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn−/− mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 21.Meyer K., Marquis J., Trüb J., Nlend Nlend R., Verp S., Ruepp M.D., Imboden H., Barde I., Trono D., Schümperli D. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum. Mol. Genet. 2009;18:546–555. doi: 10.1093/hmg/ddn382. [DOI] [PubMed] [Google Scholar]
- 22.Le T.T., Pham L.T., Butchbach M.E.R., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H.M. SMNΔ7, the major product of the centromeric survival motor neuron gene (SMN2), extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 23.Andreassi C., Jarecki J., Zhou J., Coovert D.D., Monani U.R., Chen X., Whitney M., Pollok B., Zhang M., Androphy E.J., et al. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum. Mol. Genet. 2001;10:2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- 24.Chang J.G., Hsieh-Li H.M., Jong Y.J., Wang N.M., Tsai C.H., Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl. Acad. Sci. USA. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreassi C., Angelozzi C., Tiziano F.D., Vitali T., De Vincenzi E., Boninsegna A., Villanova M., Bertini E., Pini A., Neri G., et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 26.Sumner C.J., Huynh T.N., Markowitz J.A., Perhac J.S., Hill B., Coovert D.D., Schussler K., Chen X., Jarecki J., Burghes A.H.M., et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 27.Brichta L., Hofmann Y., Hahmen E., Siebzehnrubl F.A., Raschke H., Blumcke I., Eyupoglu I.Y., Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 28.Mattis V.B., Butchbach M.E.R., Lorson C.L. Detection of human survival motor neuron (SMN) protein in mice containing the SMN2 transgene: applicability to preclinical therapy development for spinal muscular atrophy. J. Neurosci. Methods. 2008;175:36–43. doi: 10.1016/j.jneumeth.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang W.C., Yuo C.Y., Chang J.G., Chen Y.C., Chang Y.F., Wang H.Y., Ju Y.H., Chiou S.S., Jong Y.J. The effect of hydroxyurea in spinal muscular atrophy cells and patients. J. Neurol. Sci. 2008;268:87–94. doi: 10.1016/j.jns.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Grzeschik S.M., Ganta M., Prior T.W., Heavlin W.D., Wang C.H. Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann. Neurol. 2005;58:194–202. doi: 10.1002/ana.20548. [DOI] [PubMed] [Google Scholar]
- 31.Lunn M.R., Root D.E., Martino A.M., Flaherty S.P., Kelley B.P., Coovert D.D., Burghes A.H.M., Man N.T., Morris G.E., Zhou J., et al. Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism. Chem. Biol. 2004;11:1489–1493. doi: 10.1016/j.chembiol.2004.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron-Delage S., Abadie A., Echaniz-Laguna A., Melki J., Beretta L. Interferons and IRF-1 induce expression of the survival motor neuron (SMN) genes. Mol. Med. 2000;6:957–968. [PMC free article] [PubMed] [Google Scholar]
- 33.Majumder S., Varadharaj S., Ghoshal K., Monani U., Burghes A.H.M., Jacob S.T. Identification of a novel cyclic AMP response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene. J. Biol. Chem. 2004;279:14803–14811. doi: 10.1074/jbc.M308225200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Zhang M.L., Lorson C.L., Androphy E.J., Zhou J. An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: potential therapy of SMA. Gene. Ther. 2001;8:1532–1538. doi: 10.1038/sj.gt.3301550. [DOI] [PubMed] [Google Scholar]
- 35.Novoyatleva T., Heinrich B., Tang Y., Benderska N., Butchbach M.E.R., Lorson C.L., Lorson M.A., Ben-Dov C., Fehlbaum P., Bracco L., et al. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum. Mol. Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 36.Wolstencroft E.C., Mattis V., Bajer A., Young P.J., Lorson C.L. A non-sequence specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum. Mol. Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- 37.Mattis V.B., Rai R., Wang J., Chang C.W.T., Coady T., Lorson C.L. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum. Genet. 2006;120:589–601. doi: 10.1007/s00439-006-0245-7. [DOI] [PubMed] [Google Scholar]
- 38.Sakla M.S., Lorson C.L. Induction of full-length survival motor neuron by polyphenol botanical compounds. Hum. Genet. 2008;122:635–643. doi: 10.1007/s00439-007-0441-0. [DOI] [PubMed] [Google Scholar]
- 39.Hahnen E., Eyüpoglu I.Y., Brichta L., Haastert K., Tränkle C., Siebzehnrübl F.A., Riessland M., Hölker I., Claus P., Romstöck J., et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J. Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 40.Riessland M., Brichta L., Hahnen E., Wirth B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum. Genet. 2006;120:101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 41.Jarecki J., Chen X., Bernardino A., Coovert D.D., Whitney M., Burghes A.H.M., Stack J., Pollok B. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum. Mol. Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 42.Thurmond J., Butchbach M.E.R., Palomo M., Pease B., Rao M., Bedell L., Keyvan M., Pai G., Mishra R., Haraldsson M., et al. Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy. J. Med. Chem. 2008;51:449–469. doi: 10.1021/jm061475p. [DOI] [PubMed] [Google Scholar]
- 43.Singh J., Salcius M., Liu S.W., Staker B.L., Mishra R., Thurmond J., Michaud G., Mattoon D.R., Printen J., Christensen J., et al. DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem. Biol. 2008;3:711–722. doi: 10.1021/cb800120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butchbach M.E.R., Edwards J.D., Burghes A.H.M. Abnormal motor phenotype in the SMNΔ7 mouse model of spinal muscular atrophy. Neurobiol. Dis. 2007;27:207–219. doi: 10.1016/j.nbd.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monani U.R., McPherson J.D., Burghes A.H.M. Promoter analysis of the human centromeric and telometic survival motor neuron genes (SMNc and SMNt) Biochim. Biophys. Acta. 1999;1445:330–336. doi: 10.1016/s0167-4781(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 46.Echaniz-Laguna A., Miniou P., Bartholdi D., Melki J. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am. J. Hum. Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boda B., Mas C., Giudicelli C., Nepote V., Guimiot F., Levacher B., Zvara A., Santha M., LeGall I., Simonneau M. Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur. J. Hum. Genet. 2004;12:729–737. doi: 10.1038/sj.ejhg.5201217. [DOI] [PubMed] [Google Scholar]
- 48.Rouget R., Vigneault F., Codio C., Rochette C., Paradis I., Drouin R., Simard L.R. Characterization of the survival motor neuron (SMN) promoter provides evidence for complex combinatorial regulation in undifferentiated and differentiated P19 cells. Biochem. J. 2005;385:433–443. doi: 10.1042/BJ20041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan L., Battle D.J., Yong J., Gubitz A.K., Kolb S.J., Wang J., Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gabanella F., Butchbach M.E.R., Saieva L., Carissimi C., Burghes A.H.M., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;9:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H.M. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azzouz M., Le T., Ralph G.S., Walmsley L., Monani U.R., Lee D.C.P., Wilkes F., Mitrophanous K.A., Kingsman S.M., Burghes A.H.M., et al. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2004;114:1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avila A.M., Burnett B.G., Taye A.A., Gabanella F., Knight M.A., Hartenstein P., Cizman Z., DiProspero N.A., Pellizzoni L., Fischbeck K.H., et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narver H.L., Kong L., Burnett B.G., Choe D.K., Bosch-Marcé M., Taye A.A., Eckhaus M.A., Sumner C.J. Sustained improvement of spinal muscular atrophy mice treated with trichostatin A plus nutrition. Ann. Neurol. 2008;64:465–470. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pyatt R.E., Prior T.W. A feasibility study for the newborn screening of spinal muscular atrophy. Genet. Med. 2006;8:428–437. doi: 10.1097/01.gim.0000227970.60450.b2. [DOI] [PubMed] [Google Scholar]
- 56.Bail S., Kiledjian M. DcpS, a general modulator of cap-binding protein-dependent processes? RNA Biol. 2008;5:216–219. doi: 10.4161/rna.7161. [DOI] [PubMed] [Google Scholar]
- 57.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 58.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong L., Wang X., Choe D.W., Polley M., Burnett B.G., Bosch-Marcé M., Griffin J.W., Rich M.M., Sumner C.J. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J. Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vezmar S., Becker A., Bode U., Jaehde U. Biochemical and clinical aspects of methotrexate neurotoxicity. Chemotherapy. 2003;49:92–104. doi: 10.1159/000069773. [DOI] [PubMed] [Google Scholar]
- 61.Chopura V., Fox J.H., Lieberman G., Dorsey K., Matson W., Waldmeier P., Housman D.E., Kazantsev A., Young A.B., Hersch S. A small-molecule therapeutic lead for Huntington's disease: preclinical pharmacology and efficacy of C2–8 in the R6/2 transgenic mouse. Proc. Natl. Acad. Sci. USA. 2007;104:16685–16689. doi: 10.1073/pnas.0707842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X., Smith D.L., Meriin A.B., Engemann S., Russel D.E., Roark M., Washington S.L., Maxwell M.M., Marsh J.L., Thompson L.M., et al. A potent small molecule inhibits polyglutamine aggregation in Huntington's disease neurons and suppresses neurodegeneration in vivo. Proc. Natl. Acad. Sci. USA. 2005;102:892–897. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnstone R.W. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 64.Butchbach M.E.R., Burghes A.H.M. Perspectives on models of spinal muscular atrophy for drug discovery. Drug Discov. Today Dis. Models. 2004;1:151–156. [Google Scholar]
- 65.Butchbach M.E.R., Edwards J.D., Schussler K.R., Burghes A.H.M. A novel method for oral delivery of compounds to the neonatal SMNΔ7 model of spinal muscular atrophy. J. Neurosci. Methods. 2007;161:285–290. doi: 10.1016/j.jneumeth.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simard L.R., Bélanger M.C., Morissette S., Wride M., Prior T.W., Swoboda K.J. Preclinical validation of a multiplex real-time assay to quantify SMN mRNA in patients with SMA. Neurology. 2007;68:451–456. doi: 10.1212/01.wnl.0000252934.70676.ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034.1–research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young P.J., Le T.T., thiMan N., Burghes A.H.M., Morris G.E. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 69.Gabanella F., Carissimi C., Usiello A., Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]