Cytoplasmic Functions of the Tumor Suppressor p53 (original) (raw)
. Author manuscript; available in PMC: 2010 Jan 31.
Published in final edited form as: Nature. 2009 Apr 30;458(7242):1127. doi: 10.1038/nature07986
Abstract
The principal tumor suppressor protein, p53, accumulates in cells in response to DNA damage, oncogene activation, and other stresses. It acts as a nuclear transcription factor that transactivates genes involved in apoptosis, cell cycle regulation, and numerous other processes. An emerging area of research unravels additional activities of p53 in the cytoplasm, where it triggers apoptosis and inhibits autophagy. These novel functions contribute to p53’s mission as a tumor suppressor.
Transcriptional and non-transcriptional effects of p53
Approximately half of human cancers exhibit inactivating mutations of p53, and most among the remaining malignancies deactivate the p53 pathway by elevating its inhibitors, reducing its activators, or inactivating its downstream targets. p53 is best characterized as a transcription factor that binds to specific DNA sequences and transactivates a number of genes with a variety of functions including cell cycle arrest, apoptosis, changes in metabolism, and others1 (Fig. 1). In addition to this nuclear activity, p53 also possesses biological activities that are cytosolic and transcription-independent. Several years ago, Oren and colleagues2 noted that over-expression of a mutant p53, lacking most of the DNA binding domain and completely deficient in transactivation function, could nonetheless trigger apoptosis. Indeed, overexpression of a variety of transactivation-incompetent p53 mutants can efficiently induce apoptosis in human cells3. Consistent with this, Caelles and Karin4 found that apoptosis induced by stabilization of an ectopically expressed temperature sensitive mutant of p53 could proceed in the absence of RNA and protein synthesis. Similarly, activation of p53 was found to trigger apoptosis even in the absence of a nucleus5. p53-reactivating drugs that interact with oncogenic, mutant p53 protein causing it to adopt a wild-type conformation can induce apoptosis under conditions of complete transcriptional or translational blockade5, 6. Most recently, mice were generated in which the endogenous p53 gene was replaced by a chimeric p53 protein that is capable of transactivation yet lacks several domains that are required for other p53 functions7. In fibroblasts from such mice, this chimeric p53 protein was transcriptionally active and able to induce cellular senescence, but was unable to trigger apoptosis7. Altogether, these observations support the idea that a cytoplasmic pool of p53 can induce apoptosis through a transactivation-independent mechanism.
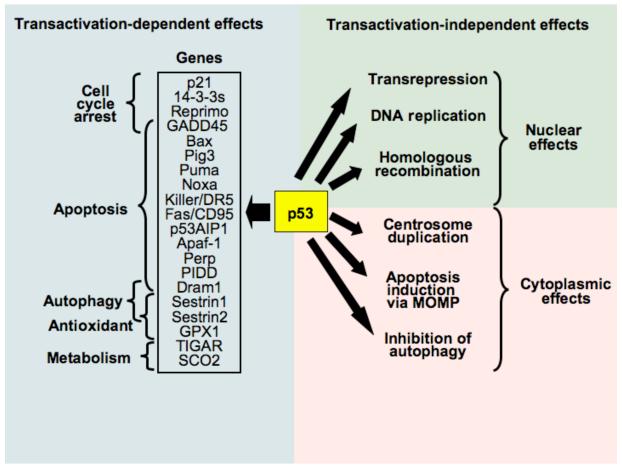
Fig. 1.
Classification of p53 activities. On the left side, some genes that are transactivated by p53 are exemplified, together with a few of the functional consequences of p53 activation. On the right side, transactivation-independent effects of p53 are listed. These can be divided into nuclear and extranuclear (cytoplasmic) p53 activities.
P53 is at the hub of numerous signaling pathways that are triggered by a range of cellular stresses including DNA damage by exogenous mutagens, oncogene activation, telomere erosion and hypoxia, all of which influence the abundance, subcellular localization, post-translational modification and/or interaction of p53 with cofactors. As a result of these context-dependent, damage-elicited alterations, p53 can facilitate the transient adaptation of cells to stressful conditions, for example by increasing DNA repair upon a transient cell cycle arrest or by enhancing the expression of ROS-detoxifying enzymes. Alternatively, p53 suppresses oncogenic potential by mediating an irreversible arrest of the cell cycle or by triggering apoptotic cell death8. In addition to its role as a tumor suppressor, p53 plays a major role in aging9, as well as in the unwarranted loss of post-mitotic cells such as heart muscle cells in infarction and neurons in stroke10. It is likely that the biological effects of p53 represent the combined activities of the nuclear and cytoplasmic protein.
Effects of cytoplasmic p53 on mitochondria
The mitochondrial membrane constitutes the battleground on which pro- and anti-apoptotic factors induce or prevent a potentially lethal permeabilization step11. Under a variety of cell death-inducing conditions, p53 rapidly moves to the mitochondria. For instance, whole body irradiation of mice causes a fraction of cellular p53 to associate with the outer mitochondrial membrane12. Similarly, ischemic damage of the rat brain triggers the translocation of p53 to the mitochondria of neurons that are particularly vulnerable to hypoxia within the CA1 area of the hippocampus10 Once at the mitochondrion, p53 induces mitochondrial outer membrane permeabilization (MOMP), thereby triggering the release of pro-apoptotic factors from the mitochondrial intermembrane space. Indeed, mitochondrion-targeted p53 can be as efficient as wild type p53 in inhibiting tumor growth12.
Multiple mechanisms have been invoked to explain how p53 triggers MOMP12. MOMP is usually inhibited by anti-apoptotic multidomain proteins of the Bcl-2 family (such as Bcl-2, Bcl-XL and Mcl-1), and is conditional on pro-apoptotic multidomain proteins from the same family (in particular Bax and Bak) that can homo-oligomerize within the outer mitochondrial membrane to form MOMP-mediating supramolecular structures. Depending on their particular affinities for multidomain Bcl-2 family proteins, a set of distinct pro-apoptotic “BH3-only” proteins can directly interact with Bax or Bak to trigger their homo-oligomerization and hence MOMP (these BH3-only proteins are referred to as “direct activators”) and/or neutralize one or more anti-apoptotic multidomain proteins (these BH3-only proteins are referred to as “sensitizers” or “derepressors”). In contrast to direct activators (in particularly the proteins Bim and Bid), sensitizers cannot trigger MOMP on their own (because they simply antagonize its inhibition) and require the input of additional stimuli for apoptosis induction13.
p53 has been suggested to act like a BH3-only protein, either as an direct activator of Bax and/or Bak, or as a sensitizer/de-repressor. Under pro-apoptotic conditions, p53 can be co-immunoprecipitated with Bcl-2, Bcl-XL and Bak12, 14, 15. In a defined system involving only recombinant proteins and synthetic membranes, p53 can trigger Bax to permeabilize liposomes through a “hit and run” mechanism, that is, through transient molecular associations16. NMR structures of p53 complexed to Bcl-2 or Bcl-XL suggest that the DNA binding domain (DBD) of p53 is involved in the docking with Bcl-2 family proteins12, 17, further supported by biochemical studies on p53 and Bak,18. Hence, oncogenic p53 mutations affecting the DBD can operate as “dual hits” and simultaneously abrogate transactivation and direct MOMP induction by p53. In addition, other studies show that the proline-rich region neighboring the DBD is important for Bax activation16 and can also associate with anti-apoptotic Bcl-2 proteins18 Further, the p53-binding interfaces within Bcl-XL and Bak may be distinct, as determined by biochemical approaches17. Since p53 has a higher affinity for Bcl-XL than for Bak19, it may first engage in molecular interactions with anti-apoptotic Bcl-2 proteins (as a sensitizer) and then with Bak and Bax (as a direct activator). Another nuclear transcription factor, Nur77, has previously been shown to bind to Bcl-2 in a way that radically changes its conformation, apparently converting Bcl-2 into a pro-apoptotic protein 20. However, the structural changes that p53 imposes on Bcl-2 or Bcl-XL are minor 12, 19, suggesting that p53 does not function in this manner.
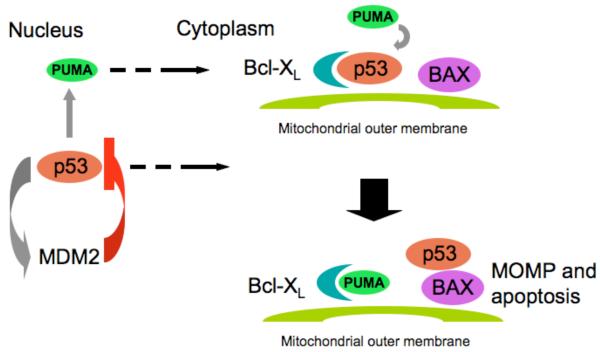
The pro-apoptotic effects of cytoplasmic p53 are not dependent on transcription, in principle. However, the control of transcription by nuclear p53 decisively contributes to the function of cytoplasmic p53. As discussed in more detail below, the p53 target MDM2 is essential for post-translational regulation of p53, without which the system would not be responsive to cellular stress. Another p53 target, PUMA, controls the sequestration of cytoplasmic p53 by the anti-apototic Bcl-XL protein, releasing p53 to activate Bax 21. Therefore, without transcription, regulated by nuclear p53, endogenous cytoplasmic p53 may not function (Fig. 2).
Fig. 2.
Interplay of the nuclear and cytoplasmic functions of p53 in apoptosis. Nuclear p53 induces the expression of MDM2, which acts to inhibit the protein through binding and ubiquitinylation. Cellular stress signals interrupt this inhibition, allowing p53 to accumulate both in the nucleus and in the cytoplasm. In the latter, p53 is sequestered by anti-apoptotic Bcl-2 proteins such as Bcl-XL. Another target of nuclear p53, PUMA, functions to disrupt the Bcl-XL-p53 interaction. The released p53 can now trigger MOMP and apoptosis through interaction with, for example, Bax.
One p53-targeted drug, pifithrin-μ, inhibits the pro-apoptotic effects of cytosolic p53, but has no apparent effect on p53-dependent transactivation22. This drug blocks the interaction of p53 with Bcl-XL22, and probably also the interaction of p53 with pro-apoptotic Bcl-2 family members (Bax and Bak), thereby accounting for its anti-apoptotic effects. Pifithrin-μ can rescue mice from otherwise lethal irradiation22, indicating that selective inhibition of the cytoplasmic p53 pathway is sufficient for radioprotection in vivo. Conversely, p53 activating drugs such as CP-31398 induce p53 translocation to mitochondria, as well as p53-dependent MOMP. This p53 translocation can be inhibited by cyclosporin A (CsA)6, pointing to a possible molecular crosstalk between the CsA inhibitable mitochondrial permeability transition pore (MPTP) and the p53 system. Reportedly, recombinant p53 may induce a more complete MOMP (with release of the pro-apoptotic factor AIF) than recombinant Bid added to purified mitochondria in vitro23, pointing to possible differences in mitochondrial permeabilization by p53 and BH3-only proteins.
Inhibition of autophagy by cytoplasmic p53
Macroautophagy (refered to as “autophagy”) consists in the sequestration and subsequent digestion of parts of the cytoplasm, allowing for the adaptation of cells to stressful conditions, as well as the removal of damaged, potentially harmful cytoplasmic organelles. Enhanced autophagy, which frequently accompanies cell death, likewise constitutes a failed attempt to adapt to stress and to survive, rather than a lethal catabolic process24. Since autophagy plays an essential role in the maintenance of genomic stability25, inhibition of autophagy is oncogenic. Accordingly, loss of only one allele of either of the two haploinsufficient autophagy genes beclin 1 and uvrag is sufficient to promote carcinogenesis, and multiple oncogenes including Bcl-2, Akt, PI3K inhibit autophagy. Similarly, the inactivation of tumor suppressor proteins such as PTEN, TSC1, TSC2 and LKB1/STK11 results in autophagy inhibition24.
Although p53 can transactivate genes that induce autophagy (such as DRAM and sestrins-1 and-2)26, 27, normal levels of p53 mediate a tonic inhibition of autophagy (Fig. 1). In fact, the deletion, depletion or pharmacological inhibition of p53 with pifithrin-α induces autophagy in mouse, human and nematode cells28. Suppression of autophagy is mediated by cytoplasmic, not nuclear p53, and physiological inducers of autophagy (such as nutrient depletion) must destroy the pool of cytoplasmic p53 to induce autophagy28. Thus, inhibition of the ubiquitin E3 ligase MDM2, which targets p53 for destruction, can suppress the induction of autophagy by starvation, rapamycin, lithium, or damage of the endoplasmic reticulum28. Cytoplasmic (but not nuclear) p53 inhibits the AMP-dependent kinase (AMPK), a positive regulator of autophagy, and activates mammalian target of rapamycin (mTOR), a negative regulator of autophagy28. How these effects are achieved, however, remains an open conundrum. Nevertheless, it is not unlikely that the transactivation-dependent metabolic effects of p53 and its cytosolic, transcription-independent inhibition of autophagy cooperate to ensure a coordinated action of p53 in cellular adaptation, such as in reprogramming metabolism towards oxidative phosphorylation.
It is tempting to speculate that the dual action of cytoplasmic p53 – inhibition of autophagy and induction of MOMP – may constitute a coordinated response for cell death induction (Fig. 3). Autophagy accounts for the removal of damaged and permeabilized mitochondria and counteracts the lethal effect of MOMP29. Therefore, autophagy inhibition by p53 may further facilitate cell death execution by MOMP. Nonetheless, it is not clear through which mechanisms cytoplasmic p53 switches from its baseline function (autophagy inhibition) to its killer activity (translocation to mitochondria and MOMP induction).
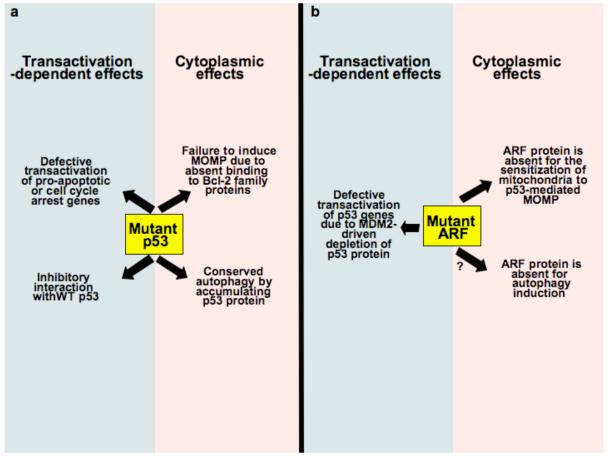
Fig. 3.
Concerted oncogenic actions of mutant p53 (a) or inactive ARF (b) in the nucleus and cytoplasm of cancer cells. Hot spot mutations of p53 affecting the DNA binding domain can abolish the transactivation of p53 genes as well as the mitochondrion-permeabilizing action of p53, yet leave intact autophagy inhibition by p53 (a). Similarly, oncogenic mutations that affect the C-terminus of ARF can lead to the depletion of p53 protein, as well as to the abolition of mitochondrion-permeabilizing and autophagy-inducing activities mediated by the ARF protein (b).
At first glance it appears paradoxical that normal levels of cytoplasmic p53 inhibit autophagy, although inhibition of autophagy is often associated with oncogenesis. This paradox is resolved by the observation that mutant p53 protein – which has lost its transactivation function and which accumulates in the cytoplasm of tumor cells – efficiently inhibits autophagy30. The structural features of p53 required for its cytoplasmic pro-apoptotic and anti-autophagic functions are clearly distinct. Indeed, deletion of the DBD does not affect autophagy inhibition by p53. Moreover, point mutations that affect the nuclear functions of p53, as well as its interaction with Bcl-2 family proteins, do not abolish its capacity to inhibit autophagy30. This may contribute to the strong oncogenic action of certain p53 mutants that is difficult to explain by the mere abolition of their tumor-suppressive capabilities.
Regulating the regulator
The nuclear versus cytoplasmic effects of p53 are determined by multiple post-translational modifications that affect its interaction with other proteins, its shuttling between the cytoplasm and the nucleus, and its biological activities. Poly(ADP)ribosylation of p53 leads to its nuclear accumulation8. In contrast, monoubiquitylation by MDM2 stimulates the nuclear export of p53, which upon arrival at mitochondrial is deubiquitylated by mitochondrial HAUSP, thus generating the apoptotically active non-ubiquitylated p53 31 Other post-translational modifications of p53 (such as phosphorylation of C-terminal serines) can stimulate nuclear export and/or mitochondrial association. Moreover, the transcription factor FOXO3a promotes p53 cytoplasmic accumulation by increasing its nuclear export, hence stimulating direct, p53-mediated MOMP induction32. This suggests that the entire context of post-transcriptional p53 modifications and protein interactions can affect the precise subcellular localization and function of p53.
A novel class of p53 activators, the tenovins, activate the tumor suppressive function of p53 by inhibiting in tumor cells the sirtuins, SIRT1 and SIRT233, which are p53 deacetylases. Similarly, a tumor suppressor, DBC1 (deleted in breast cancer 1) acts as an endogenous inhibitor of SIRT1 and as a positive regulator of p53 34. However, according to one report35, nuclear p53 is acetylated while cytoplasmic p53 is deacetylated. Knockout of SIRT1 facilitates the ROS-induced nuclear translocation of p53 and simultaneously inhibits direct MOMP induction by cytoplasmic p53, at least in mouse embryonic stem cells35. Future investigations must resolve this apparent contradiction to understand which post-transcriptional modifications of p53 determine its pro-MOMP and anti-autophagy activities.
Future directions and perspectives
Cancer cells are characterized by failing cell cycle checkpoints, reduced propensity to apoptosis and suppressed autophagy. “Hot spot” mutations of p53 within the DBD usually abolish the transactivation function of p53 (and often create dominant-negative inhibitors of wild type p53, with which they form heterotetramers) thereby preventing the expression of cell cycle-arresting, pro-apoptotic and autophagy-inducing genes. Such p53 mutations within the DBD also affect the cytoplasmic functions of p53, reducing its capacity to induce MOMP (through failure to interact with Bcl-2 family proteins)12, 15, 17, yet leaving intact its inhibitory effect on autophagy30. Whether and how such mutations perturb the cytoplasmic regulation of cell cycle checkpoints remains elusive. However, it appears that frequent p53 mutations can contribute to oncogenesis through the concerted subversion of both the nuclear and cytoplasmic programs of tumor suppression (Fig. 3). Similarly, the inactivation of the oncosuppressor Arf (which, through the activation of MDM2 leads to p53 depletion) combines transcriptional effects (due to the absence of p53) with cytoplasmic ones. Indeed, distinct splice variants of Arf can induce MOMP (though through a mechanism distinct from that of p53, involving an interaction with a specific mitochondrial receptor, p32, or an interaction with Bcl-XL) and stimulate autophagy36-38. Therefore, the net result of Arf inactivation may also be a combined subversion of apoptosis and autophagy, both at the nuclear and at the extra-nuclear levels. ARC, an apoptosis-inhibitory protein that is overexpressed in numerous cancers, is present is the nucleus, where it inhibits p53 tetramerization and stimulates its export, as well as in the cytoplasm, where it neutralizes Bax to inhibit cell death39. It remains an open question through which mechanisms many other oncogenic perturbations, such as constitutive activation of the insulin receptor pathway, may affect (p53-dependent?) tumor suppression in the cytoplasm. Understanding the extranuclear activities of p53 will likewise furnish new opportunities to pharmacologically modulate the p53 system.
Finally, p53 plays a prominent – and controversial – role in the regulation of aging and longevity9. In the nematode Caenorhabditis elegans, knockout of the p53 ortholog, cep-1, fails to cause oncogenesis, yet significantly increases both median and maximum lifespan. This gain of longevity is lost when autophagy is inhibited40. We anticipate that the investigation of whether and how p53 can participate in a longevity-increasing pathway that links apoptosis, caloric restriction, activation of sirtuins, and regulation of autophagy will yield crucial insights into the intricate relationship between tumor suppression and aging that dictates our inexorable, yet variable fate.
Acknowledgments
The authors’ own work is supported by NIH and the American Lebanese and Syrian Associated Charities (to D.R.G.), Ligue contre le Cancer, INCa, Cancéropole, ANR, ANRS and the Active p53 and Apo-Sys EU networks (to G.K.).
References
- 1.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–12. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 2.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–83. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 3.Kakudo Y, Shibata H, Otsuka K, Kato S, Ishioka C. Lack of correlation between p53-dependent transcriptional activity and the ability to induce apoptosis among 179 mutant p53s. Cancer Res. 2005;65:2108–14. doi: 10.1158/0008-5472.CAN-04-2935. [DOI] [PubMed] [Google Scholar]
- 4.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–3. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 5.Chipuk JE, Maurer U, Green DR, Schuler M. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell. 2003;4:371–81. doi: 10.1016/s1535-6108(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 6.Tang X, et al. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest. 2007;117:3753–64. doi: 10.1172/JCI32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson TM, Meade K, Pathak N, Marques MR, Attardi LD. Knockin mice expressing a chimeric p53 protein reveal mechanistic differences in how p53 triggers apoptosis and senescence. Proc Natl Acad Sci U S A. 2008;105:1215–20. doi: 10.1073/pnas.0706764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–12. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 9.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–74. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 10.Endo H, Saito A, Chan PH. Mitochondrial translocation of p53 underlies the selective death of hippocampal CA1 neurons after global cerebral ischaemia. Biochem Soc Trans. 2006;34:1283–6. doi: 10.1042/BST0341283. [DOI] [PubMed] [Google Scholar]
- 11.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 12.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–43. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 13.Letai AG. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–32. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- 14.Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6:443–50. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- 15.Mihara M, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–90. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 16.Chipuk JE, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–4. doi: 10.1126/science.1092734.This work characterizes protein-protein interactions between p53 and pro-apoptotic Bcl-2 proteins as the mechanism through which p53 can trigger the formation of supramolecular, protein-permeable conduits in the outer mitochondrial membrane.
- 17.Pietsch EC, et al. Oligomerization of BAK by p53 utilizes conserved residues of the p53 DNA binding domain. J Biol Chem. 2008;283:21294–304. doi: 10.1074/jbc.M710539200.This study uses biochemical approaches to map the interaction of p53 with Bak to a region corresponding to the analogous Bim-Bax interaction site described in Gavathiotis Nature. 2008;455:1076–1081. doi: 10.1038/nature07396.
- 18.Xu H, Tai J, Ye H, Kang CB, Yoon HS. The N-terminal domain of tumor suppressor p53 is involved in the molecular interaction with the anti-apoptotic protein Bcl-Xl. Biochem Biophys Res Commun. 2006;341:938–44. doi: 10.1016/j.bbrc.2005.12.227. [DOI] [PubMed] [Google Scholar]
- 19.Sot B, Freund SM, Fersht AR. Comparative biophysical characterization of p53 with the pro-apoptotic BAK and the anti-apoptotic BCL-xL. J Biol Chem. 2007;282:29193–200. doi: 10.1074/jbc.M705544200. [DOI] [PubMed] [Google Scholar]
- 20.Lin B, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–40. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 21.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–5. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 22.Strom E, et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–9. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 23.Wolff S, Erster S, Palacios G, Moll UM. p53’s mitochondrial translocation and MOMP action is independent of Puma and Bax and severely disrupts mitochondrial membrane integrity. Cell Res. 2008;18:733–44. doi: 10.1038/cr.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–7. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 27.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–60. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730.This article reports the discovery that cytoplasmic p53 can inhibit autophagy in several species (humans, mice and nematodes), thus revealing a novel crosstalk between autophagy and apoptosis.
- 29.Colell A, et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–97. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 30.Morselli E, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008 doi: 10.4161/cc.7.19.6751. in press. [DOI] [PubMed] [Google Scholar]
- 31.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. Embo J. 2007;26:923–34. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You H, Yamamoto K, Mak TW. Regulation of transactivation-independent proapoptotic activity of p53 by FOXO3a. Proc Natl Acad Sci U S A. 2006;103:9051–6. doi: 10.1073/pnas.0600889103.This study establishes a role for the transcription factor FOXO3a in accumulation and function of cytosolic p53.
- 33.Lain S, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454–63. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao W, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han MK, et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–51. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reef S, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–75. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–53. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy M. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem. 2008 doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foo RS, et al. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci U S A. 2007;104:20826–31. doi: 10.1073/pnas.0710017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4 doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]