Genetic control over the resting brain (original) (raw)
Abstract
The default-mode network, a coherent resting-state brain network, is thought to characterize basal neural activity. Aberrant default-mode connectivity has been reported in a host of neurological and psychiatric illnesses and in persons at genetic risk for such illnesses. Whereas the neurophysiologic mechanisms that regulate default-mode connectivity are unclear, there is growing evidence that genetic factors play a role. In this report, we estimate the importance of genetic effects on the default-mode network by examining covariation patterns in functional connectivity among 333 individuals from 29 randomly selected extended pedigrees. Heritability for default-mode functional connectivity was 0.424 ± 0.17 (P = 0.0046). Although neuroanatomic variation in this network was also heritable, the genetic factors that influence default-mode functional connectivity and gray-matter density seem to be distinct, suggesting that unique genes influence the structure and function of the network. In contrast, significant genetic correlations between regions within the network provide evidence that the same genetic factors contribute to variation in functional connectivity throughout the default mode. Specifically, the left parahippocampal region was genetically correlated with all other network regions. In addition, the posterior cingulate/precuneus region, medial prefrontal cortex, and right cerebellum seem to form a subnetwork. Default-mode functional connectivity is influenced by genetic factors that cannot be attributed to anatomic variation or a single region within the network. By establishing the heritability of default-mode functional connectivity, this experiment provides the obligatory evidence required before these measures can be considered as endophenotypes for psychiatric or neurological illnesses or to identify genes influencing intrinsic brain function.
Keywords: default mode, functional MRI, heritability, resting state networks
When not engaged in goal-directed behavior, spontaneous fluctuations in the human brain give rise to coherent and structured functional networks (1–3). Most of these resting-state networks (RSNs), as identified through connectivity analyses with functional MRI or PET, are identical to those engaged during cognitive or behavioral challenges (4). However, one network, termed the default mode (5), is thought to characterize basal neural activity, and it putatively supports self-referential or nondirected cognitive processing (6). Deactivation in the default-mode network is diminished during effortful cognitive tasks (7, 8), and it increases when one’s mind wanders (9). This connectivity, or coactivation, pattern may be intrinsic to the primate brain (4, 10), because it is present in sleeping infants (11) and anesthetized nonhuman primates (12). Aberrant default-mode connectivity has been reported in individuals with a host of neurological and psychiatric illnesses, suggesting that this intrinsic network is sensitive to pathophysiologic alterations in brain function and structure (13). Although the exact neurophysiologic mechanisms that regulate default-mode connectivity are unclear and likely differ between illnesses, there is growing evidence that genetic factors play a role. Individuals at genetic risk for Alzheimer’s disease (14) and schizophrenia (15) have abnormal default-mode connectivity. There is recent evidence that variation in the APOE gene, a risk gene for Alzheimer’s disease, influences resting state connectivity within the medial temporal cortex of healthy, young individuals (16). Together, these data provide strong circumstantial evidence that the default-mode network is influenced by genetic factors. However, the degree of genetic control over default-mode functional connectivity is unknown. Establishing the heritability of default-mode functional connectivity authorizes the use of resting-state networks as endophenotypes or intermediate phenotypes in the search for the genetic roots of illnesses that have been associated with altered default-mode connectivity (17), such as Alzheimer’s (18), autism (19), Attention Deficit Hyperactivity Disorder (ADHD) (20), and schizophrenia (21). Furthermore, identification of the genes that influence the intrinsic functional architecture of the human brain would represent a significant advance for basic neuroscience, independent of the ramifications for brain disorders.
The default-mode network includes aspects of the posterior cingulate gyrus (PCC), retrosplenial cortex and precuneus, the medial prefrontal cortex (MPF), medial and lateral parietal regions, inferior/medial temporal gyri, and cerebellar areas (7, 22). In addition to overwhelming evidence for functional connectivity between these regions (23, 24), there is increasing support for anatomic connectivity between these regions based on diffusion tensor-imaging tractography in humans (25, 26) and architectonic mapping in nonhuman primates (23). Although these findings provide anatomic plausibility for the default-mode network, they complicate inferences about potential genetic control over functional connectivity within the network. Because up to 70% of normal variation in gray-matter volume is explained by genetic factors (27) and given that white-matter tract microstructure is heritable (28, 29), it is possible that any observed genetic influences over functional connectivity within the default mode could reflect genetic control over anatomic variation rather than functional connectivity. Hence, to establish genetic influences on functional connectivity within the default mode, the genetic contributions to functional variation must be dissociated from genetic control over (gross and topographic) neuroanatomy.
Although there is considerable evidence that the brain areas within the default-mode network support basal activity or spontaneous cognitive processing as a singular system (23), the different regions that make up this network show some level of relative specialization or affinity for specific cognitive demands (30, 31). For example, the MPF may be disproportionately engaged during self-referential thought compared with other regions within the network (6). In contrast, medial temporal regions and the retrosplenial cortex may be somewhat more closely associated with autobiographical memory (32). Indeed, activity in these regions seems to be highly correlated during rest and during declarative memory tasks (33). Consistent with observations of functional coupling between pairs of regions within the default-mode network, Greicius et al. (25) recently reported robust structural connections between the PCC and MPF regions and between retrosplenial cortex and medial temporal areas. To the extent that regions within the default-mode network show relative functional specialization, it is possible that unique genetic factors influence these different regions or subnetworks. Formally testing for pleiotropic (i.e., shared genetic) effects between regions within the default-mode network will provide evidence that either regions with relative specialization are influenced by unique genetic factors or that shared genetic factors control default-mode connectivity.
The detection and estimation of the relative importance of genetic effects in humans requires information on phenotypic covariation among relatives. In this report, we employ an extended pedigree design to obtain heritability estimates for functional connectivity within the default-mode, provide evidence that the genetic factors that influence default-mode functional connectivity differ from those that control anatomic variability in the network, and show that regions within this network have both shared and unique genetic influences.
Results
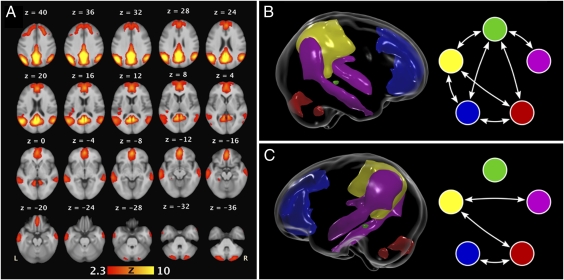
The default-mode network, as defined by the group-ICA map on 333 individuals, included eight anatomically distinct regions (Fig. 1_A_ and Table 1). The largest region included left and right precuneus and PCC. The next largest region included midline portions of the anterior cingulate, the middle frontal, and the superior frontal gyri, and it is broadly defined as the MPF. A left temporal–parietal region extended from the middle temporal gyrus through the superior temporal and supramarginal gyri to the angular gyrus and the inferior parietal lobule. A similar region within the right hemisphere was identified. Left and right cerebellar regions, constrained to the inferior semilunar lobule, were also identified. A portion of the cerebellar tonsil was included as well. The smallest region included the left parahippocampal gyrus, just anterior to the fusiform gyrus.
Fig. 1.
(A) Group-ICA map of the default-mode network derived from resting state scans of 333 individuals from large extended pedigrees. (B) Significant genetic correlations for functional connectivity between heritable regions in the default-mode network. The left parahippocampal gyrus (green) was genetically correlated with the posterior cingulate/precuneus (yellow), medial prefrontal (blue), right cerebellar (red), and right temporal-parietal (pink) regions. In addition, the posterior cingulate/precuneus, medial prefrontal, and right cerebellar regions form a circuit influenced by the same genetic factors. (C) Significant environmental correlations between these same regions.
Table 1.
Anatomy of the default-mode network
| Region | Volume* | Max Z | x† | y† | z† | Brodmann area |
|---|---|---|---|---|---|---|
| Posterior cingulate/precuneus | 82160 | 16.70 | 0 | −64 | 34 | 7/23/30/31 |
| Medial prefrontal cortex | 74888 | 8.11 | 0 | 52 | −8 | 32/9/10 |
| Left temporal–parietal region | 53672 | 12.70 | −48 | −64 | 32 | 39/22/21/19 |
| Right temporal–parietal region | 31240 | 9.88 | 52 | −60 | 30 | 39/22/21/19 |
| Left cerebellum | 5592 | 4.23 | −26 | −80 | −36 | NA |
| Right cerebellum | 5312 | 4.44 | 26 | −80 | −36 | NA |
| Cerebellar tonsil | 2496 | 4.13 | 6 | −52 | −46 | NA |
| Left parahippocampal gyrus | 456 | 2.78 | −26 | −38 | −16 | 36 |
Functional connectivity within the default-mode network as a whole was significantly heritable (h2 = 0.424 ± 0.17; P = 0.0046), suggesting that a significant proportion of the between subject variance in functional connectivity is explained by genetic factors. Furthermore, functional connectivity within six of eight individual regions in this network was significantly heritable (Table 2). Thus, genetic factors play an important role in functional connectivity.
Table 2.
Heritability estimates for regions within the default mode
| Functional connectivity | Gray-matter density | |||
|---|---|---|---|---|
| Region* | Heritability† | P value‡ | Heritability† | P value‡ |
| Posterior cingulate/precuneus | 0.423 (0.17) | 4.4 × 10−3 | 0.623 (0.16) | 6.8 × 10−5 |
| Medial prefrontal cortex | 0.376 (0.15) | 3.8 × 10−3 | 0.631 (0.15) | 5.3 × 10−6 |
| Left temporal–parietal region | 0.331 (0.19) | 3.1 × 10−2 | 0.387 (0.21) | 3.1 × 10−2 |
| Right temporal–parietal region | 0.420 (0.16) | 3.5 × 10−3 | 0.365 (0.21) | 3.4 × 10−2 |
| Left cerebellum | 0.104 (0.13) | 2.0 × 10−1 | 0.493 (0.15) | 4.9 × 10−4 |
| Right cerebellum | 0.304 (0.16) | 1.6 × 10−2 | 0.596 (0.14) | 1.6 × 10−5 |
| Cerebellar tonsil | 0.219 (0.19) | 1.1 × 10−1 | 0.271 (0.16) | 3.2 × 10−2 |
| Left parahippocampal gyrus | 0.273 (0.14) | 1.7 × 10−2 | 0.420 (0.18) | 7.5 × 10−3 |
To determine whether or not the genetic factors involved in connectivity were independent of those involved in brain anatomy, we performed bivariate genetic analysis of gray-matter density and functional connectivity. The observed heritability estimate for gray-matter density within the default mode network was h2 = 0.327 ± 0.17 (P = 0.020). However, genetic (ρg = 0.077 ± 0.38; P = 0.836) and environmental (ρe = −0.059 ± 0.21; P = 0.781) correlations between measures of functional connectivity and gray-matter density within the default-mode network were small and insignificant, suggesting that different genetic mechanisms are responsible for anatomic and functional variation within the network. Thus, our finding suggests that the genes involved in connectivity are different from those involved in brain anatomy.
To determine if the same genetic factors influence functional connectivity across nodes of the default-mode network, bivariate quantitative genetic analyses were performed between significantly heritable regions (Table 3 and Fig. 1_B_). The left parahippocampal gyrus showed significant genetic correlation with each of the heritable regions in the default-mode network. In addition, strong positive genetic correlations were identified between the posterior cingulate/precuneus region, medial prefrontal cortex, and right cerebellum. Together, these data suggest that nodes within the default-mode network are largely influenced by the same genetic factors and that the posterior cingulate/precuneus, medial prefrontal, and cerebellum may form a subnetwork influenced by distinct genetic factors.
Table 3.
Within network genetic and environmental correlations for functional connectivity
| Trait 1 | Trait 2 | Geneticcorrelation no.* | P value | Environmental correlation | P value |
|---|---|---|---|---|---|
| Left parahippocampal | Right cerebellum | 1.00 | 4.0 × 10−3 | −0.015 | 9.2 × 10−1 |
| Left parahippocampal | Right temporal–parietal | 0.953 | 5.9 × 10−3 | −0.014 | 9.3 × 10−1 |
| Left parahippocampal | Medial prefrontal cortex | 1.00 | 7.7 × 10−4 | 0.030 | 8.5 × 10−1 |
| Left parahippocampal | Posterior cingulate/precuneus | 1.00 | 2.0 × 10−2 | 0.103 | 5.0 × 10−1 |
| Right cerebellum | Right temporal–parietal | 0.497 | 1.5 × 10−1 | 0.272 | 1.4 × 10−1 |
| Right cerebellum | Medial prefrontal cortex | 0.89 | 1.6 × 10−2 | 0.386 | 2.3 × 10−2 |
| Right cerebellum | Posterior cingulate/precuneus | 1.00 | 2.5 × 10−2 | 0.305 | 9.1 × 10−2 |
| Right temporal-parietal | Medial prefrontal cortex | 0.413 | 1.7 × 10−1 | 0.399 | 3.3 × 10−2 |
| Right temporal-parietal | Posterior cingulate/precuneus | 0.309 | 4.3 × 10−1 | 0.672 | 1.2 × 10−3 |
| Medial prefrontal cortex | Posterior cingulate/precuneus | 1.00 | 4.2 × 10−3 | 0.403 | 3.7 × 10−2 |
The pattern of significant environmental correlations differed dramatically from those of the genetic correlations (Table 3 and Fig. 1_C_). Environmental correlations typically result from unmeasured aspects of the environment or correlated measurement errors. The right temporal–parietal region was significantly correlated with the posterior cingulate/precuneus and medial prefrontal cortex. Neither of these regions showed significant genetic correlations. In addition, the right cerebellum and medial prefrontal cortex had a significant environmental correlation.
Discussion
This experiment provides direct evidence that connectivity within the default-mode network, an intrinsic brain network, is influenced by genetic factors. We found that ∼40% of the between subject variance in functional connectivity within the default-mode network was under genetic control. Although neuroanatomic variation in this network was also heritable, the genetic factors that influenced default-mode functional connectivity and gray-matter density seemed to be distinct, suggesting that unique genes influence the structure and function of the network. Significant genetic correlations between all regions within the network provide evidence that the same genetic factors contributed to variation in functional connectivity throughout the default mode. In addition, strong positive genetic correlations between the posterior cingulate/precuneus region, medial prefrontal cortex, and right cerebellum suggest that these regions form a subnetwork that is influenced by genetic factors common to these regions but potentially unique from those that control the entire default-mode network. Together, these findings strongly suggest that default-mode connectivity is influenced by genetic factors that cannot be attributed to anatomic variation or a single region within the network. Establishing the heritability of default-mode functional connectivity provides critical information necessary before these measures can be appropriately used in molecular genetic studies designed to identify or functionally characterize genes influencing basal brain function. Additionally, showing significant heritability is necessary before indices of default-mode functional connectivity can be considered an intermediate phenotype or endophenotype for neurological or mental illnesses (17).
Given that blood-oxygen-level dependent (BOLD) fMRI signal indexes local variations in deoxyhemoglobin concentration that are determined by a combination of blood flow, blood volume, and oxygen metabolism (34, 35), the exact neuropsychological mechanisms responsible for the observed variation in signal that gives rise to default-mode connectivity are unknown. Furthermore, the relationship between these measures and the molecular mechanisms that govern neuronal firing patterns that are closer to gene action are complex and not fully elucidated (36). The identification of one or more genes that influence default-mode connectivity should provide a causal point in the biological chain that governs variation in BOLD signal in intrinsic brain networks across individuals. The discovery of such genes could dramatically improve our understanding for how molecular processes influence cerebral blood flow, blood volume, or oxygen metabolism. This, in turn, should provide important leads for how these processes are disrupted in illnesses associated with aberrant default-mode connectivity. The observed heritability for default-mode connectivity is the initial step in this discovery process.
Both genetic and environmental influences are critical for determining individual differences in default-mode functional connectivity. Indeed, the interaction between genetic and environmental factors that govern the brain’s intrinsic networks is likely complex, may change over the life span, and/or may be particularly prone to specific environmental stressors that predispose illness (37). The extended pedigree design applied here is specifically less likely to confound genetic factors with shared environmental factors (e.g., household-type effects) than designs relying on smaller familial configurations (36). Our approach optimally uses all available biological information including that from individuals across a broad cross-sectional age range. Although we included age as a covariate in all analyses, which corrects for a potential main effect of age, it does not control differences in genetic variance with age (potential gene times age interactions). Although it is possible to formally test for a gene times age interaction in cross-sectional familial data using substantially more complex quantitative genetic models (with a number of critical assumptions), the power to detect such interactions is substantially lower than would be observed for a longitudinal design. Therefore, we have chosen not to test for such changes in genetic variance with age in default-mode connectivity. Regardless, even in the presence of significant gene times age interaction, our approach is conservative, because the observed heritability of default-mode connectivity will simply underestimate the importance of genetic effects.
Extended pedigree designs allow for the efficient examination of pleiotropy or genetic correlation because of the effects of genes acting jointly on multiple phenotypes. Genetic correlations between regions within the default mode revealed that the parahippocampal region is linked to all other heritable regions within the network, suggesting that the same genetic factors influence default-mode functional connectivity throughout the network or that the genetic factors that control parahippocampal connectivity also influence the other nodes of the network. In addition, the posterior cingulate/precuneus, medial prefrontal cortex, and cerebellum seem to form a subnetwork or circuit influenced by genetic factors that only partially overlap with those that control the complete network. In contrast, the only significant genetic correlation for anatomic variability between default-mode regions was for the left and right temporal–parietal areas (ρg = 0.521; P = 0.045). This elaborate pattern of results suggests that partially overlapping, but not completely common, genetic factors control functional connectivity across the default-mode network. These finding are consistent with the notion that a set of genes influence default-mode function and that these genes are not common to all observed regions. This raises the possibility that although aberrant default-mode connectivity is observed in multiple illnesses, genes that influence default-mode connectivity and contribute to risk for neuropathology may differ among illnesses. Although this specific biological hypothesis can only be tested by direct interrogation of the genome, the current finding supports such a hypothesis as well as examination of resting-state data across different neurological and psychiatric illnesses. Future studies mapping and identifying the actual quantitative trait loci will likely provide insight into the genes that influence default-mode functional connectivity.
Caution should be used when interpreting the observed environmental correlations in default-mode connectivity, given that specific environmental factors (e.g., exposures to environmental toxins or experimental drug usage) were not explicitly modeled in this study. However, contrasting the pattern of environmental and genetic correlations can provide insight into the nature of these bivariate correlations. Whereas the parahippocampal region is genetically correlated with all other default-mode regions, none of the environmental correlations for this region are significant, strengthening the interpretation that genetic, rather than environmental, factors influence connectivity throughout the network. Indeed, the strongest environmental correlations are between regions with minimal evidence for pleiotropy, providing evidence that the observed genetic correlations were not biased by the intrinsic phenotypic correlation present in regions delineated with independent component analysis.
The current study, like other behavioral genetic studies conducted on twins or families, does not provide information concerning the identity of the causal genes that influence default-mode connectivity. However, it does provide substantial evidence that there are genes involved in this process and that the identification of these genes should be ultimately discoverable. Because the default-mode reflects a “baseline” system that accounts for a considerable proportion of the brain’s energy consumption (36), it is plausible that the genes that influence default-mode connectivity regulate brain metabolism, cerebral blood flow, or other aspects of basic neuronal activity (17). Identification of these genes will provide an important vantage point for understanding the brain’s intrinsic architecture and the influence that those systems have on a host of neurological and psychiatric illnesses. The data presented here represents an initial step in this discovery process (27), because establishing the heritability of default-mode connectivity is an obligate prerequisite before the launching of a costly gene-localization effort using genome-wide linkage/association strategies. Indeed, based on these results, we are increasing our sample of pedigreed subjects with resting-state data and performing high-density genotyping to perform genome-wide combined linkage/association studies of default-mode functional connectivity. The identification of even a single gene (among many) that influences the brain’s intrinsic networks would provide a causal anchor point for understanding genetic contributions to systems-level neuroscience.
To the extent that the default-mode represents one of several intrinsic neural networks critical for normal brain functioning (4, 10), our results suggest that functional imaging-based measures of these networks are sensitive to genetic mediation. These data extend other recent reports that provide heritability estimates for task-based functional MRI (38–40), because default-mode connectivity is thought to index more fundamental aspects of brain function than activation evoked by cognitive demands (2, 3). Given that default-mode connectivity is readily measureable in nonhuman primates (12), the current results also facilitate the use of resting-state imaging in genetic investigations with animal models. Evidence that default-mode connectivity is controlled by genetic influences establishes functional neuroimaging as a viable tool for investigating the genetic effects on the brain in vivo.
Methods and Materials
Participants.
Twin designs have dominated most of the previous work in brain-imaging genetics (41–43), but studies focusing on larger extended-pedigree configurations, although more difficult to obtain, have multiple benefits including (i) increased power to detect heritable effects, (ii) less confounding of genetic effects with shared environmental effects because of the inclusion of multiple households within pedigrees, and (iii) greater potential for localization and identification of the underlying causal quantitative trait loci (44). In this study, we examined 333 Mexican-American individuals from 29 large extended-pedigrees (average family size = 9 people; range = 5–32) who have participated in the Genetics of Brain Structure and Function study to date (Table S1). Participants were 63% female, ranged in age from 26 to 85 years (mean ± SD = 48.38 ± 12.9 years), had 11.37 ± 3.2 years of education, and were 95.5% right-handed. Individuals in this cohort have actively participated in genetics research for over 15 years and initially, were randomly selected from the community with the constraints that they are of Mexican-American ancestry, part of a large family, and live within the San Antonio, TX region. In the current study, individuals were excluded for MRI contraindications, history of neurological illnesses, or stroke or other major neurological event. All participants provided written informed consent on forms approved by the institutional review board at the University of Texas Health Science Center San Antonio (UTHSCSA).
Image Acquisition.
Scanning was performed at the Research Imaging Center, UTHSCSA, on a 3T Siemens Trio scanner with an eight-channel head coil. High-resolution (isotropic 800 μm) 3D TurboFlash T1-weighted anatomic images were acquired for each subject using a retrospective motion-corrected protocol (45) with the following parameters: echo time (TE)/ repetition time (TR) / time for inversion (TI) = 3.04/2,100/785 ms and flip angle = 13 °. Whole-brain, resting-state functional imaging was performed using a gradient-echo echoplanar imaging (EPI) sequence sensitive to the BOLD effect (TE/TR = 30/3,000 ms; flip angle = 90°; isotropic 1.72 mm2). The resting-state protocol included 43 slices acquired parallel to the Anterior Commissure and Posterior Commissure plane, and they were acquired over 7.5 min. During the resting-state scan, subjects were instructed to lie in dimmed light with their eyes open and try not to fall asleep.
Functional Image Analysis.
Image analysis was performed with FMRIB’s Software Library (FSL) tools (www.fmrib.ox.ac.uk/fsl) (46). Preprocessing for resting-state data included motion correction, brain extraction, spatial smoothing (5 mm FWHM Gaussian kernel), and high-pass temporal filtering (120 seconds). Each subject’s fMRI volumes were nonlinearly aligned to a common space (in Anatomical Image Analysis) using his or her high-resolution neuroanatomic scan and temporally concatenated to create a single 4D data set (333 subjects × 150 scans per subjects = 49,950 images). The concatenated data set was then decomposed into 17 components that represent large-scale patterns of functional connectivity using Independent Component Analysis as implemented in Melodic (47). Model order was estimated using the Laplace approximation to the Bayesian evidence for a probabilistic principal-component model.
A dual-regression approach (48) was used to delineate subject-specific temporal dynamics and associated subject-specific spatial maps. The dual-regression approach involves first, using the full set of group-ICA spatial maps (n = 17) in a linear model fit (spatial regression) against the separate fMRI data sets, resulting in matrices describing temporal dynamics for each component and subject, and second, using these time-course matrices in a linear model fit (temporal regression) against the associated fMRI data set to estimate subject-specific spatial maps. This procedure ultimately results in a separate estimate for each original group-ICA map and each subject.
The group-ICA default-mode network was identified using spatial correlation with a previously published default-mode map (3). The corresponding subject-specific default-mode maps generated by the dual-regression procedure were used to calculate indices for subsequent quantitative genetic analysis. Specifically, the individual (3D) subject-specific maps of the default-mode network were collected into a single 4D data set, and the first principal Eigenvector representing the subject’s connectivity was calculated within a study-specific default-mode mask (Table 1). The mask was generated with a Gaussian/gamma-mixture model fit to the intensity histogram of the group-ICA default-mode map (47), controlling the false-discovery rate (FDR) at 5% against the empirical null distribution estimated as part of the mixture-model fit. This default-mode network mask was thresholded to include only positive clusters larger than 350 mm3 and binarized. This mask included eight anatomically distinct regions (Table 1) that were separated into eight unique masks. To support between-region bivariate genetic analyses (in Anatomical Image Analysis), the first principal Eigenvector representing the subject’s connectivity with each of the eight regions in the default-mode network was calculated separately.
Anatomical Image Analysis.
To compare genetic effects between anatomic and functional aspects of the default-mode network, high-resolution structural data were analyzed with a voxel-based morphometry style analysis (49, 50). Images were brain-extracted and aligned to a common space (MNI152) using affine and nonlinear registration methods. The resulting images were averaged to create a study-specific template, and subject native-space images were then nonlinearly reregistered to this template. Registered images were segmented into tissue type, and the resulting partial-volume images underwent Jacobian modulation. Gray-matter images were then constrained to the functionally defined default-mode network (group-ICA map; in Functional Image Analysis), and the first principal Eigenvector representing between-subject gray-matter density was generated.
Quantitative Genetic Analyses.
All quantitative genetic analyses were conducted using the computer package Sequential Oligogenic Linkage Analysis Routines (SOLAR) (51). SOLAR uses maximum likelihood variance-decomposition methods to determine the relative importance of familial and environmental influences on a phenotype by modeling the covariance among family members as a function of genetic proximity (kinship). This approach can handle pedigrees of arbitrary size and complexity and thus, is optimally efficient with regard to extracting maximal genetic information. To ensure that neuroimaging traits conform to the assumptions of normality, an inverse normal transformation was applied.
Heritability (h2) represents the portion of the phenotypic variance accounted for by the total additive genetic variance (h2 = σ2g/σ2p). Phenotypes exhibiting larger covariances between genetically more similar individuals than between genetically less similar individuals have higher heritability. Within SOLAR, this is assessed by contrasting the observed covariance matrices for a neuroimaging measure with the structure of the covariance matrix predicted by kinship. Heritability analyses were conducted with simultaneous estimation for the effects of potential covariates. For this study, we included demographic covariates including age, sex, age × sex interaction, age2, age2 × sex interaction, and diagnostic status for two extremely common disorders: hypertension and diabetes. Heritability estimates were corrected for multiple comparisons at 5% FDR.
To determine if functional connectivity and anatomic variation in the default-mode network were influenced by the same genetic factors (e.g., pleiotropy), genetic correlation analyses were conducted. More formally, bivariate polygenic analyses were performed to estimate genetic (ρg) and environmental (ρe) correlations between functional and structural measures of the default-mode network and between nodes within the network with the following formula: ρp = ρg√(_h2_1_h2_2) + ρe√[(1 − _h2_1)(1 − _h2_2)], where _h2_1 and _h2_2 are the heritabilities of traits 1 and 2, respectively. The significance of these correlations was tested by comparing the log likelihood for two restricted models (with either ρg or ρe constrained to equal 0) against the log likelihood for the model in which these parameters were estimated. A significant genetic correlation (using a 5% FDR) is evidence for pleiotropy, suggesting that a gene or set of genes jointly influences both phenotypes (52). Similar analyses were conducted to test for pleiotropy between regions within the default-mode network.
Supplementary Material
Supporting Information
Acknowledgments
Financial support for this study was provided by National Institute of Mental Health (NIMH) Grants MH0708143 (to D.C.G.), MH078111 (to J.B.), MH083824 (to D.C.G.), and EB006395 (to P.K.). SOLAR is supported by NIMH Grant MH59490 (to J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 2.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 3.Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith S, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raichle ME, et al. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusnard DA, Akbudak E, Shulman GL, Raichle ME. Medial prefrontal cortex and self-referential mental activity: Relation to a default mode of brain function. Proc Natl Acad Sci USA. 2001;98:4259–4264. doi: 10.1073/pnas.071043098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greicius MD, Menon V. Default-mode activity during a passive sensory task: Uncoupled from deactivation but impacting activation. J Cogn Neurosci. 2004;16:1484–1492. doi: 10.1162/0898929042568532. [DOI] [PubMed] [Google Scholar]
- 9.Mason MF, et al. Wandering minds: The default network and stimulus-independent thought. Science. 2007;315:393–395. doi: 10.1126/science.1131295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fransson P, et al. Resting-state networks in the infant brain. Proc Natl Acad Sci USA. 2007;104:15531–15536. doi: 10.1073/pnas.0704380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent JL, et al. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- 13.Broyd SJ, et al. Default-mode brain dysfunction in mental disorders: A systematic review. Neurosci Biobehav Rev. 2009;33:279–296. doi: 10.1016/j.neubiorev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Persson J, et al. Altered deactivation in individuals with genetic risk for Alzheimer’s disease. Neuropsychologia. 2008;46:1679–1687. doi: 10.1016/j.neuropsychologia.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Whitfield-Gabrieli S, et al. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc Natl Acad Sci USA. 2009;106:1279–1284. doi: 10.1073/pnas.0809141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filippini N, et al. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci USA. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: Brain networks under genetic control. Hum Brain Mapp. 2009;30:1938–1946. doi: 10.1002/hbm.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy DP, Redcay E, Courchesne E. Failing to deactivate: Resting functional abnormalities in autism. Proc Natl Acad Sci USA. 2006;103:8275–8280. doi: 10.1073/pnas.0600674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castellanos FX, et al. Varieties of attention-deficit/hyperactivity disorder-related intra-individual variability. Biol Psychiatry. 2005;57:1416–1423. doi: 10.1016/j.biopsych.2004.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrity AG, et al. Aberrant “default mode” functional connectivity in schizophrenia. Am J Psychiatry. 2007;164:450–457. doi: 10.1176/ajp.2007.164.3.450. [DOI] [PubMed] [Google Scholar]
- 22.Damoiseaux JS, et al. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: Anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- 24.Fox MD, Snyder AZ, Zacks JM, Raichle ME. Coherent spontaneous activity accounts for trial-to-trial variability in human evoked brain responses. Nat Neurosci. 2006;9:23–25. doi: 10.1038/nn1616. [DOI] [PubMed] [Google Scholar]
- 25.Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skudlarski P, et al. Measuring brain connectivity: diffusion tensor imaging validates resting state temporal correlations. Neuroimage. 2008;43:554–561. doi: 10.1016/j.neuroimage.2008.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: Strategies for finding genes influencing brain structure and function. Hum Brain Mapp. 2007;28:488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kochunov P, et al. Genetics of DTI-derived parameters of cerebral white matter. A track-based heritability and linkage study in extended pedigree. Neuroimage. 2009;47(Suppl 1):S139. [Google Scholar]
- 29.Chiang MC, et al. Genetics of brain fiber architecture and intellectual performance. J Neurosci. 2009;29:2212–2224. doi: 10.1523/JNEUROSCI.4184-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uddin LQ, Kelly AM, Biswal BB, Xavier Castellanos F, Milham MP. Functional connectivity of default mode network components: Correlation, anticorrelation, and causality. Hum Brain Mapp. 2009;30:625–637. doi: 10.1002/hbm.20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laird A, et al. Investigating the functional heterogeneity of the default mode network using coordinate-based meta-analytic modeling. J Neurosci. 2009;29:14496–14505. doi: 10.1523/JNEUROSCI.4004-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svoboda E, McKinnon MC, Levine B. The functional neuroanatomy of autobiographical memory: A meta-analysis. Neuropsychologia. 2006;44:2189–2208. doi: 10.1016/j.neuropsychologia.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent JL, et al. Coherent spontaneous activity identifies a hippocampal-parietal memory network. J Neurophysiol. 2006;96:3517–3531. doi: 10.1152/jn.00048.2006. [DOI] [PubMed] [Google Scholar]
- 34.Buxton RB, Frank LR. A model for the coupling between cerebral blood flow and oxygen metabolism during neural stimulation. J Cereb Blood Flow Metab. 1997;17:64–72. doi: 10.1097/00004647-199701000-00009. [DOI] [PubMed] [Google Scholar]
- 35.Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: The balloon model. Magn Reson Med. 1998;39:855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- 36.Raichle ME, Mintun MA. Brain work and brain imaging. Annu Rev Neurosci. 2006;29:449–476. doi: 10.1146/annurev.neuro.29.051605.112819. [DOI] [PubMed] [Google Scholar]
- 37.Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: Joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 38.Koten JW, Jr, et al. Genetic contribution to variation in cognitive function: An FMRI study in twins. Science. 2009;323:1737–1740. doi: 10.1126/science.1167371. [DOI] [PubMed] [Google Scholar]
- 39.Matthews SC, et al. Heritability of anterior cingulate response to conflict: An fMRI study in female twins. Neuroimage. 2007;38:223–227. doi: 10.1016/j.neuroimage.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 40.Blokland GA, et al. Quantifying the heritability of task-related brain activation and performance during the N-back working memory task: A twin fMRI study. Biol Psychol. 2008;79:70–79. doi: 10.1016/j.biopsycho.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit DJ, Stam CJ, Posthuma D, Boomsma DI, de Geus EJ. Heritability of “small-world” networks in the brain: A graph theoretical analysis of resting-state EEG functional connectivity. Hum Brain Mapp. 2008;29:1368–1378. doi: 10.1002/hbm.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson PM, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- 43.Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: A review of brain imaging studies in twins. Hum Brain Mapp. 2007;28:464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1:1391–1397. doi: 10.1046/j.1538-7836.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- 45.Kochunov P, et al. Retrospective motion correction protocol for high-resolution anatomical MRI. Hum Brain Mapp. 2006;27:957–962. doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 47.Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- 48.Beckmann C, Mackay C, Filippini N, Smith S. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. Neuroimage. 2009;47(Suppl 1):S148. [Google Scholar]
- 49.Ashburner J, Friston K. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 50.Good CD, et al. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 51.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: Pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information