The Heat-Inducible Transcription Factor HsfA2 Enhances Anoxia Tolerance in Arabidopsis (original) (raw)
Abstract
Anoxia induces several heat shock proteins, and a mild heat pretreatment can acclimatize Arabidopsis (Arabidopsis thaliana) seedlings to subsequent anoxic treatment. In this study, we analyzed the response of Arabidopsis seedlings to anoxia, heat, and combined heat + anoxia stress. A significant overlap between the anoxic and the heat responses was observed by whole-genome microarray analysis. Among the transcription factors induced by both heat and anoxia, the heat shock factor A2 (HsfA2), known to be involved in Arabidopsis acclimation to heat and to other abiotic stresses, was strongly induced by anoxia. Heat-dependent acclimation to anoxia is lost in an HsfA2 knockout mutant (hsfa2) as well as in a double mutant for the constitutively expressed HsfA1a/HsfA1b (hsfA1a/1b), indicating that these three heat shock factors cooperate to confer anoxia tolerance. Arabidopsis seedlings that overexpress HsfA2 showed an increased expression of several known targets of this transcription factor and were markedly more tolerant to anoxia as well as to submergence. Anoxia failed to induce HsfA2 target proteins in wild-type seedlings, while overexpression of HsfA2 resulted in the production of HsfA2 targets under anoxia, correlating well with the low anoxia tolerance experiments. These results indicate that there is a considerable overlap between the molecular mechanisms of heat and anoxia tolerance and that HsfA2 is a player in these mechanisms.
Higher plants are aerobic organisms relying on oxygen for respiration (Bailey-Serres and Voesenek, 2008). Oxygen is required for oxygen-dependent enzymatic reactions and, most importantly, in mitochondrial respiration, where it acts as the terminal acceptor of electrons in oxidative phosphorylation that indirectly provides the plant with ATP (Perata and Alpi, 1993; Geigenberger, 2003; Voesenek et al., 2006). Shortage of oxygen (hypoxia) or a total absence of oxygen (anoxia) inhibits respiration and results in energy deficit.
Few plant species can tolerate flooding, the most common environmental cause of oxygen shortage in plants (Bailey-Serres and Voesenek, 2008). Plants species adopt different strategies to survive flooding. Some species display an “escape strategy” with rapid elongation of submerged organs in order to maintain contact with the atmosphere, allowing transport of oxygen to the submerged parts of the plant, while others tolerate low-oxygen conditions by modulating their metabolism and activating stress-tolerance pathways (Bailey-Serres and Voesenek, 2008). A breakthrough discovery in the field of plant adaptation to submergence is the identification of the rice (Oryza sativa) Submergence1A gene as a key element for rice flooding tolerance (Fukao et al., 2006; Xu et al., 2006; Perata and Voesenek, 2007). Little is known, however, about the mechanisms of anoxia tolerance in other plant species. Even flood-intolerant species such as Arabidopsis (Arabidopsis thaliana) trigger many genes generally associated with flooding tolerance (Klok et al., 2002; Branco-Price et al., 2005, 2008; Loreti et al., 2005; van Dongen et al., 2009), suggesting that the regulation of flooding tolerance and the differences between species are far more complex than previously thought.
To date, genetic evidence supports the important role of the fermentative pathway in anoxia tolerance in Arabidopsis. Arabidopsis plants devoid of alcohol dehydrogenase (ADH) or Suc synthases are less tolerant to anoxia, although an increased expression of ADH cannot confer enhanced anoxia tolerance (Ismond et al., 2003; Bieniawska et al., 2007). Overexpression of pyruvate decarboxylase, catalyzing a rate-limiting step in ethanol fermentation, has been shown to increase Arabidopsis survival (Ismond et al., 2003). Recently, a role for the transcription factor ANAC102 has been demonstrated for the Arabidopsis response to low oxygen levels during germination (Christianson et al., 2009).
Transcript profiling revealed that, besides the genes involved in the fermentative pathway, anoxia also induces several heat-related genes, including various heat shock proteins (HSPs) and heat shock factors (Hsfs; Loreti et al., 2005). HSP production under stress conditions is the most important cellular response, preventing protein aggregation, denaturation, misfolding, and degradation (Hartl, 1996; Hofmann and Feder, 1999; Swindell et al., 2007; Liberek et al., 2008), and their importance in all living organisms, including plants, has been assessed under several different stress conditions potentially threatening cellular homeostasis and leading to protein dysfunction (Vierling, 1991; Hofmann and Feder, 1999; Koh, 2002; Swindell et al., 2007; Timperio et al., 2008). The heat shock response is primarily regulated at the transcriptional level by Hsfs. In Arabidopsis, 21 Hsfs have been identified (Nover et al., 2001). Interestingly, it seems that in Arabidopsis, different from tomato (Solanum lycopersicum; Mishra et al., 2002), there is not a unique master regulator for the Hsf-mediated heat shock response. Different Hsfs have been identified as potential crucial element for Arabidopsis response not only to heat stress (Busch et al., 2005; Schramm et al., 2006, 2008; Charng et al., 2007) but also to oxidative stress (Li et al., 2005; Nishizawa et al., 2008), hydrogen peroxide (H2O2), high light (Nishizawa et al., 2006), and salt stress as well (Ogawa et al., 2007).
The possible overlap between heat and anoxia responses is supported by evidence showing that mild heat pretreatment, in addition to hypoxic treatment (Ellis et al., 1999), can confer greater tolerance to anoxia in Arabidopsis (Loreti et al., 2005). Although the combination of heat and anoxia is not relevant in an ecophysiological context, the induction of Hsfs and HSPs under anoxia and hypoxia in both Arabidopsis roots and shoots demonstrates that the heat stress signaling pathway is activated under conditions of oxygen shortage at normal temperatures (Banti et al., 2008). A correlation between HSP mRNA induction by anoxia and low-oxygen tolerance has also been demonstrated (Banti et al., 2008). This evidence led to the speculation that HSPs, and possibly other heat shock-related genes, might be important in the physiological response to anoxia (Loreti et al., 2005; Banti et al., 2008).
In this study, we investigated the role of heat-induced genes in Arabidopsis tolerance to anoxia. We used a transcriptomic approach to identify the common elements in both the heat and anoxia responses, highlighting the repression of several anaerobic genes in the heat-anoxia combined treatment and the strong induction of several heat-related genes and transcription factors. We identified HsfA2 as an important transcription factor that confers anoxia tolerance to Arabidopsis seedlings.
RESULTS
Transcriptomic Patterns of Anoxia and Heat Stress Are Very Similar
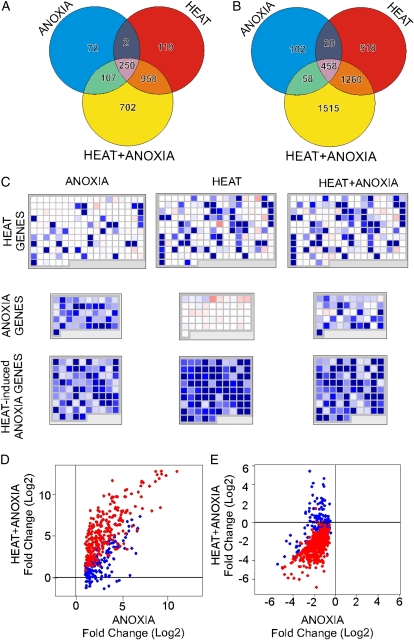
The induction of several heat-related genes under anoxia suggests that the anoxia response may partly overlap with the heat response (Loreti et al., 2005). In order to analyze the responses to heat, anoxia, and combined heat + anoxia, we performed a transcript profiling experiment using Arabidopsis seedlings. We identified 6,141 differentially regulated genes (genes showing a change of ≥ 2-fold or ≤−2-fold in a hybridization signal, namely the ratio between the intensity in the treated spot versus the control, and a false discovery rate [FDR] of ≤ 0.05; two biological replicates for each treatment; Fig. 1, A and B). Among these, 2,210 were up-regulated following three different treatments (Fig. 1A; Supplemental Table S1) and 3,931 were down-regulated (Fig. 1B; Supplemental Table S2). Four hundred thirty-one genes were induced by anoxia, heat up-regulated 1,329 genes, and the combined heat + anoxia treatment resulted in 2,017 up-regulated genes. We observed a considerable overlap between the transcriptomic patterns of the three treatments. The heat + anoxia treatment up-regulated 702 genes that were not regulated by either heat or anoxia. Several of these genes were found to be heat regulated under various experimental conditions (35% when comparing our results with the data set of Larkindale and Vierling [2008]), while others were also actually modulated by either heat or anoxia in our experiments, although the statistical significance of their change in expression did not pass the threshold used to select differentially regulated genes. Analysis of data using MapMan (Thimm et al., 2004; Fig. 1C) revealed that some heat-induced genes (“heat genes” as identified by MapMan) were induced by anoxia, but both heat and the combined heat + anoxia treatments were much more effective, inducing a larger number of genes (Fig. 1C). A mild heat pretreatment (e.g. 38°C, 90 min) can induce acclimation to anoxia in Arabidopsis (Loreti et al., 2005; Banti et al., 2008). Therefore, we were interested in evaluating the effects of heat pretreatment on the expression of the genes induced by anoxia. Anoxia induced two groups of genes: those exclusively induced by anoxia (“anoxia genes”) and genes induced by anoxia but also responsive to heat (“heat-induced anoxia genes”). While the expression of heat-induced anoxia genes was enhanced by the heat pretreatment, the induction of anoxia genes was reduced by the pretreatment (Fig. 1C). In order to expand this analysis to a larger number of genes, we identified a list of genes modulated by anoxia, and their expression was compared in plants only treated under anoxia and under heat + anoxia (Fig. 1, D and E). Statistical analysis showed that there was a significant general increase in the expression of the anaerobic gene cluster when seedlings had been previously acclimated by a heat pretreatment (Wilcoxon signed rank test, P < 2.2e-16). This result suggests that cross-acclimation may be due to an enhanced anaerobic response. However, the heat pretreatment inevitably induces genes related to the heat response. In fact, genes within the anaerobic cluster that were also induced by heat (red dots in Fig. 1D) showed enhanced mRNA levels in the combined treatment (Wilcoxon signed rank test, P < 2.2e-16). On the other hand, genes induced by anoxia but not by heat (“real” anoxia-dependent genes; blue dots in Fig. 1D) showed a significant reduction in gene expression (Wilcoxon signed rank test, P = 2.280e-11). A similar but opposite pattern was observed for down-regulated genes (Wilcoxon signed rank test, P = 4.088e-07; Fig. 1E). Genes repressed by both treatments (red dots) showed a stronger repression by the combined treatment (Wilcoxon signed rank test, P < 2.2e-16), whereas the opposite happened for genes exclusively repressed by anoxia (blue dots; Wilcoxon signed rank test, P < 2.2e-16).
Figure 1.
Transcriptome analysis of anoxia-, heat-, and heat + anoxia-treated Arabidopsis seedlings. A and B, Venn diagrams showing genes with differential expression (A, up-regulated; B, down-regulated). Genes were selected on the basis of their fold changes (≥ 2-fold or ≤−2-fold) and their FDR (≤ 0.05). C, MapMan representation of the effects of anoxia, heat, and heat + anoxia on genes annotated as “heat responsive” or “anoxia responsive.” D and E, Effects of the heat pretreatment on the expression levels of anoxic genes. Genes induced (D) or repressed (E) by anoxia were plotted on the basis of their fold change following the anoxic treatment (horizontal axis) and the heat + anoxia treatment (vertical axis). Red dots represent genes induced or repressed by either anoxia or heat, while blue dots represent genes that were not found to be heat responsive. Relative expression levels are shown as fold change values (log2).
Heat Pretreatment Dampens the Anaerobic Response
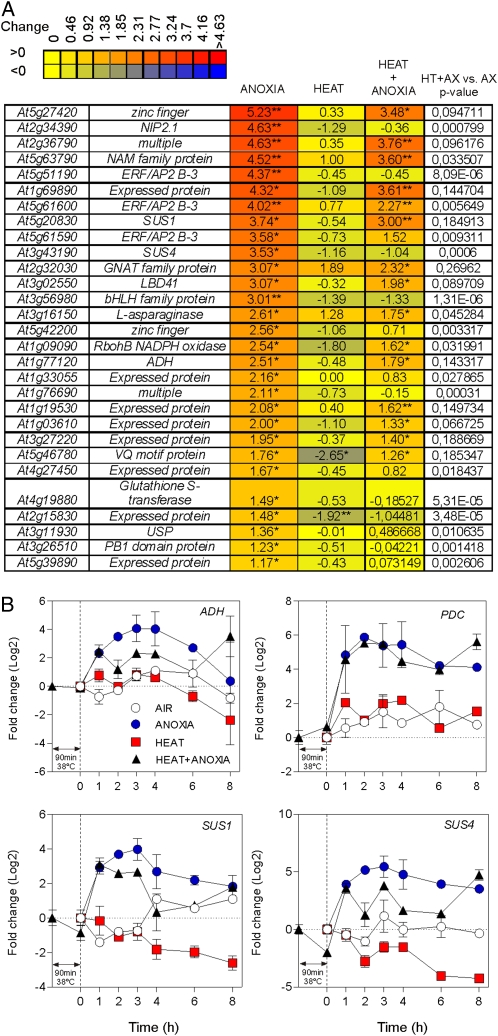
Heat pretreatment appears to dampen the induction of several anaerobic genes under anoxia (Fig. 2A). Examples of genes showing this pattern included SUCROSE SYNTHASE1 (SUS1; At5g20830), SUS4 (At3g43190), and ADH (At1g77120), as shown in Figure 2A, but not PYRUVATE DECARBOXYLASE1 (PDC1; At4g33070; Supplemental Table S1). Real-time reverse transcription-PCR (qPCR) analysis confirmed the dampening effect of the heat pretreatment on the expression of anaerobic genes (Fig. 2B). Interestingly we also found reduced ADH activity and anaerobic production of ethanol in the heat-pretreated samples (Supplemental Fig. S1). The increased expression of ADH, SUS1, SUS4, and PDC at the 8-h time point in acclimated plants (Fig. 2B, heat + anoxia), in comparison with nonacclimated plants, is probably due to the increased viability of the acclimated seedlings (Loreti et al., 2005; Banti et al., 2008).
Figure 2.
Effects of anoxia, heat, and combined heat + anoxia treatments on the expression of selected anaerobic genes. A, Heat map showing the expression of anaerobic genes showing a reduced expression in the combined heat + anoxia treatment. Expression data were visualized using Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgibin/ntools_heatmapper_plus.cgi). The fold change is shown as log2. Statistical significance for the comparison “treatment versus control (air, 23°C)” is reported: ** FDR ≤ 0.01, * FDR ≤ 0.05. The FDR P value for the comparison “combined heat + anoxia treatment versus anoxia” is shown in the table (HT + AX versus AX P value). B, Time courses of gene expression following anoxia, heat, and combined heat + anoxia treatments. The fold change (log2) was measured by qPCR using time = 0 (air) as a reference.
HsfA2 Is Strongly Up-Regulated by Both Heat and Anoxia
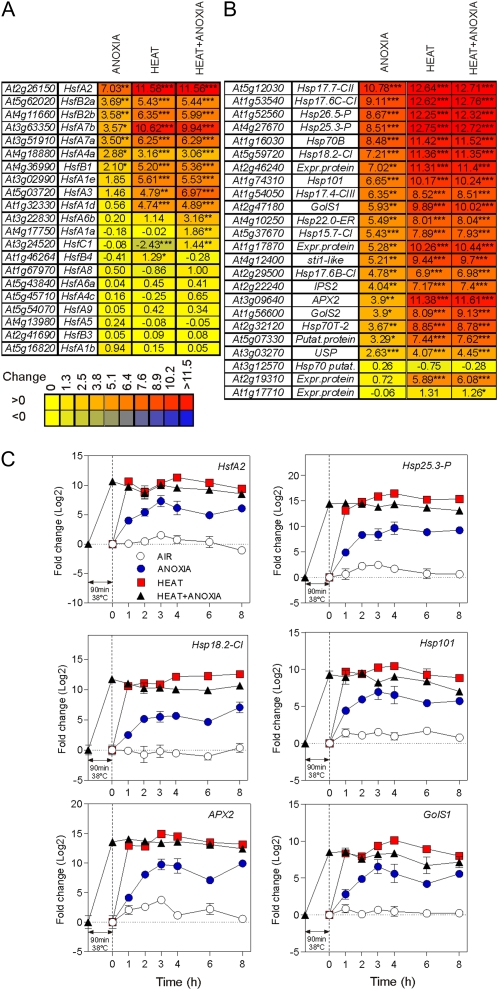
To understand which transcriptional activator is involved in the induction of heat-related genes under anoxia, we searched our microarray data sets for abiotic stress-related transcription factors (Supplemental Fig. S2). We found several transcription factors strongly induced by both heat and anoxia, including HsfA2, the most highly induced transcription factor among the seven Hsfs up-regulated by anoxia (Fig. 3A). Interestingly, several putative HsfA2 targets (Nishizawa et al., 2006; Schramm et al., 2006) are also induced by anoxia (Fig. 3B). Among them are several heat shock proteins and genes involved in oxidative stress responses, such as Ascorbate Peroxidase2 (APX2) and Galactinol Synthases (GolS; Nishizawa et al., 2008). An analysis of the expression of Hsp25.3-P, Hsp18.2-CI, APX2, GolS1, and Hsp101 over several hours of treatment under anoxia, heat, or heat + anoxia revealed that their expression pattern closely mirrored HsfA2 (Fig. 3C).
Figure 3.
Effects of anoxia, heat, and combined heat + anoxia treatments on the expression of _Hsf_-related genes. A, Heat map showing the effects of anoxia, heat, and combined heat + anoxia treatments on the expression of Hsf genes. Expression data were visualized using Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgibin/ntools_heatmapper_plus.cgi). The output of the software is shown. The fold change is shown as log2. Statistical significance is reported: *** FDR ≤ 0.001, ** FDR ≤ 0.01, * FDR ≤ 0.05. B, Heat map showing the effects of anoxia, heat, and combined heat + anoxia treatments on the expression of putative targets of HsfA2. Details are as in A. C, Time courses of gene expression following anoxia, heat, and combined heat + anoxia treatments. The fold change (log2) was measured by real-time qPCR using time = 0 (air) as a reference.
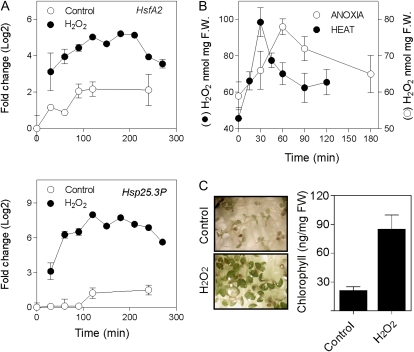
HsfA2 Is Likely Up-Regulated by Heat- or Anoxia-Induced H2O2
HsfA2 and several HSPs are H2O2-regulated genes in Arabidopsis (Vanderauwera et al., 2005). Both HsfA2 and its target Hsp25.3-P are induced by an H2O2 treatment (Fig. 4A; Nishizawa et al., 2006). The production of H2O2, therefore, may be the trigger for HsfA2 and HSP expression following either heat or anoxic treatment (Blokhina et al., 2001; Volkov et al., 2006). H2O2 is quickly produced after the onset of both treatments (Fig. 4B), supporting the hypothesis of a mechanistic overlap in the physiological response to heat and anoxia. Interestingly, treating Arabidopsis seedlings with H2O2 results in enhanced anoxia tolerance (Fig. 4C).
Figure 4.
Effects of H2O2 on the expression of HsfA2 and Hsp25.3P. A, Expression of HsfA2 and Hsp25.3P following treatment with 5 mm H2O2 for the times indicated. Seedlings were grown in liquid medium for 4 d in the dark. H2O2 (5 mm) was added for the time shown. The fold change (log2) was measured by qPCR using time = 0 as a reference. B, H2O2 production under anoxia and heat (38°C). C, Effects of 5 mm H2O2 on anoxia tolerance. Seedlings were grown in liquid medium for 4 d in the dark. H2O2 (5 mm) was added (2-h treatment) to the wells containing the seedlings followed by rinsing in fresh medium and transfer to anoxia (26 h). The photographs show the seedlings recovering (5 d) after the anoxic treatment. The chlorophyll content in replicated (n = 3) samples is shown in the histogram. FW, Fresh weight.
HsfA2 Overexpression Confers Tolerance to Anoxia
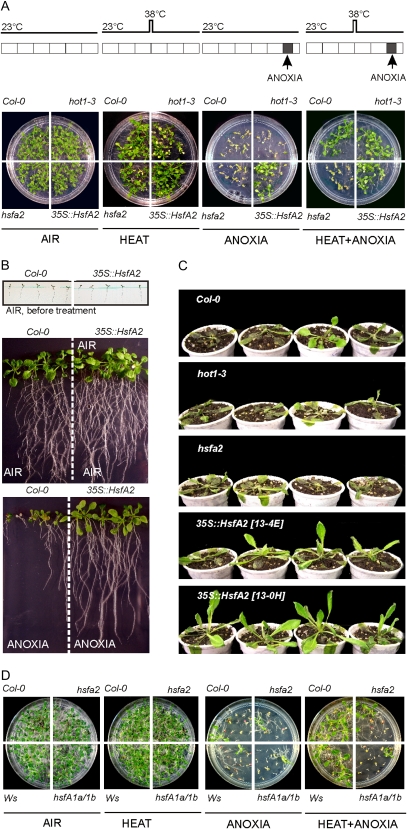
HsfA2 is required for long-term acquired thermotolerance (Charng et al., 2007) and for salt and osmotic stress tolerance (Ogawa et al., 2007). To verify the role of HsfA2 under anoxia, we isolated a knockout mutant for HsfA2 (hsfA2) and produced an _HsfA2_-overexpressing line (35S::HsfA2). We tested the anoxia tolerance of hsfA2, 35S::HsfA2, and the hot1-3 mutant. hot1-3 is deficient in Hsp101 and negatively affected in both basal and acquired thermotolerance (Fig. 5A; Hong and Vierling, 2000; Queitsch et al., 2000). Interestingly, 35S::_HsfA2_-overexpressing lines survived a 13-h-long anoxic treatment, while the wild-type (Columbia ecotype [Col-0]), hsfA2, and hot1-3 seedlings died (Fig. 5A). hsfA2 and hot1-3 seedlings, however, were as tolerant as the wild type even when anoxia treatments were sublethal for the wild type (data not shown). hsfA2 seedlings showed lower anoxia tolerance than the wild type when heat pretreated, but only after a long-term (2 d) recovery from the heat treatment followed by anoxia (Fig. 5A). This indicates that while both HsfA2 and Hot1 are required for acquired thermotolerance (Hong and Vierling, 2000; Queitsch et al., 2000), only HsfA2 is required for heat-induced anoxia tolerance. The tolerance of 35S::HsfA2 to anoxia is particularly evident when seedlings were grown on vertical plates. 35S::HsfA2 seedlings cannot be distinguished from the wild type before the treatment and grow normally in air (Fig. 5B, air before treatment). After anoxia only, 35S::HsfA2 seedlings recover and grow at a rate comparable to the aerobic seedlings, while the Col-0 seedlings display stunted growth (Fig. 5B, anoxia). Anoxia-treated 35S::HsfA2 seedlings produce more ethanol than Col-0 seedlings (Supplemental Fig. S3), a likely consequence of the ability of 35S::HsfA2 seedlings to better tolerate anoxia. Interestingly, 3-week-old 35S::HsfA2 pot-grown plants also showed better tolerance (survival, 100%; chlorophyll content, 220 ± 22 ng mg−1 fresh weight) to submergence when compared with Col-0 (survival, 50%; chlorophyll content, 150 ± 21 ng mg−1 fresh weight), hsfA2 (survival, 40%; chlorophyll content, 145 ± 23 ng mg−1 fresh weight), and hot1-3 (survival, 40%; chlorophyll content, 144 ± 24 ng mg−1 fresh weight; Fig. 5C).
Figure 5.
Role of HsfA2 in anoxia tolerance. A, Tolerance assay in Col-0, hot1-3, hsfA2, and 35S::HsfA2. A schematic representation of treatments is shown. Each box represents 1 d. The gray box represents the anoxic treatment (13 h). Photographs were taken 1 week after the end of the treatments. The results shown refer to 35S::HsfA2 line 13-OH. Comparable results were obtained using the independent 35S::HsfA2 line 13-4E. B, Tolerance assay in Col-0 and 35S::HsfA2 seedlings grown on vertical plates. The anoxic treatment was 13 h long. Photographs were taken before treatment and 2 weeks after the end of the anoxic treatment. The results shown refer to 35S::HsfA2 line 13-OH. Comparable results were obtained using the independent 35S::HsfA2 line 13-4E. C, Tolerance assay in Col-0, hot1-3, hsfA2, and 35S::HsfA2 plants grown in pots and subjected to 24 h of complete submergence in the dark. The results shown refer to both 35S::HsfA2 lines 13-OH and 13-4E. D, Tolerance assay in hsfA2, its wild type (Col-0), the double mutant hsfA1a/1b, and its wild type (Wassilewskija [Ws]). A schematic representation of the treatments is shown in A. Photographs were taken 1 week after the end of the treatments.
The expression of most HsfA2-regulated genes appears also to be dependent on HsfA1a/HsfA1b (Busch et al., 2005). Although HsfA1a/HsfA1b are not induced by either heat or anoxia (Fig. 3A), these two HSFs appear to cooperate with HsfA2 in the heat response (Schramm et al., 2006). Therefore, we checked whether HsfA1a/HsfA1b are required for the heat-induced anoxia tolerance and found that the double mutant hsfA1a/1b displayed the inability to tolerate anoxia when heat pretreated (Fig. 5D).
HsfA2 Overexpression Up-Regulates Several Heat-Related Genes
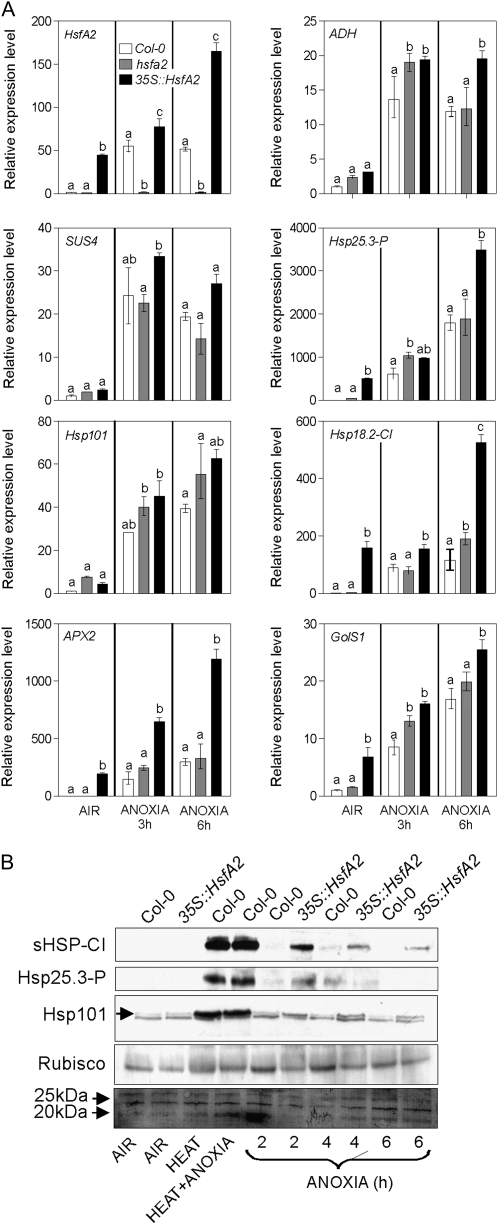
We analyzed the expression of anaerobic and heat-related genes in the 35S::HsfA2 and hsfA2 plants (Fig. 6A). As expected, hsfA2 and 35S::HsfA2 lines showed lower and higher HsfA2 mRNA levels, respectively. Interestingly, 35S::HsfA2 seedlings showed increased amounts of HsfA2 mRNA following exposure to anoxia. This may be due to the endogenous transcripts, whose level increased following the onset of the stress. ADH and SUS4 are induced by anoxia, irrespective of the genotype. A higher expression of both ADH and SUS4 in the 35S::HsfA2 seedlings is likely due to the higher anoxia tolerance of this genotype, preventing the decline in expression of these genes toward the end of the anoxic treatment. Hsp25.3-P, Hsp18.2-CI, APX2, and GolS1 were clearly up-regulated in 35S::HsfA2 seedlings, both in air and, mirroring HsfA2 patterns, also 6 h after the onset of anoxia (Fig. 6A). Hsp101 did not appear to be up-regulated in the 35S::HsfA2 line. The absence of a significant reduction in the anoxic expression of HsfA2 targets in the knockout line suggests that other Hsfs, besides HsfA2, might be involved in the regulation of these genes under anoxia.
Figure 6.
Effects of HsfA2 on the expression of anaerobic and heat-related genes and proteins. A, Expression of HsfA2, ADH, SUS4, HSP25.3-P, HSP101, HSP18.2-CI, APX2, and GolS1 in Col-0, hsfA2, and 35S::HsfA2 seedlings. Relative expression levels are shown as fold change values (1 = Col-0, air). Differences among genotypes within each treatment (air, 3 h of anoxia, and 6 h of anoxia) were evaluated by two-way ANOVA (Bonferroni posttest P < 0.05). Bars represent means of at least three measurements ± sd. B, Immunoblotting of proteins extracted from 4-d-old dark-grown seedlings treated under anoxia. The antibodies used recognized HSP25.3-P, sHSP-CI, and HSP101 (the specific bands are indicated by the arrows). Blots were stained with Ponceau-S and probed with an antibody recognizing the large subunit of Rubisco to confirm even loading and transfer. Treatments were as follows: air, heat (90 min at 38°C), heat + anoxia (90 min at 38°C followed by 6 h under anoxia), and anoxia (2–6 h).
HSPs Are Present in the Anoxic 35S::HsfA2 Seedlings But Not in the Wild Type
Translational efficiency under anoxia can be greatly impaired, a likely consequence of a low ATP content and posttranscriptional regulation mechanisms (Branco-Price et al., 2005, 2008). This may result in a lack of correlation between the mRNA and corresponding protein levels. To verify whether the increased HSP mRNA levels resulted in correspondingly increased protein levels, we checked three anoxia-induced HSPs (small HSP class I [sHSP-CI], Hsp25.3-P, and Hsp101) by immunoblotting. The results showed that heat and heat + anoxia strongly increase these HSP protein levels (Fig. 6B). Under anoxia, the wild-type seedlings do not appear to accumulate HSPs. We only managed to detect faint sHSP-CI and Hsp25.3-P signals in Col-0 after 4 h under anoxia (Fig. 6B). Despite the increased HSP mRNA steady state in 35S::HsfA2 (Fig. 6A), these plants did not accumulate the corresponding proteins under aerobic conditions (Fig. 6B). However, when transferred to anoxia, 35S::HsfA2 seedlings showed a clear increase in the levels of sHSP-CI and Hsp25.3-P (Fig. 6B), while only a slight increase in Hsp101 was observed. Therefore, HSPs do not accumulate under anoxia but are produced in the anoxic 35S::HsfA2 plants.
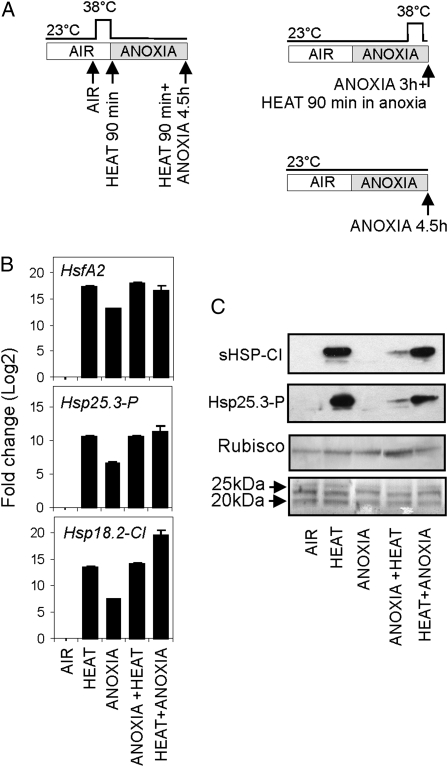
The level of sHSP-CI and Hsp25.3-P proteins is high in heat-pretreated seedlings transferred to anoxia, while it is low in anoxia-treated seedlings (Fig. 6B). This suggests that under anoxia, HSPs fail to accumulate despite the presence of high levels of the corresponding transcripts. We tested whether anoxia negatively affects HSPs transcripts and protein stabilities using actinomycin D and cycloheximide (Supplemental Fig. S4, A and B). The results showed that the Hsp18.2-CI and Hsp25.3-P mRNAs induced by a heat treatment are degraded at a slower rate under anoxia when compared with the effects of an aerobic incubation following the heat treatment (Supplemental Fig. S4C). Anoxia, however, enhances HSP degradation through a mechanism requiring de novo protein synthesis, since cycloheximide stabilizes HSPs under anoxia (Supplemental Fig. S4D). We then compared the effects of a heat treatment applied before anoxia or “during anoxia” to evaluate their consequences of HSP protein accumulation (Fig. 7A). Seedlings treated at 38°C before a 4.5-h anoxia treatment (heat + anoxia) accumulate HSP mRNAs at a level comparable to that of the heat-only treatment (Fig. 7B). The level of HSP proteins is only slightly lower in the heat + anoxia samples when compared with the heat-only treatment (Fig. 7C). When the heat treatment is applied during anoxia (anoxia + heat), the level of HSP mRNAs is higher than under anoxia only, showing the additive effect of the two stress treatments and indicating that anoxia does not interfere with the triggering effect of heat at the mRNA level (Fig. 7B). However, HSP proteins fail to accumulate at significant levels (anoxia + heat; Fig. 7C), indicating that, despite a mRNA level comparable to that of the heat treatment (compare heat with anoxia + heat; Fig. 7A), the HSP proteins fail to accumulate (Fig. 7B). The level of HSP proteins in the heat + anoxia treatment (heat treatment before anoxia) is only slightly lower than in the heat-only sample, suggesting that 4.5 h under anoxia had a moderate effect on HSP protein stability. On the other hand, a heat treatment on seedlings that already experienced 4.5 h under anoxia (heat treatment during anoxia) was only effective at the mRNA level (Fig. 7B) but produced a modest HSP protein accumulation (Fig. 7C).
Figure 7.
Effects of anoxia on HSP mRNA and protein accumulation. A, Schematic representation of treatments. B, Expression of HSFA2, HSP25.3-P, and HSP18.2-CI in Col-0 seedlings. Relative expression levels are shown as fold change values (log2; using “air” as a reference). C, Immunoblotting of proteins extracted from 4-d-old dark-grown seedlings treated under anoxia. The antibodies used recognized HSP25.3-P and sHSP-CI. Blots were stained with Ponceau-S and reprobed with an antibody recognizing the large subunit of Rubisco to confirm even loading and transfer.
DISCUSSION
Mild heat treatment results in acquired anoxia tolerance (Loreti et al., 2005). The molecular basis of this phenomenon is unknown, and there are several possible hypotheses. First, heat may induce several stress-related transcription factors, thus activating a large number of signaling pathways conferring tolerance to anoxia. A second possibility is that heat pretreatment enhances tolerance by dampening the anoxic responses of the seedlings, allowing them to enter into a quiescent state preserving cellular viability. Third, the increased tolerance could be due to the activation of a functional overlap between the heat and the anoxia acclimation mechanisms.
The first hypothesis is reasonable, since we observed a strong induction of the genes belonging, for example, to the ERF, MYB, WRKY, NAC, and HSF families (Supplemental Fig. S2). However, none of the transcription factors listed in Supplemental Table S1 is known to positively affect anoxia tolerance. The dampening of the anoxic response as part of a “low oxygen quiescence syndrome” (Bailey-Serres and Voesenek, 2008) is also feasible, since heat pretreatment reduces the expression of several anaerobic genes (Fig. 2) and also slows down the ethanol production (Supplemental Fig. S1). However, the higher production of ethanol observed in the 35S::HsfA2 seedlings (Supplemental Fig. S3), which were also more tolerant to anoxia (Fig. 5A), suggests that the dampening effect of heat over the anaerobic response may simply reflect the hierarchies of stress response regulatory systems.
However, the activation of a heat-dependent anoxia acclimation mechanism(s) that shares similarities with acquired thermotolerance pathways would seem to be possible, so we turned our attention to HsfA2 (Fig. 3). HsfA2 plays an important role in acquired thermotolerance and also appears to play a role in abiotic stress tolerance. Indeed, some studies have reported the role of HsfA2 in oxidative stress (Li et al., 2005), high light conditions and chilling (Nishizawa et al., 2006, 2008), and salt/osmotic stress tolerance, suggesting a role in multiple stress tolerance (Ogawa et al., 2007). The results obtained through a genetic approach demonstrate that the hsfA2 mutant cannot cross-acclimate to anoxia through mild heat pretreatment, in contrast to the wild type (Fig. 5). Assuming that HsfA2 is important for anoxia tolerance, we would also have expected a lower anoxia tolerance in the hsfA2 plants when a sublethal anoxic treatment was applied, but this was not the case (data not shown). However, expression of HsfA2 targets in hsfA2 explains this result and is most likely due to possible redundancy among the 21 Arabidopsis HSFs (Lohmann et al., 2004). Seven _Hsf_s (Fig. 3A) were induced under anoxia and may have compensated for the absence of HsfA2 in the hsfA2 mutant. Differences between the wild type and hsfA2 in heat-dependent acquired anoxia tolerance were observed only after a long (2-d) recovery from the heat treatment, in line with the role of HsfA2 in acquired thermotolerance (Charng et al., 2007). It has been demonstrated that only after a long-term recovery from heat pretreatment is it possible to observe differences both in heat tolerance and expression of _HsfA2_-dependent HSPs, such as Hsp25.3-P (Charng et al., 2007). Additionally, as previously suggested for heat stress (Schramm et al., 2006), HsfA2 appears to cooperate with HsfA1a/1b in the mechanism of heat-induced anoxia tolerance (Fig. 5D). Both HsfA2 and HsfA1a/1b are indeed involved in the mechanism of acquired thermotolerance (Wunderlich et al., 2003; Charng et al., 2007).
35S::HsfA2 seedlings and plants showed enhanced tolerance to anoxia and to submergence, respectively (Fig. 5). The enhanced anoxia tolerance could be explained either as a real tolerance to the lack of oxygen or a better ability to recover during the postanoxic phase. The results obtained with the submergence experiments, which showed healthier 35S::HsfA2 plants during and immediately after the end of the submergence (Fig. 5C), suggest that HsfA2 overexpression protects the plants during the stress treatment rather than from the consequences of reaeration. These results demonstrate that HsfA2 is not only responsible for the heat-dependent acclimation to anoxia but also plays a role in the low oxygen-tolerance response in Arabidopsis.
HsfA2 is the most H2O2-responsive Hsf (Miller and Mittler, 2006). It is likely that H2O2 is the trigger that induces HsfA2 under anoxia, since H2O2 is produced at the onset of anoxia (Fig. 4B). Exogenous H2O2 treatment induces HsfA2 and confers anoxia tolerance (Fig. 4, A and C). Signaling by reactive oxygen species under oxygen deprivation (Bailey-Serres and Chang, 2005) and protection from oxidative stress are important under anoxia (Blokhina et al., 2002). HSPs themselves may be involved in protecting the cell from oxidative stress (Timperio et al., 2008). GolS1 is induced by HsfA2 and is important for plant acclimation to oxidative stress (Nishizawa et al., 2008). Galactinol, produced by GolS, may well play a role under anoxia, as de novo galactinol biosynthesis from Suc would be required, and Suc is known to enhance Arabidopsis tolerance to low oxygen (Banti et al., 2008). Genes regulated by HsfA2 include several sHSPs (Nishizawa et al., 2006; Schramm et al., 2006). sHSPs are overrepresented under several stress conditions, underlining the putative importance of their ATP-independent role in stabilizing proteins and membranes undergoing conformational disruption (Swindell et al., 2007). This ATP-independent function may be particularly important under anoxic conditions, since it reduces the energy-dependent de novo protein synthesis.
Despite the anoxic induction of HsfA2 at levels close to those observed after heat treatment (Fig. 3), Arabidopsis seedlings are highly susceptible to anoxia. Heat-pretreated or 35S::HsfA2 plants, on the other hand, are highly tolerant to anoxia. This can be explained by the failure of wild-type plants to induce the HsfA2 targets at the protein level (Fig. 6B). Indeed, while HsfA2 target mRNAs are induced under either heat or anoxia, we only detected a faint protein band corresponding to sHSP-CI and Hsp25.3-P in the 4-h anoxia-treated Col-0 plants (Fig. 6B). Plants able to withstand anoxic treatment (heat pretreated or overexpressing HsfA2), on the other hand, showed HSP accumulation under anoxia (Fig. 6B). The lack of expression of HSPs at the protein level in the anoxic samples is somewhat surprising, since HSP mRNAs are loaded onto polyribosomes under low oxygen (Branco-Price et al., 2008). This suggests that the translation of HSP mRNA is controlled by a complex regulatory mechanism. This hypothesis is reinforced by the evidence of a lack of HSP protein accumulation even in the 35S::HsfA2 aerobic seedlings, despite the strong expression of the respective HSP mRNAs. The HSP proteins only accumulated when the 35S::HsfA2 plants were exposed to anoxia, indicating that a “stress” signal is required for the translation of the HSP mRNAs. The failure to induce HSP accumulation under anoxia in wild-type plants is possibly due to the slower timing of the HsfA2 expression and its targets under anoxia compared with the expression following heat and heat + anoxia (Fig. 3C). In the 35S::HsfA2 plants, HsfA2 and its target mRNAs are already present in air, which is likely to lead to their rapid translation as soon as anoxia is established, before the ability to produce and accumulate HSP proteins declines (Fig. 6B).
In this study, we demonstrated the importance of HsfA2 in protecting Arabidopsis plants under low-oxygen conditions. The overexpression of HsfA2 transactivates its target genes early enough to enable them to be translated even under anoxic conditions. It is likely that both HSPs and genes involved in the protection against oxidative stress are responsible for the acquisition of anoxia tolerance. These results indicate that there is a considerable overlap between the molecular mechanisms of heat and anoxia tolerance and that HsfA2 is a major player in these mechanisms.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0, its hot1-3 mutant (kindly provided by Dr. Elizabeth Vierling), HsfA2 (At2g26150) T-DNA insertion line SALK_008978 (obtained from the Nottingham Arabidopsis Stock Centre; code N508978), the double mutant hsfA1a/HsfA1b (abbreviated hsfA1a/1b; kindly provided by Dr. Friedrich Schöffl), and the hsfA1a/1b wild type (Wassilewskija) were used in this study. Transgenic plants overexpressing HsfA2 (35S::HsfA2) were obtained by infiltration with the floral dip method (Clough and Bent, 1998). Cloning and construction of the expression cassette for HsfA2 were performed using Gateway technology (Karimi et al., 2002; Invitrogen). For gene expression experiments, seeds were sterilized with diluted bleach (10 min of incubation in 1.7% sodium hypochlorite, rinsed, and washed seven times in sterile water) and incubated in 2.5 mL of liquid growing medium (Murashige and Skoog half-strength solution on six-well plates) containing 90 mm Suc. The plates were incubated in the dark at 4°C for 2 d (vernalization, to promote homogeneous germination) and then transferred to 23°C for 4 d in the dark before the treatment. Anoxic treatments were carried out in the dark. An enclosed anaerobic workstation (Anaerobic System model 1025; Forma Scientific) was used to provide an oxygen-free environment for seedling incubation. This chamber uses palladium catalyst wafers and desiccant wafers to maintain strict anaerobiosis to less than 10 μ g mL−1oxygen (according to the manufacturer's specifications). High-purity N2 was used to initially purge the chamber, and the working anaerobic gas mixture was N2:H2 with a ratio of 90:10. Three independent, replicated experiments were performed for each experimental condition. Each independent experiment consisted of four replicated seedling cultures pooled before RNA extraction. Agar plates were used to evaluate anoxia tolerance (Fig. 5). They were prepared using a solid growing medium (Murashige and Skoog half-strength solution containing 1% agar and 90 mm Suc). The plates were then transferred to the light for the postanoxic recovery (14-h/10-h photoperiod at 150 μ mol photons m−2 s−1). Treatments were performed by transferring the plates containing the 7-d-old (vertical plates) or 5-d-old (horizontal plates) Arabidopsis seedlings to 38°C for 90 min immediately before the anoxic treatments. The submergence treatments shown in Figure 5C were performed using 14-d-old Arabidopsis plants grown in pots and subjected to 24-h complete submergence in the dark. The tolerance experiment reported in Figure 4 (effect of H2O2 treatment of tolerance) was performed using Arabidopsis seedlings (dark germinated, 4 d old) grown in liquid growing medium (Murashige and Skoog half-strength solution on six-well plates) containing 90 mm Suc. H2O2 (5 mm) was added 2 h before the anoxia treatment. Seedlings were rinsed in fresh Murashige and Skoog half-strength solution to remove H2O2 immediately before the anoxia treatment. All the treatments started at 9 am.
qPCR
RNA was extracted from the whole seedlings. The total RNA was extracted with an RNAqueous kit (Ambion) according to the manufacturer's instructions and then subjected to a DNase treatment using a TURBO DNA-free kit (Ambion). Two micrograms of each sample was reverse transcribed into cDNA with a high-capacity cDNA archive kit (Applied Biosystems). qPCR amplification was carried out with an ABI Prism 7000 sequence detection system (Applied Biosystems) using Ubiquitin10 (At4g05320) as an endogenous control. SYBR Green probes for each gene were used. The primers used are reported in Supplemental Table S3. PCR was carried out using 50 ng of cDNA and SYBR PCR master mix (Applied Biosystems) following the manufacturer's protocol. Relative quantitation of each single gene expression was performed using the comparative threshold cycle method, as described in the ABI PRISM 7700 Sequence Detection System User Bulletin No. 2 (Applied Biosystems).
RNA Isolation, Copy RNA Synthesis, and Hybridization to Affymetrix GeneChips
Total RNA was extracted from seedling samples using an Ambion RNAqueous kit (Ambion). Two biological replicates for each experimental condition were used. RNA quality was assessed by agarose gel electrophoresis and spectrophotometry. RNA was processed for use on Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays as described previously (Loreti et al., 2005). Hybridization, washing, staining, and scanning procedures were performed by Genopolis (University of Milano-Bicocca), as described in the Affymetrix technical manual. Microarray analysis was performed using R/Bioconductor (Gentleman et al., 2004). Expression measures were obtained using GeneChip Robust Multi-Array (Wu and Irizarry, 2005), a multiarray analysis method that estimates probe set signals taking into account the physical affinities between probes and targets. Normalization was carried out using a quantile method (Bolstad et al., 2003). To reduce the number of noninformative genes, two different filters were applied: the first removes the probe sets presenting an Affymetrix absent call for both conditions; the second eliminates the probe sets showing, as the maximum signal in the two conditions, a value of less than or equal to the 95th percentile of the overall absent call signal distribution. To identify a statistically reliable number of differentially expressed genes between the two conditions, a linear model was used (Wettenhall and Smyth, 2004). To assess the differential expression, an empirical Bayesian method (Smyth, 2004) was used to moderate the se of the estimated log-fold changes. To control P values in a multiple testing problem context, a Benjamini-Hochberg correction of the FDR (Reiner et al., 2003) was applied (adjusted P ≤ 0.05), leading to a list of differentially expressed probe sets. Microarray data sets were deposited in a public repository with open access (accession no. GSE16222; http://www.ncbi.nlm.nih.gov/projects/geo). The differentially expressed genes were selected by applying a filter that considers both the fold change (≥ 2 or ≤−2) in a hybridization signal (namely the ratio between the intensity in the treated spot versus the control) and the FDR (≤ 0.05). Scatterplots and the statistical analyses were performed using R and BRB array software for a multidimensional scaling comparison. In order to build gene expression maps, MapMan software (Thimm et al., 2004) from http://gabi.rzpd.de/projects/MapMan/ was used. Heat shock-related genes were used as defined by MapManx BINs. For anoxia custom maps, a consensus of up-regulated genes (≥ 2-fold) from Loreti et al. (2005), Branco-Price et al. (2005, 2008), and this study (Supplemental Table S4) was used.
H2O2 Quantitation
Liquid-grown whole seedlings were sampled immediately after treatment and homogenized in a HEPES buffer, pH 7.5, 1 mm EDTA. The resulting extract was then centrifuged for 10 min at 14,000_g_ at 4°C. A 2:1 volume of chloroform:methanol was added to the supernatant. The mixture was vortexed and subsequently centrifuged for 3 min at 10,000_g_ at 4°C. The aqueous phase was then used for the analyses. H2O2 abundance was quantified according to Guilbault et al. (1967) and the subsequent modification by Muhlenbock et al. (2007), in a reaction volume of 3 mL containing 50 mm HEPES, pH 7.5, 50 mm homovanilic acid, 4 _μ_m peroxidase, and an aliquot of extract. Fluorescence was then measured at 425 nm after excitation at 315 nm, 30 min after the beginning of the reaction. Samples were kept strictly in the dark. H2O2 was determined as a limiting factor in the oxidation mediated by homovanilic acid, calculated on the basis of the standard curve obtained with different amounts of H2O2.
ADH Assay
ADH activity was assayed after anoxia (6 h), heat (90 min), and heat (90 min) followed by anoxia (6 h) in 4-d-old dark-grown seedlings. Soluble proteins were extracted using 100 mm HEPES buffer, pH 7.0, and 5 mm dithiothreitol. Protein amount was estimated using the Bio-Rad Protein Assay, based on the method of Bradford (1976). ADH activity was monitored using the spectrophotometric assay (_A_340) as described by Russell et al. (1990).
Ethanol Assay
The seedlings used for the ethanol production experiment were grown in liquid medium contained in petri dishes sealed with Parafilm. Ethanol evaporation under these conditions was negligible. Samples were assayed through coupled enzymatic assay methods, measuring the increase in _A_340, as described previously (Banti et al., 2008).
Chlorophyll Assay
Chlorophyll quantitation was performed on plant material that was frozen immediately after the treatments. The plant material was weighed, ground in liquid nitrogen, and extracted with 80% (v/v) acetone. Chlorophyll was quantified photometrically as described by Lichtenthaler (1987) and Porra (2002).
Immunoblotting
Samples (about 500 mg) were ground in liquid nitrogen. An extraction buffer, described by Siddique et al. (2008), was added in a 1:2 ratio (plant tissue:buffer) Total protein content was quantified with a BCA Protein Assay (Pierce). SDS-PAGE was performed on a 10% Criterion polyacrylamide gel (Bio-Rad Laboratories). Blotting on an Amersham Hybond-P polyvinylidene difluoride membrane was performed with a Novablot electrophoretic transfer system (Amersham Pharmacia Biotech). Immunoblotting was performed using Immun-Star HRP Chemiluminescent Detection Kits (Bio-Rad Laboratories). Antibodies against Hsp101, Hsp25.3-P, and sHSP-CI were purchased from Agrisera (product codes AS07 253, AS08 285, and AS07 254, respectively). Ponceau-S staining and immunoblotting using the antibody against the large subunit of Rubisco (AS03037) were used to confirm even loading and transfer (Agrisera).
Microarray data from this article were submitted to the public National Center for Biotechnology Information Gene Expression Omnibus database (GEO accession no. GSE16222).
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Figure S1. Effects of anoxia, heat, and combined heat + anoxia treatments on ADH activity and ethanol production.
- Supplemental Figure S2. Heat map of transcription factors strongly up-regulated under anoxia.
- Supplemental Figure S3. Ethanol production in anoxic Col-0 and 35S::HsfA2 seedlings.
- Supplemental Figure S4. Stability of HSP transcripts and proteins under aerobic and anoxic conditions.
- Supplemental Table S1. List of up-regulated genes.
- Supplemental Table S2. List of down-regulated genes.
- Supplemental Table S3. List of primers used in this work.
- Supplemental Table S4. MapMan mapping file for anoxia genes reported in Figure 1C.
Supplementary Material
[Supplemental Data]
Acknowledgments
We thank Dr. Elizabeth Vierling for kindly providing us with the hot1-3 seeds and Dr. Friedrich Schöffl for kindly providing us with the hsfA1a/hsfA1b seeds.
References
- Bailey-Serres J, Chang R. (2005) Sensing and signaling in response to oxygen deprivation in plants and other organisms. Ann Bot (Lond) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LA. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Banti V, Loreti E, Novi G, Santaniello A, Alpi A, Perata P. (2008) Heat acclimation and cross-tolerance against anoxia in Arabidopsis. Plant Cell Environ 31: 1029–1037 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Barratt DHP, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Blokhina O, Virolainen E, Fagerstedt K. (2002) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot (Lond) 91: 173–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina OB, Chirkova TV, Fagerstedt KV. (2001) Anoxic stress leads to hydrogen peroxide formation in plant cells. J Exp Bot 52: 1–12 [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bradford M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 7: 248–254 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJH, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot (Lond) 96: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Charng Y, Liu H, Liu N, Chi W, Wang C, Chang S, Wang T. (2007) A heat-inducible transcription factor, HsfA2, is required for extension for acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for _Agrobacterium_-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P. (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbault GG, Kram DN, Hackley E. (1967) A new substrate for fluorimetric determination of oxidative enzymes. Anal Chem 39: 271–272 [DOI] [PubMed] [Google Scholar]
- Hartl FU. (1996) Molecular chaperones in cellular protein folding. Nature 381: 571–580 [DOI] [PubMed] [Google Scholar]
- Hofmann GE, Feder ME. (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61: 243–282 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E. (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismond KP, Dolferus R, De Pauw M, Dennis ES, Good AG. (2003) Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol 132: 1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for _Agrobacterium_-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES. (2002) Expression profile analysis of low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh I. (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53: 225–229 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Vierling E. (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen Q, Qijun C, Xinqi G, Bishu Q, Naizhi C, Shouming X, Jia C, Xuechen W. (2005) AtHsfA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci China Ser C Life Sci 48: 540–550 [DOI] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Zietkiewicz S. (2008) Chaperones in control of protein disaggregation. EMBO J 27: 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F. (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271: 11–21 [DOI] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mittler R. (2006) Could heat shock factors function as hydrogen peroxide sensors in plants?. Ann Bot (Lond) 98: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Schärf KD. (2002) In the complex family of the heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenbock P, Plaszczyca M, Plaszczyca M, Mellerowicz E, Karpinski S. (2007) Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 19: 3819–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need?. Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T. (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased thermotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Perata P, Alpi A. (1993) Plant response to anaerobiosis. Plant Sci 93: 1–7 [Google Scholar]
- Perata P, Voesenek LA. (2007) Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends Plant Sci 12: 43–46 [DOI] [PubMed] [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. (2000) Heat Shock Protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Russell DA, Wong DML, Sachs MM. (1990) The anaerobic response of soybean. Plant Physiol 92: 401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P. (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60: 759–772 [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P. (2008) A cascade transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53: 264–274 [DOI] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf KD. (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. (2004) MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–937 [DOI] [PubMed] [Google Scholar]
- Timperio AM, Egidi MG, Zolla L. (2008) Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J Proteomics 71: 391–411 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F. (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- Voesenek LA, Colmer TD, Pierik R, Millenaar FF, Peeters AJM. (2006) How plants cope with complete submergence. New Phytol 170: 213–226 [DOI] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Mullineaux PM, Schöffl F. (2006) Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol Biol 61: 733–746 [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. (2004) limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20: 3705–3706 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. (2005) Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol 12: 882–893 [DOI] [PubMed] [Google Scholar]
- Wunderlich M, Werr W, Schöffl F. (2003) Generation of dominant-negative effects on the heat shock response in Arabidopsis thaliana by transgenic expression of a chimaeric HSF1 protein fusion construct. Plant J 35: 442–451 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]