p53-Responsive MicroRNAs 192 and 215 Are Capable of Inducing Cell Cycle Arrest (original) (raw)
. Author manuscript; available in PMC: 2010 Mar 11.
Abstract
microRNAs provide a novel layer of regulation for gene expression by interfering with the stability and/or translation of specific target mRNAs. Overall levels of microRNAs are frequently down-regulated in cancer cells, and reducing general microRNA processing increases cancerogenesis in transgenic models, suggesting that at least some microRNAs might act as effectors in tumor suppression. Accordingly, the tumor suppressor p53 up-regulates miR-34a, a microRNA that contributes to apoptosis and acute senescence. Here, we used array hybridization to find that p53 induces two additional, mutually related clusters of microRNAs, leading to the up-regulation of miR-192, miR-194, and miR-215. The same microRNAs were detected at high levels in normal colon tissue but were severely reduced in many colon cancer samples. On the other hand, miR-192 and its cousin miR-215 can each contribute to enhanced CDKN1A/p21 levels, colony suppression, cell cycle arrest, and cell detachment from a solid support. These effects were partially dependent on the presence of wild-type p53. Antagonizing endogenous miR-192 attenuated 5-fluorouracil–induced accumulation of p21. Hence, miR-192 and miR-215can act as effectors as well as regulators of p53; they seem to suppress cancerogenesis through p21 accumulation and cell cycle arrest.
Introduction
The tumor suppressor p53 acts as a transcription factor and regulates the expression of numerous genes, leading to cell cycle arrest, apoptosis, and senescence (1). Although p53 can also induce the intrinsic pathway of apoptosis independently of transcription (2), the identification of p53-responsive genes remains central to our understanding of tumor suppression. Such p53 target genes can be classified as those that primarily induce cell cycle arrest (e.g., the cyclin-dependent kinase inhibitor CDKN1A/p21) and those that mediate apoptosis (e.g., Puma and Noxa). p53 activity is induced by DNA damage (e.g., through the application of chemotherapeutics). More recently, a small molecule was identified that specifically activates p53 without obvious genotoxicity (3). Nutlin-3 disrupts the interaction between p53 and its principal antagonist Mdm2. Consequently, Mdm2-mediated ubiquitination and degradation of p53 is abolished, and active p53 accumulates to induce its target genes. Besides its therapeutic perspectives, Nutlin-3 also represents an important tool to activate p53 in experimental systems, without targeting other transcription factors that would respond to DNA damage.
Recent discoveries identified a new class of genes, encoding RNA species without protein coding function but with regulatory activities. Through several enzymes and transporters, microRNAs are processed from their precursors and exported to the cytoplasm, where they associate with a complex of auxiliary proteins and with a specific set of target mRNAs through (sometimes imperfect) base pairing. As a consequence, target mRNAs are degraded and/or their translation is inhibited (4, 5). Thus, microRNAs provide an additional mechanism to determine the extent of individual gene expression, comparable with transcriptional regulation.
Currently, a few hundred microRNA species were detected in metazoan tissues at levels that make them likely candidates for gene regulation. Some of them were found disproportionally expressed in cancer cells, and a few were shown to regulate cancer-relevant gene products (6). Notably, however, the overall levels of mature microRNAs seem to be reduced in at least a subset of tumors when compared with corresponding normal tissue (7), perhaps as a result of down-regulated argonaute proteins (8). In agreement, transgenic mouse models with silenced expression of microRNA processing factors were reported to have increased susceptibility to cancer (9). These findings point to a role of microRNAs in tumor suppression.
This raises the question what particular microRNA species can suppress tumor development. One approach to answer this question consists in the identification of microRNAs that are regulated through known tumor-suppressive pathways, assuming that such microRNAs are representing effectors that determine the biological response to such pathways. Accordingly, miR-34a and other types of the miR-34 species were identified as p53-regulated microRNAs by several groups (10–20), and subsequent analysis revealed that this microRNA species can contribute to apoptosis and/or acute senescence. miR-34a affects the levels and/or translation of a multitude of mRNAs (20), and it is currently not clear whether individual target mRNAs can be held responsible for the overall biological response to this microRNA. In any case, miR-34a appears as a p53 target gene that mediates some of the biological effects elicited by p53.
Among protein-coding mRNA species, p53 activates hundreds of target genes, and many of them carry out important functions in the context of the p53 response in at least a subset of cell species. We therefore reasoned that miR-34a may not represent the only p53-responsive species among all microRNAs. In an effort to obtain a more complete set of p53-responsive microRNAs, we hybridized microarrays with small RNA from Nutlin-3–treated cells and found that the two clusters encoding miR-192, miR-194, and miR-215 were p53 responsive, in addition to miR-34a. The same clusters are down-regulated in colon cancer relative to normal colon tissue, further supporting the idea that they might be part of a tumor-suppressing program. Functional analysis revealed that the new p53-responsive microRNAs were capable of inducing p21 expression and cell cycle arrest in a p53-dependent manner, suggesting that they are capable of activating p53. Accordingly, antagonizing miR-192 attenuated the accumulation of p53 and p21 in response to 5-fluorouracil (5-FU).
Materials and Methods
Cell culture and drug treatment
U2OS, SJSA, HT29, and A549 cells were maintained in DMEM; Lovo, DLD-1, and HCC-2998 in RPMI 1640; and HCT116 cells in McCoy’s medium. Media were supplemented with 10% fetal bovine serum. Nutlin-3 (Sigma-Aldrich) was dissolved in DMSO as a stock solution of 20 mmol/L, and Adriamycin (=doxorubicin; purchased from Sigma-Aldrich) was dissolved in double-distilled water (ddH2O) as a stock solution of 2 mg/mL (=3.4 mmol/L). Camptothecin (Sigma-Aldrich) was dissolved in DMSO as a stock solution of 260 µmol/L for microRNA induction experiments and of 2.6 µmol/L for cotreatment in combination with microRNA overexpression experiments. 5-FU (Sigma-Aldrich) was dissolved in DMSO as a stock solution of 300 mmol/L. Corresponding amounts of DMSO alone were added in control experiments. In experiments involving nucleic acid transfection and drug treatment, the cells were first transfected, incubated for 24 h, and then treated with the chemotherapeutic drug.
MicroRNA microarray analysis to identify p53-responsive micro-RNAs
SJSA cells were treated with Nutlin-3 at a concentration of 8 µmol/L for 24 h or with DMSO alone. RNA samples enriched for small RNA molecules were isolated by using the mirVana RNA Isolation kit (Ambion) according to the manufacturer’s protocol. Synthetic control RNA molecules contained in mirVana microRNA Labeling kit (Ambion) were added, and 40 µg of small RNA molecules from each condition were tailed using the mirVana microRNA Labeling kit. Half of each volume was labeled with the Cy3 fluorescent dye, and the other half was labeled with a Cy5 reactive dye (Cy3 and Cy5 dyes were purchased from Amersham) to perform a dye swap. The Nutlin-3 sample RNA labeled with Cy3 and the DMSO sample RNA labeled with Cy5 were cohybridized on three microarrays, and the dye-swapped labeled RNA of both conditions was cohybridized on another three microarrays. Microarrays were prepared by robotic spotting of DNA oligonucleotide probes complementary to human microRNA sequences (mirVana microRNA Probe Set, Ambion) on glass slides that had been epoxy coated (CodeLink).
After overnight incubation, the hybridized microarrays were washed four times with increasing dilutions of SDS and SSC and scanned with the G2505B Microarray Scanner (Agilent Technologies). Data were normalized to let-7 gene family members.
RNA extraction and quantitative reverse transcription-PCR analysis
Total RNA, including microRNA, was isolated by using the mirVana RNA Isolation kit. To detect the p21 mRNA expression, the isolated RNA was DNase treated and reverse transcribed using iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer’s instructions. iQ SYBR Green Supermix (Bio-Rad) was used for real-time PCR applications. The primer set for p21 quantitative reverse transcription-PCR (qRT-PCR) is 5′-TAGGCGGTTGAATGAGAGG and 5′-AAGTGGGGAGGAGGAAGTAG. PCR conditions were as follows: 2 min at 95°C followed by 40 cycles of 95°C for 15 s, 56°C for 30 s, and 72°C for 45 s. Stem-loop qRT-PCR for mature microRNAs was done using the Taqman MicroRNA assays (Applied Biosystems) as described (21). Gene expression profiles were normalized to RNU6B or miR-16, calculated using the 2−ΔΔ_C_t method.
Chromatin immunoprecipitation
Cells were treated with 300 nmol/L camptothecin for 24 h. Chromatin was cross-linked with proteins by 1% (v/v) formaldehyde. The cross-linking was stopped by adding glycine to a final concentration of 0.125 mol/L. After washing twice in PBS, the cells were scraped into Tris-EDTA collection buffer [10 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 1 mmol/L EDTA] and pelleted. The pellets were resuspended in SDS-Tris-EDTA lysis buffer [50 mmol/L Tris (pH 8.0), 10 mmol/L EDTA, 1% (w/v) SDS]. The probes were sonicated 25 times for 30 s with a Bioruptor sonicator (Diagenode) and pelleted. The supernatant was diluted with dilution buffer [17 mmol/L Tris (pH 8.0), 167 mmol/L NaCl, 1.2 mmol/L EDTA, 1.1% (v/v) Triton X-100, 0.01% (w/v) SDS]. Antibodies (1 µg) against p53 (Santa Cruz Biotechnology) were added and incubated overnight at 4°C. Anti-HA antibody (Santa Cruz Biotechnology) was used as negative control under the same condition. Protein A-Sepharose was added for 2 h. Immunoprecipitates were washed with Bio-Rad Micro Bio-Spin Columns. After brief centrifugation, the pellet was resuspended in 10% Chelex (Bio-Rad) and digested with proteinase K for 30 min at 55°C. Proteinase K was then inactivated by incubation at 95°C for 10 min. The probes were then pelleted, and 2 µL of the supernatant were used for PCR. The following primers were used for quantitative PCR (qPCR): 5′-AGCAGGCTGTGGCTCTGATT-3′ and 5′-CCAGCCTCTTCTATGCCAGA-3′ for known p53-binding site on the CDKN1A/p21 gene (p21 −2283; positive control), 5′-TTGTTCAATGTATCCAAAAGAAACA-3′ and 5′-TGAGATAAAGCTTCTTCCCTTAAAAA-3′ for the presumptive binding site close to the miR-194-1/miR-215 gene, and 5′-CATAAGCCTGCGTCAGATCA-3′ and 5′-CCTGTGTTGGGTTGACAGTG-3′ for the hMT-RNR2 gene (negative control). The reaction mix consisted of 2 µL template, 12.5 µL of 2× PCR Master Mix (Bio-Rad), 1.5 µL forward primer, 1.5 µL reverse primer, and 7.5 µL ddH2O. qPCR was performed as described above for microRNA qRT-PCR. Conditions were as follows: 10 min at 95°C followed by 40 cycles of 95°C for 15 s and 60°C for 60 s.
Colony formation assay
U2OS and H1299 cells were transfected with miR-vectors, a generous gift of R. Agami (The Netherlands Cancer Institute, Amsterdam, the Netherlands; ref. 22), in six-well plates. At 24 h after transfection, cells were trypsinized, counted, and seeded in flasks. Cultures were maintained for 2 wk with blasticidin (5 µg/mL), and cells were then fixed, stained, and photographed.
Immunoblot analysis
Proteins were separated on SDS-polyacrylamide gels and transferred to nitrocellulose. This was followed by an incubation with antibodies dissolved in PBS containing 5% milk powder and Tween 20 (0.05%). The following primary antibodies were used: mouse anti-Mdm2 (2A10, generous gift of A.J. Levine, Institute for Advanced Study, Princeton, NJ), mouse anti-p53 (DO-1, Santa Cruz Biotechnology), mouse anti-p21 (EA10, Calbiochem), mouse anti-Puma (EP512Y, Abcam), mouse anti-Noxa (114C307, Abcam), mouse anti-poly(ADP-ribose) polymerase-1 (Ab-2, Oncogene), rabbit anti-cleaved caspase-3 (5A1, Cell Signaling), and rabbit anti-β-actin (ab8227, Abcam). Primary antibodies were detected by a peroxidase-coupled secondary antibody (Jackson) and chemiluminescence (Pierce).
Transient transfection of cells with microRNAs
HCT116 cells were transfected with 5 or 10 nmol/L of precursor microRNA molecules (Ambion) by using HiPerFect Transfection Reagent (Qiagen) after 24 h of seeding. A549 and DLD-1 cells were reverse transfected in six-well plates. Briefly, microRNA was diluted in 250 µL medium without serum and incubated for 5 min. Meanwhile, 5 µL Lipofectamine 2000 (Invitrogen) was mixed with 250 µL medium without serum and incubated for 5 min. After 5-min incubation, the diluted microRNA was combined with the diluted Lipofectamine 2000, and mixed and incubated for 20 min at room temperature. The microRNA-Lipofectamine 2000 complex was added to each well. Then, counted cells were added directly on the top of the complex, and mixed well and incubated for indicated days.
MicroRNA knockdown
Scramble locked nucleic acid (LNA) or LNA oligonucleotides against miR-192 (anti-miR-192) were obtained from Exiqon A/S. A549 cells were reverse transfected with 100 nmol/L anti-miR-192 by using Lipofectamine 2000 as described above.
Crystal violet staining
Crystal violet stock solution consisted of 0.1% crystal violet and 25% methanol. After transfection and drug treatment, the cells were incubated on ice for 5 min and then rinsed once with ice-cold PBS followed by incubation with ice-cold methanol for 10 min. Subsequently, the crystal violet stock solution was applied at room temperature for 10 min. After repeated rinsing with PBS, the plates were air dried and scanned with an Epson Perfection 4990 Photo Scanner in Positive Film mode. The blue pixel rate was determined in each case using the computer software Adobe Photoshop 9.
Fluorescence-activated cell sorting analysis
Attached cells, combined with the floating cells, were harvested. Cells were fixed in 70% ethanol at 4°C overnight. Cells were then washed, rehydrated, and resuspended in PBS containing 0.5 mg/mL RNase A and 30 µg/mL propidium iodide. Samples were analyzed by FACScan Flow Cytometry System. Cell cycle analysis was carried out by the ModFit software.
Cell images
Cells were photographed using the Zeiss Axiovert 40 CFL microscope in combination with a Canon PowerShot A620 camera.
Staining for senescence-associated β-galactosidase
Cells were transfected with pre-miR molecules or treated with camptothecin (20 nmol/L) for 120 h in total. Staining for senescence-associated β-galactosidase (SA-β-Gal) was performed with the Senescence β-Galactosidase Staining kit purchased from Cell Signaling. After overnight staining, the plates were washed with PBS and photographed. The amount of blue color staining was quantified by digital analysis of the images (Adobe Photoshop).
Patient material
Biological material from primary tumors was obtained directly from surgery. All tumors were classified according to the International Union Against Cancer tumor-node-metastasis staging system. Informed written consent was obtained from all patients, and research protocols were approved by the ethical committee of Aarhus County. The tumor biopsies used for expression profiling were immediately lysed in a guanidinium thiocyanate solution and snap frozen in liquid nitrogen. RNA was extracted from the samples using a standard Trizol RNA extraction method (Invitrogen). The quality of the RNA samples was assessed using an Agilent Bioanalyzer (criteria: 28S/18S > 1 and RIN > 5). For this study, we only included patients with stage II colon cancers. Clinical and histopathologic variables for the patients included in this study are listed in Supplementary Table S3.
MicroRNA microarray expression profiling and data analysis from patient samples
Microarrays were produced using a LNA-based oligonucleotide probe library (miRCURY LNA array ready to spot v.7.1; Exiqon). Oligonucleotides were spotted in duplicates in a 10 µmol/L phosphate buffer on CodeLink microarray glass slides (GE Healthcare) using a VersArray ChipWriter Pro system (Bio-Rad) as previously described (22). Sample RNA (2 µg) was directly labeled with Hy3 using the miRCURY LNA array labeling kit (Exiqon). As reference, we used a pool of RNA extracted from bladder, prostate, and colon tumors. For each experiment, we labeled 2 µg of the reference RNA with Hy5 using the LNA array labeling kit (Exiqon). Hybridization and washing of the microarray slides were performed as recommended by Exiqon. Scanning was performed using a ScanArray 4000 scanner (GSIL umonics). Following scanning of the microarrays, we used TIGR Spotfinder 2.23 software to generate raw intensity data, which were LOWESS (global) normalized using TIGR MIDAS 2.19 software (23). Average log2 ratios were calculated from the normalized data based on the two measurements of each microRNA. We assessed the significance of the expression differences between normal mucosa and microsatellite stable [MSS; and unstable microsatellite (MSI)] cancer using the nonparametric Mann-Whitney U test.
Results
p53 induces the expression of specific microRNA species
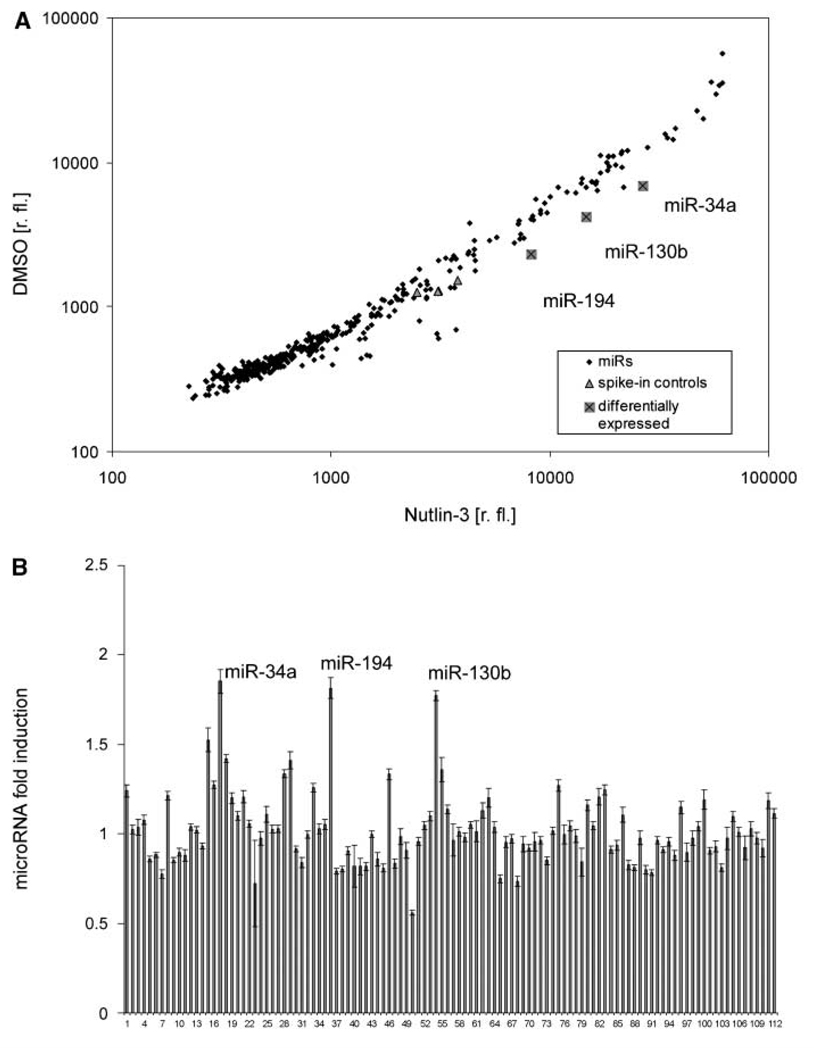
We first established a robust yet specific system to identify p53-regulated microRNA species. SJSA cells (formerly called OsA-CL) are derived from an osteosarcoma and massively overexpress the principal antagonist of p53, the Mdm2 oncoprotein, as a result of MDM2 gene amplification (23). These cells were treated with Nutlin-3a, a well-characterized pharmacologic antagonist of the Mdm2-p53 interaction with little p53-independent effects on gene expression (3). The induction of known p53-responsive genes by this treatment (e.g., CDKN1A/p21 and PIG3) was verified by immunoblot analysis and/or qRT-PCR (Supplementary Fig. S1). Subsequently, we purified small RNA species from SJSA cells that were treated with Nutlin-3a or the DMSO solvent alone followed by the labeling of these RNAs with fluorescent dyes and hybridization of an oligonucleotide array representing known microRNAs. The signal intensities corresponding to ~400 microRNA species were monitored, comparing the two conditions in three independent experiments with two dye swaps each. Consequently, several microRNAs were found with consistently higher expression levels in response to Nutlin-3a (Fig. 1; Supplementary Table S1). Most notably, besides the previously described miR-34a, we identified miR-194 as a robust p53-responsive microRNA species.
Figure 1.
microRNA expression profiling. RNA samples enriched for small RNA molecules, extracted from SJSA cells treated with 8 µmol/L Nutlin-3 or DMSO alone, were conjugated with Cy3 or Cy5 dyes and hybridized to three microarrays. A dye swap was performed on another three microarrays. A, the raw signal level for each probe is presented in relative fluorescence units (r. fl.), calculated as an average from three microarrays (see Supplementary Table S2 for raw signal values). Rhombi represent all analyzed microRNAs, triangles represent spiked-in synthetic control RNA molecules, and crossed squares represent human microRNAs that showed a notable differential expression. B, fluorescent signals of spots corresponding to human microRNAs with a relative fluorescence over 1,000 were normalized according to let-7 gene family members. The relative induction is presented for each microRNA. Six microarrays, including dye swap, were analyzed. Columns, mean for each microRNA; bars, SE. The names of microRNAs represented by the columns correspond to the order shown in Supplementary Table S1.
p53 induces the expression of miR-192, miR-194, and miR-215
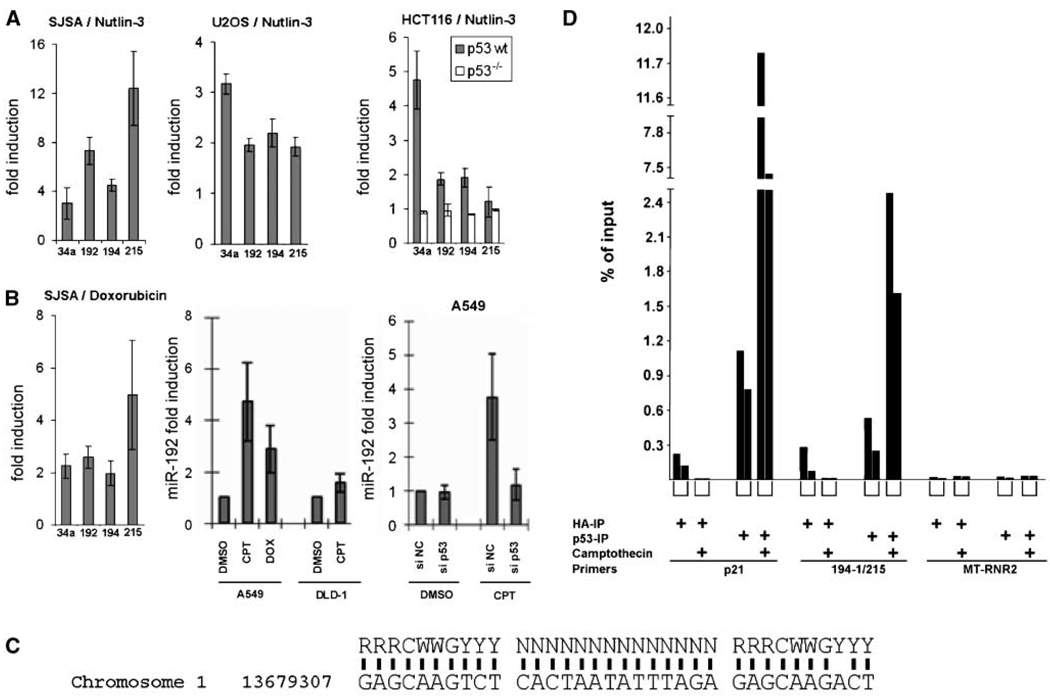
To confirm the responsiveness of selected microRNAs to p53 under various conditions, cell lines with varying p53 status were treated with Nutlin-3a (Fig. 2_A_) or genotoxic agents (Fig. 2_B_) followed by qRT-PCR to monitor microRNA levels. In these assays, the response of miR-130b to p53 was somewhat variable (data not shown), despite its differential fluorescence signal on the arrays (Fig. 1), whereas miR-194 was consistently induced. We therefore focussed our analysis on the latter microRNA species. Importantly, however, miR-194 can be derived with identical sequence from two different microRNA clusters (i.e., the miR-194-2/miR-192 cluster on chromosome 11 and the miR-194-1/miR-215 cluster on chromosome 1); strikingly, the partners of miR-194 in each cluster, miR-192 and miR-215, are closely related to each other (depicted in Supplementary Fig. S2), further increasing the similarity of the two clusters. miR-192 and miR-215 were not detected above background fluorescence on our arrays, leaving open the question whether they are p53 inducible. We therefore analyzed the expression of miR-194 along with miR-192 and miR-215 in response to p53 by qRT-PCR, and we used miR-34a as a positive control. All four microRNA species were robustly and comparably induced in various cell lines (SJSA, U2OS, and HCT116) by Nutlin-3a (Fig. 2_A_) and DNA damage (Fig. 2_B_). Comparison of HCT116 cells [carrying wild-type (wt) p53] with an isogenic derivative that carries a targeted deletion of the TP53 gene (24) revealed that the induction of microRNAs was strictly dependent on p53 (Fig. 2_A_). Accordingly, miR-192 was induced by camptothecin in A549 (p53 wt) but not DLD-1 (p53 mutant) cells (Fig. 2_B_), and the induction of miR-192 by camptothecin in A549 cells (p53 wt) was abolished by small interfering RNA (siRNA) to p53 (Fig. 2_B_). In an effort to identify the mechanism(s) leading to the induction of the two microRNA clusters, we searched for putative p53-binding sites in the genome surrounding the two microRNA clusters, applying an appropriate computer algorithm, P53MH (25). We found a sequence element with close similarities to a consensus p53-binding site 2.7 kb upstream of the miR-194-1/miR-215 cluster (Fig. 2_C_). Chromatin immunoprecipitation (ChIP) with antibodies to p53 revealed that this site indeed associated with p53, and this association was further increased when p53 levels were augmented through the genotoxic agent camptothecin (Fig. 2_D_). As expected, an even larger proportion of DNA from the strongly p53-responsive CDKN1A/p21 promoter associated with p53, whereas the control amplification of a MT-RNR2 gene portion yielded very little background signals. Thus, p53 associates with a DNA region in proximity of the miR-194-1/miR-215 cluster, providing a possible explanation for its ability to induce the levels of the corresponding microRNAs. We also cloned a 3-kb genomic fragment upstream of the miR-194 sequence into a luciferase reporter plasmid, but this did not lead to a p53-inducible signal (data not shown). However, it is currently unknown where the transcription of the corresponding pri-microRNA starts. Analogous analysis of microRNA promoters that are repressible by c-Myc has revealed that many transcriptional start sites frequently start more than 50 kb upstream of the region corresponding to the mature microRNA (26). Traditional approaches to identify the 5′ ends of such transcripts are hampered by their rapid processing. Like in these analogous cases, it seems that other regulatory elements of transcription, in addition to the identified binding site, might contribute a large part of p53 responsiveness. In any case, the net result is a robust induction of miR-192/miR-194/miR-215 transcription that entirely depends on the presence and activity of p53.
Figure 2.
Induction of microRNA expression through p53. A, qRT-PCR analysis of miR-34a, miR-192, miR-194, and miR-215 in SJSA, HCT116 p53 wt, and p53−/−, as well as U2OS cells treated with 8 µmol/L Nutlin-3 for 24 h. Fifty nanograms of extracted RNA enriched for small RNA molecules were subjected to qRT-PCR to quantify miR-34a, miR-192, miR-194, and miR-215, with miR-16 used for internal normalization. Columns, relative microRNA expression levels (“fold induction”) as calculated from the results of three independent experiments; bars, SE. B, SJSA cells (p53 wt) were treated with the doxorubicin, and the relative levels of microRNAs (fold induction) were then determined by qRT-PCR. A549 (p53 wt) or DLD-1 (p53 mutant) were treated with the indicated drugs [camptothecin (CPT), 200 nmol/L; doxorubicin (DOX), 100 ng/mL] and/or siRNA as detailed in Materials and Methods. The relative levels of miR-192 (fold induction) were then determined by qRT-PCR. C, presumptive p53-binding site on chromosome 1, 2.7 kb upstream of the miR-194-1/miR-215 cluster, with comparison with the known consensus sequence bound by p53 (43). D, ChIP of p53 with the genomic fragment comprising the sequence element shown in C. A549 cells were treated with camptothecin (300 nmol/L) for 24 h. Cells were harvested and then subjected to ChIP analysis with antibodies directed against p53 or the HA tag as a control. Precipitated DNA was analyzed by qPCR with primers amplifying the known 5′ p53-binding site of the CDKN1A/p21 promoter and the putative binding site near the miR-194-1/miR-215 gene cluster. A fragment of the MT-RNR2 gene was used as negative control. Columns, percentage of precipitated DNA compared with the amount of input. The results from two independently performed experiments are shown in each case, with brackets below the columns representing equivalent conditions.
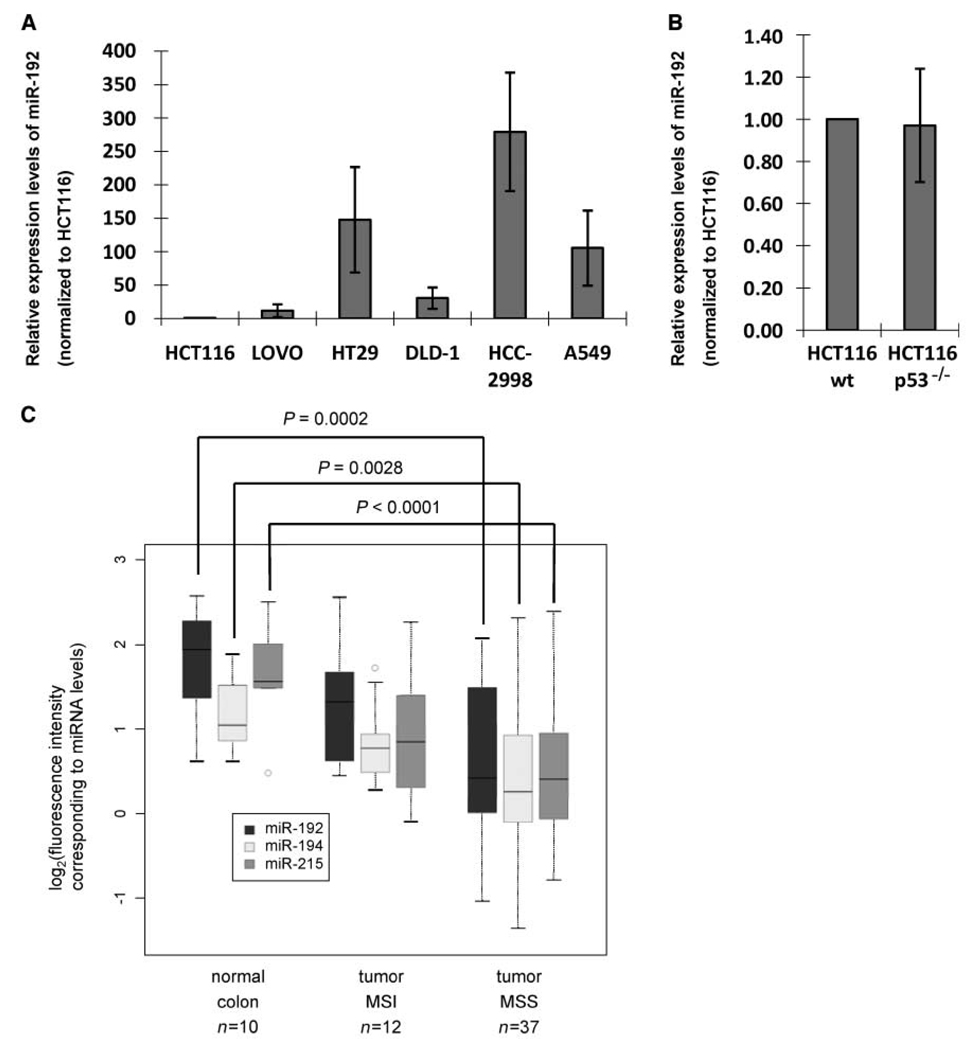
The levels of miR-192, miR-194, and miR-215 are high in normal colon tissue but lower in colon cancer cells
miR-192, miR-194, and miR-215 are mostly expressed in colon and liver (7, 27). When studying their levels in several cell lines, some but not all colon cancer–derived cell lines showed high expression (Fig. 3_A_). HCT116 cells contained comparatively low levels of miR-192 (Fig. 3_A_), and this was not affected by a deletion of the p53 gene (Fig. 3_B_), in contrast to the situation in the same cell lines but after treatment with Nutlin-3 (Fig. 2_A_). In addition, the lung adenocarcinoma–derived cell line A549 (p53 wt) contained high levels of these microRNAs, and these were further induced by DNA-damaging agents (Fig. 2_B_). As ~50% of colon cancers have lost or inactivated p53 (28), we reasoned that at least in a subset of colon carcinomas, the microRNAs under study may be down-regulated relative to normal mucosa. Indeed, microRNA profiling of 49 stage II colon cancers and 10 normal mucosa samples using spotted LNA-based microarrays revealed that the microRNAs encoded from both clusters were down-regulated in cancer tissue relative to normal mucosa. The down-regulation was more pronounced in cancers that were MSS and less prominent in cancers with MSI (Fig. 3_C_; Supplementary Fig. S3; Supplementary Tables S2–S4). Interestingly, p53 mutations are less common in MSI tumors than in MSS tumors (29). Noteworthy, the abundance of all three microRNAs was positively correlated to each other (Pearson correlation coefficient: range, 0.73–0.83), supporting the notion of a common regulation, possibly in part by p53. However, the available samples did not allow the simultaneous detection of p53 mutations or the analysis of common pathways that can suppress wt p53 functions. Therefore, it remains very possible that the down-regulation of the microRNAs under study in the tumor samples is partially due to alterations unrelated to the p53 status. Because the transition from normal through immortalized to fully transformed cell phenotype can be recapitulated in culture for breast cancer but not colon cancer, we are unable to assess the stage(s) of cancerogenesis where miR-192 is lost. In any case, however, all three microRNAs are down-regulated in a significant subset of colon cancers. It is therefore tempting to speculate that these microRNA species might carry out a tumor-suppressive function, and possible mechanisms for this were further examined in subsequent experiments.
Figure 3.
miR-192 levels in colon and colorectal cancer. A, RNA samples were extracted from different colorectal cancer cell lines (HCT116, Lovo, HT29, DLD-1, and HCC-2998) and also from the lung adenocarcinoma cell line A549. miR-192 levels were quantified by qRT-PCR as described in Materials and Methods. B, RNA samples were extracted from HCT116 p53 wt or p53−/− cells. miR-192 levels were quantified by qRT-PCR as described in Materials and Methods. C, microRNA expression levels from patient material were determined by array hybridization. The log2 of hybridization intensities were determined for each sample, grouped according to its origin: normal colon mucosa, colon cancer with MSI, and colon cancer with MSS, as shown in details in Supplementary Fig. S3 and Supplementary Tables S2 to S4. The data from this study are summarized in a box plot format, including the level of significance that distinguishes the MSS tumors from normal tissue for each microRNA.
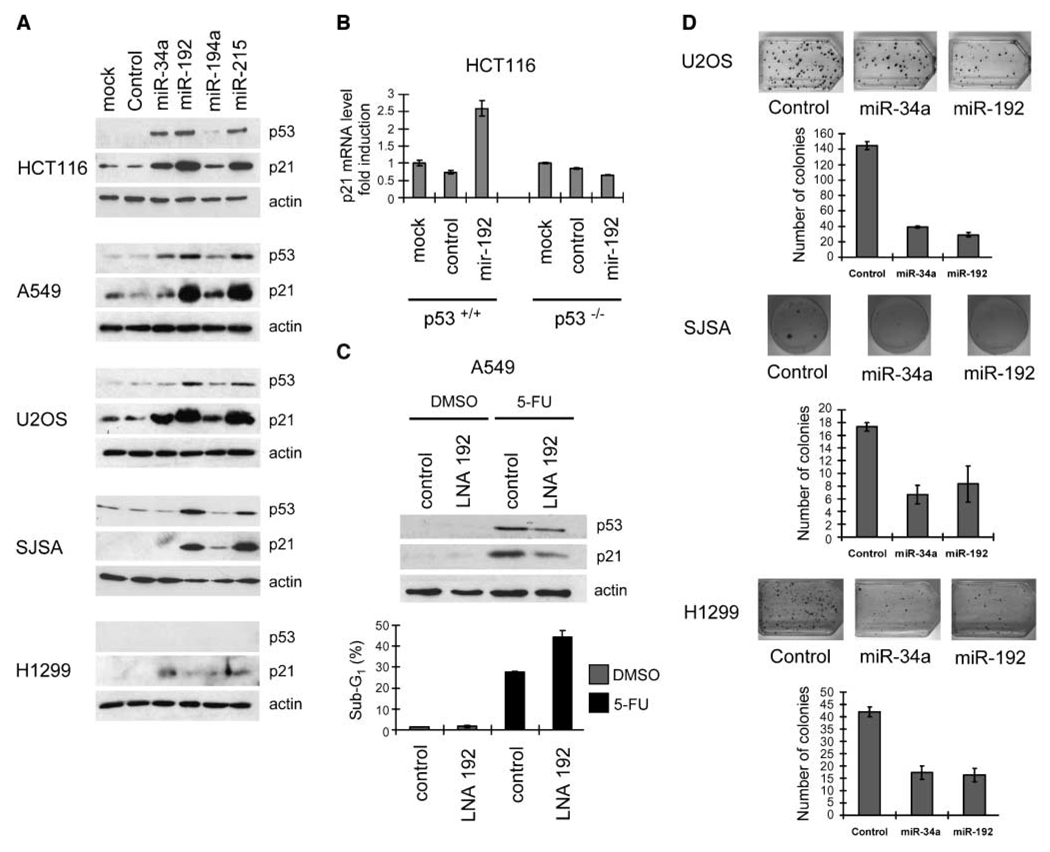
p53-responsive microRNAs are capable of enhancing p21 levels through p53, but they can suppress cell clone formation in a partially p53-independent manner
First, we asked whether the newly identified p53-responsive microRNAs might feedback on the activity of p53. To test this, miR-192, miR-194, miR-215, and miR-34a were introduced by transfection into HCT116, A549, U2OS, and SJSA cells, all carrying wt p53, as well as the p53−/− H1299 cells, followed by the detection of p53 and its target gene product p21 using immunoblot analysis. We found that miR-192 and miR-215 induced the expression of p21 in all cells that carried wt p53, whereas miR-34a and miR-194 did so only to a lesser extent; p21 and p53 were detected weakly or not at all in H1299 cells (Fig. 4_A_). In HCT116 cells with wt p53, but not with a p53−/− status, miR-192 increased the levels of p21 mRNA and protein (Fig. 4_B_; Supplementary Fig. S4), strongly suggesting that miR-192 activates p53. Proapoptotic p53 target gene products accumulated less or not at all (Supplementary Fig. S4), arguing that miR-192 mostly activates the cell cycle arrest function of p53. To determine whether endogenous miR-192 also makes a detectable contribution to p21 induction, we antagonized miR-192 in A549 cells using antisense LNA oligonucleotides (and a scrambled negative control LNA) while inducing a p53 response with 5-FU. Strikingly, antagonizing miR-192 attenuated the accumulation of p53 and p21 (Fig. 4_C_), in agreement with the concept of miR-192 as an internal amplifier of p21 induction by p53. On the other hand, 5-FU–induced cell death was enhanced by LNA-antagonizing miR-192 perhaps because the antiapoptotic activity of p21 (30–32) was now compromised (Fig. 4_C_; Supplementary Fig. S5). Next, we sought to determine whether p53 target microRNAs might interfere with clonogenic survival. To this end, U2OS cells (wt p53) were transfected with expression vectors for miR-192 or miR-34a (22) followed by selection of stable transfectants with blasticidin. The number of cell colonies obtained in this way revealed that miR-192 suppressed cell clone formation to an extent comparable with miR-34a (Fig. 4_D_). The same observation was made when using SJSA cells (Fig. 4_D_), the cell line with wt p53 that we had originally used to identify p53-responsive microRNAs (Fig. 1). We next addressed the question whether colony suppression is due to the induction of p53 by carrying out the analogous clonogenic assays in H1299 cells, a cell line that lacks endogenous p53 (HCT116 cells could not be used in this assay because they were most susceptible to a disruption of cell adhesion, as further explained below). Somewhat surprisingly, miR-192 still revealed colony suppression about as efficiently as miR-34a (Fig. 4_D_), strongly suggesting that both microRNAs are capable of interfering with cell clone formation by mechanisms that are at least in part independent of p53.
Figure 4.
p21 induction and colony suppression by p53-responsive microRNAs. A, cells of the indicated lines were transfected with the indicated microRNAs (10 nmol/L) and incubated for 48 h; then, cell lysates were subjected to immunoblot analysis to detect p53, p21, or β-actin (input control). B, HCT116 cells, p53 wt or p53−/−, were transfected with either control microRNAs or miR-192 (10 nmol/L) and incubated for 48 h, and p21 mRNA levels were quantified by qRT-PCR. Columns, mean values obtained from three independent experiments, normalized to the mock-transfected p53 wt cells; bars, SE. C, A549 cells were transfected with either scrambled LNA or anti-miR-192 LNA (100 nmol/L) and then (24 h after transfection) treated with 500 µmol/L 5-FU or the DMSO solvent alone. After 24 h of 5-FU treatment, the cells were harvested and cell lysates were subjected to immunoblot analysis. In parallel, flow cytometry was performed after transfection and 48 h of incubation with 500 µmol/L 5-FU or DMSO alone. The cells were then trypsinized, fixed with ethanol, and stained with propidium iodide. The percentages of cells with a DNA content below 2N (sub-G1, gated to the total number of cells) are indicated in each case. The columns are based on the raw data shown in Supplementary Fig. S5 and indicate the mean values obtained from three independent experiments; bars, SE. D, U2OS, H1299, or SJSA cells were transfected with control miR-vector or miR-vec-34a or miR-vec-192. The cultures were maintained for 2 wk with blasticidin (5 µg/mL) to select stable transfectants, and the cells were then fixed, stained with crystal violet, and photographed. The numbers of colonies were determined by a person unaware of the identity of the samples, and the results from three experiments are shown in the column diagrams. Columns, mean; bars, SE.
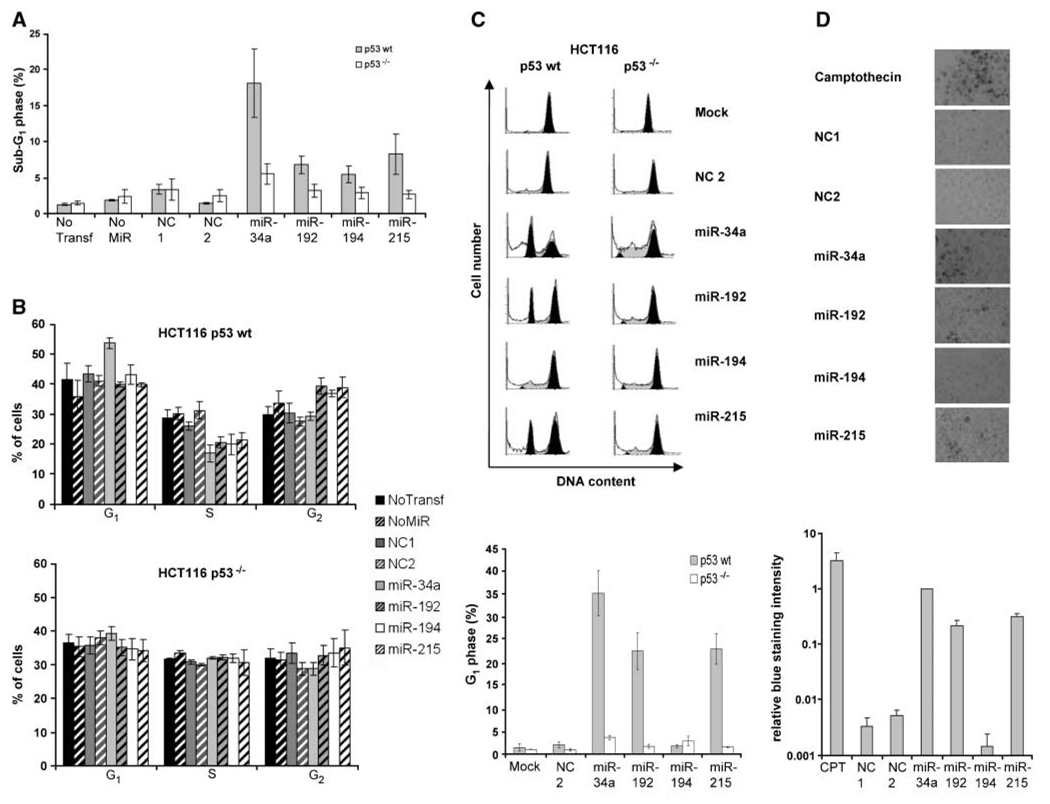
miR-192 and miR-215 are capable of inducing cell cycle arrest, but they trigger cell death only with lower efficiency than miR-34a
To further explore the mechanism(s) that allows miR-192 to interfere with colony formation, we used flow cytometry to analyze how miR-192, miR-194, and miR-215, in comparison with miR-34a, affect the progression through the cell cycle and the corresponding distribution of DNA content. Importantly, none of the novel p53-responsive microRNAs induced the appearance of cells in a sub-G1 fraction (indicative of cell death) to an extent comparable with miR-34a, although some sub-G1 cells were observed with miR-192 and miR-215 (Fig. 5_A_; Supplementary Fig. S6). Only in HCT116 cells that lack p21 due to a targeted disruption of the gene (24), miR-192 could induce apoptosis to a moderate degree (Supplementary Fig. S7). miR-192 and its close relative, miR-215, were both capable of inducing a moderate but reproducible accumulation of HCT116 cells with a DNA content of 4N, corresponding to G2 or M phases of the cell cycle (Fig. 5_B_; Supplementary Fig. S6). miR-194 showed a similar effect. The effects of all microRNAs under study on apoptosis and cell cycle distribution were largely (although not completely) dependent on p53, as revealed by the parallel analysis of HCT116 cells with a p53 knockout (Fig. 5_A_ and C; Supplementary Fig. S6). Using a nocodazole trap, we found that miR-192 and miR-215, as well as miR-34a but not miR-194, were efficiently retaining a portion of a HCT116 cell population in the G1 phase of the cell cycle (Fig. 5_B_). This was not observed in HCT116 cells lacking p53 (Fig. 5_B_) nor in HCT116 cells lacking p21 (Supplementary Fig. S7), whereas A549 cells (wt p53) were also retained in G1 by miR-192 and miR-215 (Supplementary Fig. S8). Hence, miR-192 and miR-215 can arrest cells in G1 by mechanisms that are at least partially dependent on functional p53 and p21. In summary, we conclude that the induction of apoptosis, while being well documented for miR-34a (10–12), cannot fully account for the ability of miR-192 to suppress the formation of cell clones. Rather, miR-192 and miR-215 induce cell cycle arrest in G1 and G2-M (in an accompanying article by Georges and colleagues, arrest in G2 rather than M is further specified). To follow-up on this comparison between the novel p53-responsive microRNAs and miR-34a, we addressed their ability to induce acute senescence, previously described for miR-34a (17). HCT116 cells were transfected with control RNA or the microRNAs under study followed by SA-β-Gal assay. As shown in Fig. 5_D_, miR-192 and miR-215 were capable of inducing senescence to some extent but not as efficiently as miR-34a. Again, we conclude that miR-192 and miR-215 suppress cell clone formation by mechanisms that differ from miR-34a.
Figure 5.
Cell cycle arrest and senescence induced by p53-responsive microRNAs. A, extent of apoptosis after transfection of HCT116 p53 wt or p53−/− cells with the indicated pre-miR molecules and 48 h of incubation. The cells were trypsinized, fixed with ethanol, and stained with propidium iodide. The percentages of cells with a DNA content below 2N (sub-G1, gated to the total number of cells) are indicated in each case. An example of the raw data obtained by flow cytometry is shown in Supplementary Fig. S6. B, cells were prepared as in A. The diagrams indicate the percentage of cells in G1, S, and G2 phases of the cell cycle after transfection with the indicated pre-miR molecules. Flow cytometry diagrams are shown in Supplementary Fig. S6. C, HCT116 p53 wt or p53−/− cells transfected with 10 nmol/L of the indicated microRNAs were incubated for 2 d, and cells were then treated with nocodazole (100 ng/mL) for another 18 h. Cell cycle profiles were monitored as described in Materials and Methods. The diagram shows the percentages of cells with a DNA content below 2N (sub-G1, gated to the total number of cells). D, microRNAs were transfected as pre-miR molecules in HCT116 p53 wt cells. One well was left without transfection (no transfection) as a negative control, and one well was treated with low-dose camptothecin treatment (20 nmol/L) as a positive control. SA-β-Gal was stained after 4 d of incubation. Blue staining intensity of three pictures for every condition was estimated by Adobe Photoshop software. Staining intensity was normalized to miR-34a staining intensity.
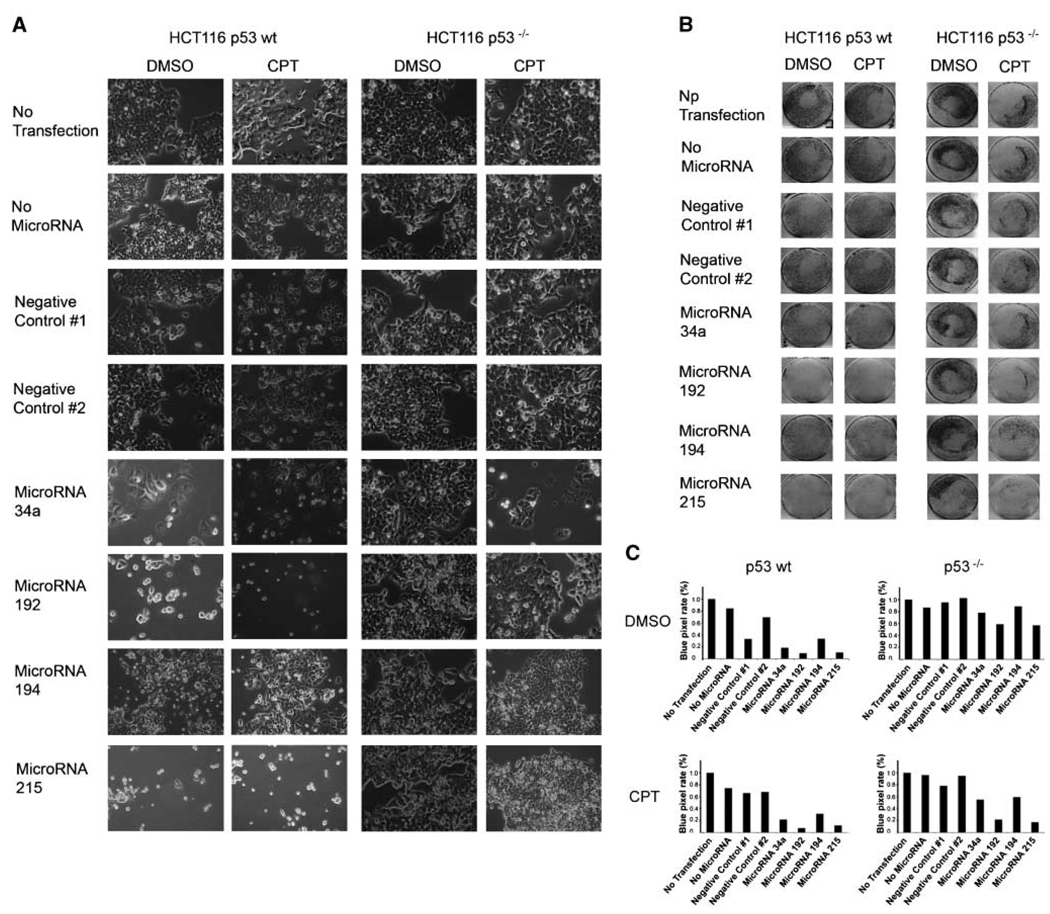
miR-192 and miR-215 can interfere with cell adhesion
While carrying out the experiments described above, we noticed that transfection with miR-192 and miR-215 frequently induced a morphologic change in HCT116 cells, as documented in Fig. 6_A_. In response to these microRNAs, the cells assumed a round shape and partially detached from the plate. This was further aggravated when the cells were additionally treated with camptothecin. However, in contrast to the apoptotic cells seen on transfection with miR-34a, the cells that had taken up miR-192 and miR-215 frequently retained a normal size and undamaged surface, as if they were only detaching without dying immediately. To quantify this phenomenon, the detached cells were washed off the plate followed by crystal violet staining (Fig. 6_B_). For quantification, the plates were scanned, and blue color intensity (pixel rate) was determined (Fig. 6_C_). Indeed, it was found that miR-192 and miR-215 were capable of inducing cell detachment, whereas miR-34a and miR-194 did so only to a lesser extent. Detachment was not observed in transfected but otherwise untreated HCT116 cells lacking p53. When the cells were additionally exposed to camptothecin, however, miR-192 and miR-215 led to cell detachment in the presence and, to some extent, even in the absence of p53. We conclude that miR-192 and miR-215 can interfere with cell adhesion and that this phenomenon is partially but not fully dependent on functional p53.
Figure 6.
Cell detachment induced by miR-192 and miR-215. HCT116 cells with or without a targeted disruption of TP53 were seeded at 60,000 per well in six-well plates and transfected with pre-miR molecules to express miR-34a, miR-192, miR-194, and miR-215. In addition, two molecules that are not occurring in nature were transfected (Negative Control #1 and Negative Control #2), and one well for each condition was treated with transfection reagent but without transfected nucleic acid (No MicroRNA) and one was cultured without nucleic acid and without transfection reagent (No Transfection). One day after transfection, camptothecin was added to a final concentration of 20 nmol/L, and negative controls were performed with DMSO alone. A, images were taken to show cell morphology after 48 h of incubation. B, cells were stained with crystal violet solution after another 48 h of camptothecin or DMSO incubation. Images were taken to show the entire wells and the remaining cells within them. C, blue pixel rates of the images shown in Fig. 4_B_ were determined using the Photoshop software and are indicated by columns for each condition.
Discussion
The application of a direct pharmacologic activator of p53 allowed us to identify two clusters of microRNAs expressing three distinct microRNA species. The same microRNAs were also up-regulated by DNA damage in a p53-dependent fashion, but they were down-regulated in colon cancer. The functional analysis of miR-192 and miR-215 revealed that they can induce the accumulation of p53 and its target gene product p21, and antagonizing endogenous miR-192 attenuated this accumulation. Accordingly, miR-192 and miR-215 suppressed proliferation and cell adhesion in a partially but not completely p53-dependent manner.
Given the numerous searches for p53-responsive genes, it seems surprising that miR-192/miR-194/miR-215 was not identified earlier with regard to their inducibility by p53, especially because, in some systems under study, they were more strongly induced than miR-34a. We speculate that this is due to the use of a specific Mdm2 inhibitor, rather than DNA damage, for activation of p53, in combination with a cell line (SJSA) that contains high baseline amounts of Mdm2 due to gene amplification (23). Most previous studies searching for p53-responsive microRNAs used a DNA-damaging protocol to induce p53 in cells with a normal MDM2 gene dosage, and our results also show that, under such circumstances, miR-34a is up-regulated more profoundly than miR-192/miR-194/miR-215 (Fig. 1_A_). However, DNA damage activates transcription factors other than p53 (e.g., E2F1 or FOXO family members), and this might modify the transcriptional program of p53 alone. Moreover, it is known that many p53-responsive protein-encoding genes are induced only under specific circumstances (e.g., as a function of DNA damage–induced posttranslational modifications of p53). It is therefore tempting to speculate that miR-34a might belong to the apoptosis-specific class of p53-responsive genes, such as Puma, Noxa, or AIP, whereas miR-192/miR-194/miR-215 is a novel representative of the cell cycle regulatory class, similar to p21, GADD45, or _14-3-3_σ.
The mechanism(s) that allows miR-192 and miR-215 to induce p53 activity seem to involve a spectrum of target mRNAs. Obvious candidate mRNAs, such as those encoding Mdm2, MdmX (33), or ARF-BP1 (34), did not turn out to be targeted by miR-192/miR-215 according to our unpublished observations. Instead, an accompanying article (35) identifies numerous target mRNAs for both microRNA species, highly enriched for gene products that govern cell cycle regulation, although none of these mRNAs has previously been described to encode a direct regulator of p53. We therefore favor the hypothesis that these target mRNAs, when regulated in their entity, may alter cell cycle progression in a way that ultimately affects p53 activity (e.g., by increased numbers of illegitimate checkpoint transitions). Along the same line, we cannot be certain whether p53 activation is the primary effect induced by miR-192/miR-215 or whether it only represents a secondary phenomenon that enhances initially p53-independent biological effects. In agreement with this notion, miR-192/miR-215 seems to reduce tumor cell proliferation even in the absence of p53, albeit with reduced efficiency.
For some tumor-suppressive functions studied here (i.e., apoptosis and senescence), the effects of miR-192/miR-215 were much weaker than those of miR-34a, raising the suspicion that miR-34a is the major effector microRNA for p53-induced cell death. However, loss of HCT116 cell adhesion was induced almost exclusively by miR-192/miR-215, in contrast to miR-34a. Detachment can have many reasons, including cell death, aberrant mitosis, loss of cell polarity, dysfunctional cytoskeleton, or reduced expression of one or several adhesion molecules. It should therefore not be regarded as a specific, monocausal phenotype. Interestingly, however, the accompanying study (35) found the DLG5 mRNA is one of the strongest targets of miR-192/miR-215, and DLG5 can interfere with cell adhesion through the reduction of cadherin transport to the cell surface (36). Disrupting cell adhesion might kill cells (e.g., by anoikis; ref. 37). On the other hand, the process of invasion and metastasis involves the loosening of existing cell-matrix contacts and the formation of new ones. It is tempting to speculate that miR-192/miR-215 might contribute to the elimination of anchorage-dependent cells in response to p53 activation, thereby avoiding tumorigenesis at an early stage.
Our finding that miR-192, miR-194, and miR-215 are lost in colorectal cancer while being highly expressed in normal colon further supports the idea that these microRNAs might carry out a tumor suppressor function. However, the available data also indicate that their expression is not exclusively driven by p53. p53 is a ubiquitously expressed protein, whereas the microRNA clusters under study are specifically found in the liver and colon, as revealed by previous studies (38–40). In addition, miR-192 expression was observed in glomeruli of the kidney (41). It remains to be determined whether the levels of miR-192, miR-194, and miR-215 also drop in tumor cells from liver and/or kidney, as they seem to do during colon carcinogenesis. In tissue-specific settings, the microRNAs under study might also carry out functions in addition to mediating the p53 response. Interestingly, it was recently shown that miR-194 is induced by the transcription factor HNF1-α when colonic epithelial cells undergo the transition from a stem cell–like to a more differentiated phenotype (42). Together with the presence of miR-192/miR-215 and miR-194 in the same clusters, this implies that miR-192 or miR-215, or both, might also be expressed under such conditions. Hence, the ability of the two latter microRNAs to induce cell cycle arrest might also contribute to the differentiation of colon epithelial cells.
Supplementary Material
1
2
3
Acknowledgments
Grant support: EU 6th Framework Program (Integrated Project Active p53), German Research Foundation, German Cancer Aid/Dr. Mildred Scheel Stiftung, Wilhelm Sander Stiftung, Statens Sundhedsvidenskabelige Forskningsråd of Denmark, Danish Cancer Society (Kræftens Bekæmpelse), and Novonordisk Fonden. C.J. Braun is a recipient of a stipend provided by the Studienstiftung des Deutschen Volkes. X. Zhang is a member of the Goöttingen Graduate School for Neurosciences and Molecular Biosciences.
We thank N. Chau and S. Georges for sharing with us their data on miR-192 before publication, R. Agami for the gift of microRNA expression plasmids, A. Scheel for help with diagrams, and C. Hippel and A. Dickmanns for excellent technical assistance.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 2.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17:631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 4.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 9.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 10.Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 11.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 13.He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 14.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Raver-Shapira N, Oren M. Tiny actors, great roles: microRNAs in p53’s service. Cell Cycle. 2007;6:2656–2661. doi: 10.4161/cc.6.21.4915. [DOI] [PubMed] [Google Scholar]
- 16.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 19.Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 20.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 23.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 24.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 25.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci U S A. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barad O, Meiri E, Avniel A, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soussi T, Ishioka C, Claustres M, Beroud C. Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nat Rev Cancer. 2006;6:83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- 29.Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 30.Bunz F, Hwang PM, Torrance C, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wendt J, Radetzki S, von Haefen C, et al. Induction of p21CIP/WAF-1 and G2 arrest by ionizing irradiation impedes caspase-3-mediated apoptosis in human carcinoma cells. Oncogene. 2006;25:972–980. doi: 10.1038/sj.onc.1209031. [DOI] [PubMed] [Google Scholar]
- 32.Hemmati PG, Normand G, Verdoodt B, et al. Loss of p21 disrupts p14 ARF-induced G1 cell cycle arrest but augments p14 ARF-induced apoptosis in human carcinoma cells. Oncogene. 2005;24:4114–4128. doi: 10.1038/sj.onc.1208579. [DOI] [PubMed] [Google Scholar]
- 33.Marine JC, Jochemsen AG. Mdmx and Mdm2: brothers in arms? Cell Cycle. 2004;3:900–904. [PubMed] [Google Scholar]
- 34.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 35.Georges SA, Biery MC, Kim S-Y, et al. Coordinated regulation of cell cycle transcript by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 36.Nechiporuk T, Fernandez TE, Vasioukhin V. Failure of epithelial tube maintenance causes hydrocephalus and renal cysts in Dlg5−/− mice. Dev Cell. 2007;13:338–350. doi: 10.1016/j.devcel.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossmann J. Molecular mechanisms of “detachment-induced apoptosis—anoikis”. Apoptosis. 2002;7:247–260. doi: 10.1023/a:1015312119693. [DOI] [PubMed] [Google Scholar]
- 38.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with “antagomirs”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 39.Tang X, Gal J, Zhuang X, Wang W, Zhu H, Tang G. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-β-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hino K, Fukao T, Watanabe M. Regulatory interaction of HNF1-α to microRNA-194 gene during intestinal epithelial cell differentiation. Nucleic Acids Symp Ser (Oxf) 2007;51:415–416. doi: 10.1093/nass/nrm208. [DOI] [PubMed] [Google Scholar]
- 43.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3