The cellular geometry of growth drives the amino acid economy of Caenorhabditis elegans (original) (raw)
Abstract
The nematode Caenorhabditis elegans grows largely by increases in cell size. As a consequence of this, the surface: volume ratio of its cells must decline in the course of postembryonic growth. Here we use transcriptomic and metabolomic data to show that this change in geometry can explain a variety of phenomena during growth, including: (i) changes in the relative expression levels of cytoplasmic and membrane proteins; (ii) changes in the relative usage of the twenty amino acids in expressed proteins, as estimated by changes in the transcriptome; and (iii) changes in metabolite pools of free amino acids. We expect these relations to be universal in single cells and in whole multicellular organisms that grow largely by increases in cell size, but not those that grow by cell proliferation.
Keywords: Caenorhabditis elegans, growth, gene expression profiling, metabolomics
1. Introduction
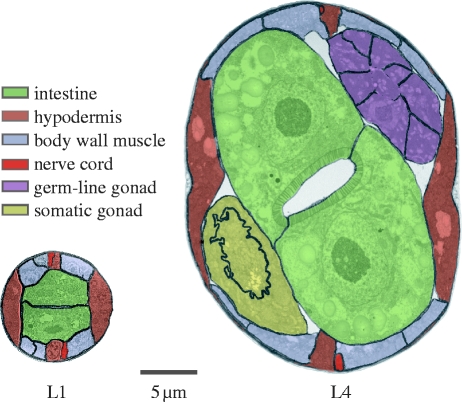
As animals grow they acquire more cells of approximately constant size (Levi 1925; Wilson 1925; Tessier 1939). This commonplace of metazoan biology means that the ratio of cell surface to cell volume must remain approximately constant regardless of how large an animal becomes. Peculiarly, this is not true for Caenorhabditis elegans. In this nematode, most postembryonic tissues grow without proportional increases in cell number. This can be seen in figure 1, which shows cross-sections of a hatchling (L1) and a late larval (L4) worm in which the cell membranes have been marked. Between these stages, the worm increases approximately 32-fold in volume yet the total number of cells increases only by approximately 3.5-fold. The number of cells in the hypodermis increases by only 2.5×, the nervous system by 1.2×, body wall muscles by 1.2×, while the pharynx and the gut do not increase in cell number at all (Sulston & Horvitz 1977; Sulston et al. 1983). Only two tissues undergo much postembryonic proliferation: the somatic gonad and the germ-line, both of which begin as two cells in the L1, but have 143 and approximately 1000 cells, respectively by young adulthood (Kimble & Crittenden 2005). In fact, these counts underestimate the decrease in cell surface: volume ratio since some tissues, such as the hypodermis and vulva, undergo cell fusion to form syncytia (Sulston & Horvitz 1977; Podbilewicz 2006). Even the ‘cells’ of the germ-line have less membrane than might be expected relative to their cytoplasmic volume since they, too, are connected in a syncytium that communicates via a central cavity, the rachis.
Figure 1.
Worms grow mostly by increases in cell size. Transverse mid-body sections with cell membranes outlined. L1, approximately 15 hours post hatching; L4, approximately 46 hours post hatching. The hypodermis mostly remains a single, large, syncytium (some hypodermal cells, not visible here, are added to the head and tail); the intestine does not increase in cell number. Body wall muscle boundaries are not visible at this magnification and are inferred: the L1 has 81 body wall muscles and gains an additional 14 during postembryonic development intercalated among those already present; in the L1, 1–2 are typically visible in each quadrant; in the L4, typically 2–3. The major proliferative tissues during postembryonic development are the germ-line and the somatic gonad. The germ-line is a partial syncytium as shown by the absence of cell membranes flanking the central rachis. The somatic gonad tissues visible here belong to the spermatheca–uterus valve and a junctional core cell, both of which are also syncytial. Individual neurons are not shown, but only a few are added during postembryonic development; excretory cells are also not shown, but remain constant in number. Cross-sections courtesy of Nichol Thompson and Dave Hall.
The result of these peculiar features is that the worm grows rather like a balloon. More accurately, many of its cells do in that they expand in volume many times during the course of postembryonic growth without dividing. We hypothesize, therefore, that the demand for cell membrane components must decline relative to the demand for cytoplasmic components during the course of postembryonic growth. This hypothesis assumes that intracellular membranes such as endoplasmic reticulum and nuclear envelopes scale no more than isometrically with cytoplasmic volume. But even the increase of nuclear membranes will probably show negative allometry with worm volume since several tissues such as the hypodermis and the gut increase their genomic content by endoreduplication, forming large, polyploid nuclei bound by a single membrane (Hedgecock & White 1985; Flemming et al. 2000). Indeed, endoreduplication drives the growth of these tissues (Lozano et al. 2006). The decrease in the cellular surface: volume ratio, and hence the membrane: cytoplasm ratio, is one of the most striking features of the worm's growth. Here, we interrogate transcriptomic and metabolomic data to study the impact of the ‘balloon-effect’ on the dynamics of protein usage, amino acid supply and demand during growth.
2. Material and methods
(a). Strains and culture
We cultured wild-type worms strain N2 (Bristol) for 1H nuclear magnetic resonance (NMR) spectroscopy at 20°C on 90 mm NGM agar plates seeded with 50 µl Escherichia coli strain OP50 (Brenner 1974) and supplemented with 5 µg ml−1 nystatin (Sigma Aldrich). We synchronized populations by bleaching, and collected samples of approximately 2000 worms from the larval stages 1–4 and young adulthood (66 hours after bleaching) using a 50 µm Nitex filter (Sefar Ltd) to separate the adults. We quick-froze the samples (0.5 ml volume) in liquid nitrogen and stored them at −80°C. We then ground the tissue in a liquid nitrogen-cooled mortar, and directly added 2 ml of ice-cold methanol to the frozen ground tissue to give a final extract concentration of 80 per cent methanol. The mortars were then rinsed with a further volume of 80 per cent methanol and the combined extracts cleared by centrifugation (10 min, 16 000_g_), dried in a rotary vacuum concentrator (6 hours; 45°C), rehydrated in 660 µl of 30 per cent NMR buffer (100% D2O, 0.02% TSP, 0.1 M phosphate pH 7.0) and centrifuged again. We then transferred 600 µl samples to 5 mm NMR tubes.
(b). 1H NMR spectroscopy
We measured the relative concentrations of amino acids by 1H NMR spectroscopy, using a Bruker Avance DRX600 spectrometer with 14.1 T magnet and resulting proton resonance frequency of 600 MHz, equipped with a 5 mm broad-band inverse probe (Bruker BioSpin, Rheinstetten, Germany). Samples were held at room temperature while on the autosampler and at 300 K during acquisition. Data were acquired essentially as previously described (Beckonert et al. 2007). Eight dummy scans and 128 scans were collected per sample. We then processed the data and quantified metabolite concentrations using Chenomx NMR Suite v. 5.0 (Chenomx, Edmonton AB, Canada) for assisted manual fitting of metabolites (Weljie et al. 2006). We fitted 14 standard amino acids to the data. We did not fit histidine because the high pH sensitivity of the imidazole peaks meant that they were subject to large chemical shift differences; tryptophan was present below detection limits; and proline, serine, cysteine and asparagine were not readily fitted in the one-dimensional spectra, because of a combination of relatively low concentrations and extensive peak overlap. We normalized the data by dividing each profile by a single normalization factor, the median fold change across all compounds relative to a reference profile; in this case, a median of all profiles (Dieterle et al. 2006).
(c). Analysis of transcriptomic data
We examined a previously published transcriptomic dataset that gives the relative expression levels of 17 869 genes in the four larval and young adult stages of C. elegans (Jiang et al. 2001). This dataset that is based on two-channel microarrays, gives the expression level of each gene, at each stage, relative to its expression in a mixed-age population of worms. This approach allows the relative expression of genes to be compared across life stages. We exploited this to divide the genes into two classes—‘late’ and ‘early’—depending upon whether the relative expression of the gene increased or decreased as the worm aged. To obtain this division into late and early genes, we first assigned times in hours to each of the five life stages using standard times for the life cycle at 20°C. We then correlated the expression level of each gene against time using four different models—linear, log, power and exponential—and for each model, took the sign ‘+’ or ‘−’ of the correlation. We used the four different regression models because of uncertainty as to how gene expression changed with age. Indeed, we found that no model fitted the data much better than any other: the linear model gave the best fit for 5024 genes; the log for 4376; the power for 3577; and the exponential for 4892. Given this uncertainty, and to increase robustness, we excluded any gene in which the sign of the correlation was not identical under all four models. Under this criterion, 5 per cent of the genes were rejected, and the remaining 95 per cent assigned as either late or early.
We also determined which genes were associated with the cytoplasm and which with the various cellular membranes (plasmalemma, nuclear, endoplasmic reticulum, etc.) by referring to the cellular component division of the Gene Ontology database as of 12 May 2008. We considered all GO categories containing 10 or more genes, giving a coverage of almost 98 per cent of the roughly 55 per cent of C. elegans genes that have been given an assignation in the cellular component division of GO. We now had four sets of genes: late; early; ‘cytoplasmic’; and ‘membrane’. To determine the amino acid composition of each set—and of the intersection of sets, e.g. ‘late cytoplasmic’—we used build 189 of the C. elegans genome (ftp://ftp.sanger.ac.uk/pub/databases/wormpep/wormpep189) to provide a series of vectors, each containing the counts for the 20 amino acids. Each vector was then standardized so that it summed to 1 across the 20 amino acids. The shifts in amino acid usages between sets—e.g. late/early—were then obtained by dividing the relevant vectors. These vectors were log-transformed before being used in Pearson's regression analysis. We report not only Pearson's _p_-values, but also the corresponding _p_-values from a Spearman's rank test. The non-parametric statistic provides added reassurance in the light of the relatively complex derivation of the vectors in our regressions. In no case are results significant by one test non-significant by the other.
3. Results
The geometry of worm growth predicts differential age-specific expression of cytoplasmic and membrane proteins. As the worm grows, demand for, and production of, all cell components naturally increases. However, because of the special balloon-like features of C. elegans, the demand for membrane components should decrease relative to the demand for cytoplasmic components. To test this idea, we examined previously published transcriptomic data that gives the relative expression levels of 17 869 genes in four larval stages (L1–L4) and young adults (Jiang et al. 2001), and studied the ‘age-response’ of these genes, asking whether their relative expression increases (late genes) or decreases (early genes) with age, discarding those that had an unclear age-response (see §2 for details). We then classified the remaining genes into cellular compartments using Gene Ontology categories so as to identify membrane-related genes (3860) and cytoplasm-related genes (334) with the remainder being discarded (see §2 for details). We then asked, for each of the two GO classes, how many genes increased and decreased with age. We found that the number of cytoplasmic genes that increased with age was 2.37× greater than the number that decreased while for membrane genes the same ratio was 0.62; the difference between these ratios is highly significant (_p_=5.4×10−30 _χ_2-test). Assuming that gene expression reflects protein levels this result suggests that the relative production of cytoplasmic proteins, increases with age while that of membrane proteins falls, which is consistent with a balloon-effect.
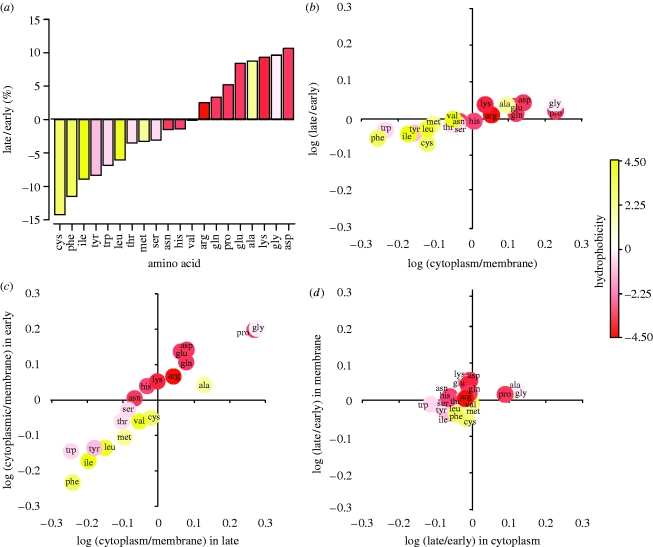
Amino acid usage is driven by geometry rather than age per se. When we compare the early and the late genes we find a striking difference in amino acids that they use (figure 2a). Why is this? We showed that as the worm grows there is a change in the kinds of proteins—cytoplasmic or membrane—that it makes. Furthermore, it has long been known that the proteins of some intracellular compartments are especially rich in certain kinds of amino acids (Cedano et al. 1997). For example, many membrane-related proteins are rich in hydrophobic amino acids in order to keep them anchored in the lipid bilayer. Perhaps, then, this compartmental shift accounts for the temporal shift in amino acid use. Consistent with this idea, we find that the change in amino acid use between cytoplasmic and membrane proteins explains 77 per cent of the change in use between late and early proteins (figure 2b), (_r_2=0.77, _p_=1.7×10−7, one-tailed Pearson correlation, _p_=1.2×10−8, one-tailed Spearman rank). Or, to put it another way, almost four-fifths of the change in amino acid use of worms as they grow can be explained by the different amino acid content of cytoplasmic versus membrane proteins. This same difference also accounts for the shift in the hydrophobicity (Kyte & Doolittle 1982) of the amino acids used in early versus late proteins seen in figure 2a (_p_=0.012, two-tailed Pearson correlation, _p_=0.011, two-tailed Spearman rank) since in a multiple correlation analysis in which age-response—log (late/early)—is the dependent, and hydrophobicity and compartment—log (cytoplasmic/membrane)—are independent variables, hydrophobicity is no longer significant (two-tailed partial regression p values: compartment _p_=0.000018, hydrophobicity _p_=0.50, interaction _p_=0.65; and without interaction, compartment _p_=0.000010, hydrophobicity _p_=0.36).
Figure 2.
The consequences of the changing cellular geometry of the worm for its amino acid economy. (a) Change in amino acid proportions in late versus early proteins. Based on all proteins with values in Jiang et al. (2001). (b) Association between the change in amino acid proportions in late versus early proteins with the change in cytoplasmic versus membrane proteins. Correlation based on all proteins with values in Jiang et al. (2001; _y_-axis values) and all proteins with values in the Gene Ontology database (_x_-axis values), i.e. not just the intersection of the two sets. (c) Association between the change in amino acid proportions in cytoplasmic proteins versus membrane proteins when late (_x_-axis) and early (_y_-axis) proteins are considered separately. (d) Association between the change in amino acid proportions in late proteins versus early proteins when cytoplasmic (_x_-axis) and membrane (_y_-axis) proteins are considered separately. Analyses in (c,d) are based on the intersection of the Jiang et al. (2001) and Gene Ontology sets. For statistics, see text.
Although the correlation between the shift in amino acid usages seen in the compartment and age-responses is striking, it does not follow that the former actually drives the latter. Amino acid usage might vary across ages for some altogether different, but hidden reason. An alternative explanation, for example, is that selection for metabolic efficiency varies with age and has shaped the age-specific composition of proteins. Metabolic efficiency is a particularly strong candidate hypothesis as it is known generally to shape protein composition (Akashi & Gojobori 2002; Swire 2007). But it does not explain the age-response in this dataset (for proof, see electronic supplementary material information IV). However, although we can reject metabolic efficiency, it is not possible more generally—given that our data are not experimental but entirely correlative—to exclude definitively the agency of some hidden variable or to establish the direction of causality. Nevertheless, we think that cellular geometry is the driver. In part, this is because, as mentioned above, membrane proteins have the amino acid composition that they do for good functional reasons—to keep them anchored in the lipid bilayer—and so there is simply no need to invoke other unknown age-dependent processes to explain their composition. But it is also because a closer look at our data provides further evidence in support of our explanation.
First, while there is a very large shift in amino acid usage between cytoplasmic and membrane proteins, the shift between early and late proteins is much smaller (the former exceeding the latter by 4.4×, figure 2b). Looking at this another way, when we compare cytoplasmic with membrane proteins, we find that the relative frequency of the typical amino acid differs by 30 per cent; but when we compare late and early proteins we find that this difference is 7 per cent. Second, we can control directly for the effects of age-response and compartment. We control for the effects of age-response by calculating the shifts in amino acid usage between cytoplasmic and membrane proteins within each of the early or late subsets (figure 2c). These shifts are highly correlated, which implies that the difference in amino acid usage between cytoplasmic and membrane proteins is largely independent of age-response (_r_agecontrolled2=0.88, _p_=4.9×10−10 Pearson, _p_=1.6×10−10 Spearman). Analogously, we control for the effects of compartment by calculating the shifts in amino acid usage between early and late proteins within each of the cytoplasmic or membrane subsets (figure 2d). These shifts, by contrast, are only very weakly correlated, which implies that the difference in amino acid usage between these late and early proteins largely depends on compartment (_r_compartmentcontrolled2=0.19, _p_=0.028, _p_=0.021). In order to examine the robustness of these results, we resampled our dataset 100 000 times and found that _r_agecontrolled2 was constant and high while _r_compartmentcontrolled2 was variable and low (see figure S1 in the electronic supplementary material). Given these results, we conclude that the changing geometry of the growing worm, rather than its increasing age, predicts the amino acid composition of the proteins that it is manufacturing at any time. Indeed, there is little evidence for an age effect independent of the changing geometry.
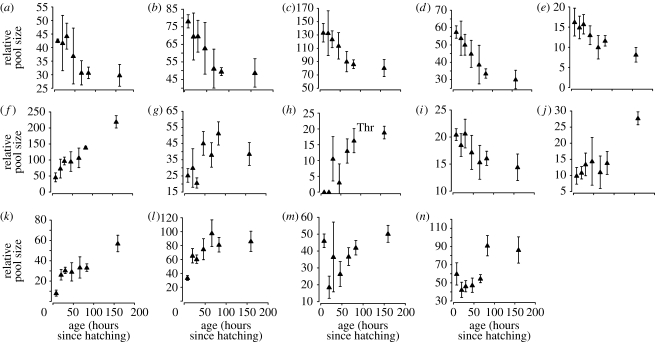
The geometry of growth predicts amino acid pool sizes. If, during growth, the amino acid composition of the worm's proteins changes, then so too must the demand for particular amino acids. Is this change in demand reflected in the pool sizes of amino acids? To answer this question, we measured the relative pool sizes of 14 standard amino acids using 1H NMR spectroscopy. (The remaining six were not estimated, see §2.) We sampled worms at seven ages: each of the four larval stages, young adulthood (66 hours after bleaching eggs), middle age (83 hours) and old age (159 hours). All 14 amino acids showed strong changes with age, usually in a consistent direction (figure 3). Once again, we used four different regression models—linear, log, power and exponential—and for each amino acid estimated the slope of relative concentration against time. We only fitted the first five ages because we wanted to compare our results directly to Jiang et al.'s transcriptomic data.
Figure 3.
Dynamics of free pool size in 14 standard amino acids during the course of postembryonic growth. Relative pool sizes measured using 1H NMR spectrophotometry and semi-automated fitting of individual metabolites. ((a) Ile; (b) Val; (c) Leu; (d) Phe; (e) Met; (f) Ala; (g) Gly; (h) Thr; (i) Tyr; (j) Asp; (k) Gln; (l) Glu; (m) Lys; (n) Arg).
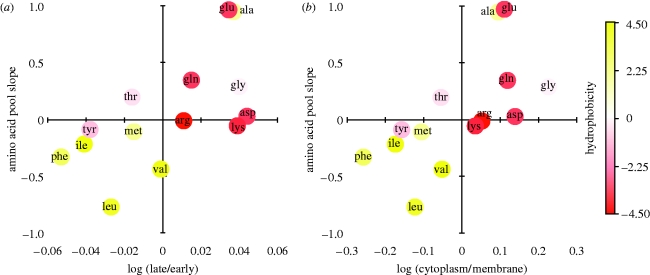
We have already shown how amino acid usage changes during growth (figure 2b). In fact, the _y_-axis values of this plot are estimates, albeit imperfect ones, of changes in amino acid demand during growth. Similarly, the slopes describing how the relative pool sizes of amino acids change during growth are, potentially, estimates of changes in their supply. Figure 4a shows that the change in supply and demand for the 14 amino acids is positively correlated (linear: _r_2=0.39; _p_=0.0087, _p_=0.0061; see the electronic supplementary material for other models). This suggests that amino acid pool sizes are dynamically regulated in response to the changing demands of protein synthesis. Figure 4b shows that this regulation seems to be driven by the increasing proportion of cytoplasmic—and decreasing proportion of membrane proteins—produced as the worm grows since the change in supply of the amino acids is also positively correlated with log (cytoplasmic/membrane) as in the _x_-axis of figure 2b (linear: _r_2=0.41; _p_=0.0067, _p_=0.00092; see the electronic supplementary material for other models). In other words, just as the changing cellular geometry of the growing worm predicts the amino acid composition of the proteins that it is manufacturing at any time, it also predicts the size of its amino acid pools.
Figure 4.
Association between changes in free amino acid concentration with the change in amino acid proportions: (a) in late versus early proteins; (b) in cytoplasmic versus membrane proteins. For further details of models and statistics, see text.
4. Discussion
We have found that the cellular geometry of growth has a substantial impact on the kind of proteins that the worm produces in the course of its growth, and hence on its amino acid economy, a phenomenon we have termed the balloon-effect. In fact, we find that the balloon-effect explains 77 per cent of the variation in amino acid usage of the growing worm. Indeed, this effect overwhelms any independent effect of age. Although the balloon-effect arises from the peculiarities of the worm's growth, we predict that it will be found in several other circumstances: first, in the growth of the many small bodied animal phyla that also have syncytial tissues and are thought to grow largely in the absence of cell proliferation (van Cleave 1932; Flemming et al. 2000); second, in dipteran larvae, which similar to C. elegans, grow largely by increases in cell size (Trager 1935); and third, in polyploid cells—whether they are naturally polyploid cells of normally diploid animals (e.g. mammalian megakaryocytes), polyploid tumorigenic cells (e.g. DLD1 colorectal cells), abnormally polyploid strains of unicellular organisms (e.g. yeast), or polyploid species (e.g. the giant bacterium Epulopiscium; Galitski et al. 1999; Raslova et al. 2007; Mendell et al. 2008). The balloon-effect may even be detectable in the doubling of size that takes place during the normal cell cycle of any prokaryote or eukaryote. Conversely, we expect to see a deflating-balloon effect during the early embryogenesis of animals in which a single large oocyte becomes divided into successively smaller blastomeres. We are currently testing some of these predictions using available gene profiling data (Galitski et al. 1999; Arbeitman et al. 2002; Storchova et al. 2006; Koutsos et al. 2007; Raslova et al. 2007).
Our results also bear on the interpretation of studies that compare the transcriptomes, proteomes or metabolomes of different mutants, environmental treatments or species of nematodes. All dwarf C. elegans mutants that have been studied in any detail so far—whether they disrupt cytoskeletal components, extracellular components or intercellular signals—show failures of cell growth rather than cell proliferation (Morita et al. 1999, 2002; Suzuki et al. 1999; Flemming et al. 2000; Nystrom et al. 2002; Patel et al. 2002; Maduzia et al. 2005; Fung et al. 2007). Plastic responses of body size to changes in diet, and even evolved differences among species and strains of nematode, are also either entirely or largely due to changes in cell size (Flemming et al. 2000; Tain et al. 2008). Given this, we expect that future studies of these treatments will show a balloon-effect. Specifically, we predict that for treatments that cause dwarfing (or gigantism), membrane-related transcripts, proteins and amino acids will show systematic increases (or decreases) relative to wildtype. Indeed, given that body size phenotypes are extremely common—Wormbase currently lists more than 400 loci that, when inactivated, have growth phenotypes and that is probably a gross underestimate—the balloon-effect may confound the interpretation of omic data in many mutants.
One of our most striking findings is that the concentrations of free amino acids track changes in demand that arise from protein synthesis during growth. In a way, this association is surprising. That is because our measure of demand—the number and kinds of proteins that the worm makes—is limited in several ways. First, Jiang et al.'s transcriptomic data, being based on two-channel microarrays, yields information on temporal, but not static, variation in transcript levels. In effect, then, our analysis assumes that the absolute expression level of all genes is the same. Second, our measure of protein production does not take into account post-transcriptional levels of regulation such as mRNA recruitment from the nucleus, levels of polysomal occupancy of transcripts and alterations in protein turnover (Beyer et al. 2004; Castrillo et al. 2007). Third, we assume that the protein: lipid ratio of membranes remains constant over ontogeny. Our analysis suggests that these unaccounted sources of variance are roughly evenly distributed across our different classes of protein, for despite these unaccounted sources, we are able to explain 41 per cent of the change in amino acid pool levels in the metabolome by our simple balloon model.
But our findings are also surprising for the light that they shed upon the amino acid economy of the growing worm. If changes in amino acid demand are met by changes in the rate of amino acid production, then amino acid pool sizes might remain constant. Alternatively, if production rate is constant then increases in the demand for an amino acid would result in a decreased pool size, and vice versa. We, however, find that worms show a third pattern: apparently altering pool sizes to match consumption. Why do they do this? One possible explanation is that optimal translational efficiency depends on relative amino acid concentrations. This could occur if a mismatch between the relative concentration of an amino acid and its cognate tRNA results in a reduced rate of tRNA charging. An analogous argument has been used to explain the positive correlation between tRNA copy number and codon usage visible in eukaryote genomes (Berg & Kurland 1997; Percudani et al. 1997; Kanaya et al. 1999).
Testing causal hypotheses such as this will require a general account of the growth of C. elegans. This goal requires integrating metabolomic (Blaise et al. 2007; Atherton et al. 2008), proteomic (Dong et al. 2007) and transcriptomic (Jiang et al. 2001) data. Studies of cellular growth in microbes are much more advanced than for any animal, and it is worth considering how our study compares to the various approaches that have been used in E. coli and yeast. Two general approaches to microbial growth may be distinguished: regulatory accounts that seek to explain metabolite dynamics in terms of the expression of particular enzymes, pathways, or else global regulators (e.g. Castrillo et al. 2007) and flux accounts that seek to explain metabolite dynamics in terms of the optimal combination of fluxes that maximize growth under a given set of conditions (Covert et al. 2001; Fong et al. 2003; Duarte et al. 2004; Nanchen et al. 2006; Andersen et al. 2008). Our approach differs from both. Where regulatory accounts interrogate the transcriptome to reveal the reasons for changes in the supply of metabolites, we do so to reveal the reasons for changes in their demand, i.e. we are interested in the function of amino acid pools. Flux accounts, by contrast, are concerned with demand for they explicitly assume cells of a particular composition. However, they typically also assume a steady-state and, as such, apply only to growth in chemostats. Even non-steady-state flux analyses do not consider proteome composition and hence miss this functional level of explanation (Antoniewicz et al. 2007; Baxter et al. 2007). Our results highlight that, unlike a population of microbes in a chemostat, the worm's cellular composition, and hence the demands on its metabolism, change continuously during growth. Or, to put it more simply, C. elegans' metabolome has an ontogeny. We note, however, that similar ontogenetic changes should not be found in vertebrates and other animals that grow largely by increases in cell number. In this sense, at least a human child resembles a microbial culture more than it does a larval worm.
Acknowledgments
We thank Dave Hall and the Center for Caenorhabditis elegans Anatomy (Albert Einstein College of Medicine) for supplying the cross-sections in figure 1 and assisting in their interpretation. The N2 strain was obtained from the Caenorhabditis Genetics Center (University of Minnesota). Both the Center for Caenorhabditis elegans Anatomy and the CGC are supported by the National Institutes of Health—National Center for Research Resources.
References
- Akashi H., Gojobori T.2002Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA 99, 3695–3700 (doi:10.1073/pnas.062526999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M.R., Nielsen M.L., Nielsen J.2008Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol. Syst. Biol 4, article 178.doi:10.1038/msb.2008.12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz M.R., Kraynie D.F., Laffend L.A., Gonzalez-Lergier J., Kelleher J.K., Stephanopoulos G.2007Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab. Eng 9, 277–292 (doi:10.1016/j.ymben.2007.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman M.N., et al. 2002Gene expression during the life cycle of Drosophila melanogaster. Science 297, 2270–2275 (doi:10.1126/science.1072152) [DOI] [PubMed] [Google Scholar]
- Atherton H.J., Jones O.A., Malik S., Miska E.A., Griffin J.L.2008A comparative metabolomic study of NHR-49 in Caenorhabditis elegans and PPAR-alpha in the mouse. FEBS Lett 582, 1661–1666 (doi:10.1016/j.febslet.2008.04.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter C.J., Liu J.L., Fernie A.R., Sweetlove L.J.2007Determination of metabolic fluxes in a non-steady-state system. Phytochemistry 68, 2313–2319 (doi:10.1016/j.phytochem.2007.04.026) [DOI] [PubMed] [Google Scholar]
- Beckonert O., Keun H.C., Ebbels T.M., Bundy J., Holmes E., Lindon J.C., Nicholson J.K.2007Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc 2, 2692–2703 (doi:10.1038/nprot.2007.376) [DOI] [PubMed] [Google Scholar]
- Berg O.G., Kurland C.G.1997Growth rate optimised tRNA abundance and codon usage. J. Mol. Biol 270, 544–550 (doi:10.1006/jmbi.1997.1142) [DOI] [PubMed] [Google Scholar]
- Beyer A., Hollunder J., Nasheuer H.-P., Wilhelm T.2004Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics 3, 1083–1092 (doi:10.1074/mcp.M400099-MCP200) [DOI] [PubMed] [Google Scholar]
- Blaise B.J., Giacomotto J., Elena B., Dumas M.E., Toulhoat P., Segalat L., Emsley L.2007Metabotyping of Caenorhabditis elegans reveals latent phenotypes. Proc. Natl Acad. Sci. USA 104, 19 808–19 812 (doi:10.1073/pnas.0707393104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S.1974The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo J.I., et al. 2007Growth control of the eukaryote cell: a systems biology study in yeast. J. Biol 6, 4.doi:10.1186/jbiol54) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedano J., Aloy P., Perez-Pons J.A., Querol E.1997Relation between amino acid composition and cellular location of proteins. J. Mol. Biol 266, 594–600 (doi:10.1006/jmbi.1996.0804) [DOI] [PubMed] [Google Scholar]
- Covert M.W., Schilling C.H., Famili I., Edwards J.S., Goryanin I.I., Selkov E., Palsson B.O.2001Metabolic modeling of microbial strains in silico. Trends Biochem. Sci 26, 179–186 (doi:10.1016/S0968-0004(00)01754-0) [DOI] [PubMed] [Google Scholar]
- Dieterle F., Ross A., Schlotterbeck G., Senn H.2006Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem 78, 4281–4290 (doi:10.1021/ac051632c) [DOI] [PubMed] [Google Scholar]
- Dong M.Q., Venable J.D., Au N., Xu T., Park S.K., Cociorva D., Johnson J.R., Dillin A., Yates J.R., III2007Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 (doi:10.1126/science.1139952) [DOI] [PubMed] [Google Scholar]
- Duarte N.C., Herrgard M.J., Palsson B.O.2004Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 14, 1298–1309 (doi:10.1101/gr.2250904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming A.J., Shen Z.Z., Cunha A., Emmons S.W., Leroi A.M.2000Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc. Natl Acad. Sci. USA 97, 5285–5290 (doi:10.1073/pnas.97.10.5285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S.S., Marciniak J.Y., Palsson B.O.2003Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J. Bacteriol 185, 6400–6408 (doi:10.1128/JB.185.21.6400-6408.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung W.Y., Fat K.F., Eng C.K., Lau C.K.2007_crm-1_ facilitates BMP signaling to control body size in Caenorhabditis elegans. Dev. Biol 311, 95–105 (doi:10.1016/j.ydbio.2007.08.016) [DOI] [PubMed] [Google Scholar]
- Galitski T., Saldanha A.J., Styles C.A., Lander E.S., Fink G.R.1999Ploidy regulation of gene expression. Science 285, 251–254 (doi:10.1126/science.285.5425.251) [DOI] [PubMed] [Google Scholar]
- Hedgecock E.M., White J.G.1985Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol 107, 128–133 (doi:10.1016/0012-1606(85)90381-1) [DOI] [PubMed] [Google Scholar]
- Jiang M., Ryu J., Kiraly M., Duke K., Reinke V., Kim S.K.2001Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 98, 218–223 (doi:10.1073/pnas.011520898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S., Yamada Y., Kudo Y., Ikemura T.1999Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238, 143–155 (doi:10.1016/S0378-1119(99)00225-5) [DOI] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L.2005Germline proliferation and its control. WormBook, pp. 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsos A.C., et al. 2007Life cycle transcriptome of the malaria mosquito Anopheles gambiae and comparison with the fruitfly Drosophila melanogaster. Proc. Natl Acad. Sci. USA 104, 11 304–11 309 (doi:10.1073/pnas.0703988104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F.1982A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 157, 105–132 (doi:10.1016/0022-2836(82)90515-0) [DOI] [PubMed] [Google Scholar]
- Levi G.1925Wachstum und Körpergröbe: Die strukturelle Grundlage der Körpergröbe bei vollausgebildeten und in Wachstum beigriffenen Tieren. Ergb. Anat. Entwicklungsgesch. 26, 86–352 [Google Scholar]
- Lozano E., Saez A.G., Flemming A.J., Cunha A., Leroi A.M.2006Regulation of growth by ploidy in Caenorhabditis elegans. Curr. Biol 16, 493–498 (doi:10.1016/j.cub.2006.01.048) [DOI] [PubMed] [Google Scholar]
- Maduzia L.L., Roberts A.F., Wang H., Lin X., Chin L.J., Zimmerman C.M., Cohen S., Feng X.H., Padgett R.W.2005_C. elegans_ serine–threonine kinase KIN-29 modulates TGFβ signaling and regulates body size formation. BMC Dev. Biol 5, 8.doi:10.1186/1471-213X-5-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.E., Clements K.D., Choat J.H., Angert E.R.2008Extreme polyploidy in a large bacterium. Proc. Natl Acad. Sci. USA 105, 6730–6734 (doi:10.1073/pnas.0707522105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Chow K.L., Ueno N.1999Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 126, 1337–1347 [DOI] [PubMed] [Google Scholar]
- Morita K., et al. 2002A Caenorhabditis elegans TGFβ, DBL-1, controls the expression of LON-1, a PR-related protein, that regulates polyploidization and body length. EMBO J 21, 1063–1073 (doi:10.1093/emboj/21.5.1063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanchen A., Schicker A., Sauer U.2006Nonlinear dependency of intracellular fluxes on growth rate in miniaturized continuous cultures of Escherichia coli. Appl. Environ. Microbiol 72, 1164–1172 (doi:10.1128/AEM.72.2.1164-1172.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom J., Shen Z.Z., Aili M., Flemming A.J., Leroi A., Tuck S.2002Increased or decreased levels of Caenorhabditis elegans lon-3, a gene encoding a collagen, cause reciprocal changes in body length. Genetics 161, 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.N., Knight C.G., Karageorgi C., Leroi A.M.2002Evolution of germ-line signals that regulate growth and aging in nematodes. Proc. Natl Acad. Sci. USA 99, 769–774 (doi:10.1073/pnas.012511099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R., Pavesi A., Ottonello S.1997Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol 268, 322–330 (doi:10.1006/jmbi.1997.0942) [DOI] [PubMed] [Google Scholar]
- Podbilewicz B.2006Cell fusion. WormBook, pp. 1–32 http://wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raslova H., et al. 2007Interrelation between polyploidization and megakaryocyte differentiation: a gene profiling approach. Blood 109, 3225–3234 (doi:10.1182/blood-2006-07-037838) [DOI] [PubMed] [Google Scholar]
- Storchova Z., Breneman A., Cande J., Dunn J., Burbank K., O'Toole E., Pellman D.2006Genome-wide genetic analysis of polyploidy in yeast. Nature 443, 541–547 (doi:10.1038/nature05178) [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R.1977Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol 56, 110–156 (doi:10.1016/0012-1606(77)90158-0) [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N.1983The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol 100, 64–119 (doi:10.1016/0012-1606(83)90201-4) [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yandell M., Roy P., Krishna S., Savage-Dunn C., Ross R., Padgett R., Wood W.1999A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126, 241–250 [DOI] [PubMed] [Google Scholar]
- Swire J.2007Selection on synthesis cost affects interprotein amino acid usage in all three domains of life. J. Mol. Evol 64, 558–571 (doi:10.1007/s00239-006-0206-8) [DOI] [PubMed] [Google Scholar]
- Tain L.S., Lozano E., Saez A.G., Leroi A.M.2008Dietary regulation of hypodermal polyploidization in C. elegans. BMC Dev. Biol 8, 28.doi:10.1186/1471-213X-8-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier G.1939Biometrie de la cellule. Tabulae Biologicae 19, 1–64 [Google Scholar]
- Trager W.1935The relation of cell size to growth in insect larvae. J. Exp. Zool 7, 489–508 (doi:10.1002/jez.1400710308) [Google Scholar]
- van Cleave H.J.1932Eutely or cell constancy in its relation to body size. Q. Rev. Biol 7, 59–67 (doi:10.1086/394396) [Google Scholar]
- Weljie A.M., Newton J., Mercier P., Carlson E., Slupsky C.M.2006Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem 78, 4430–4442 (doi:10.1021/ac060209g) [DOI] [PubMed] [Google Scholar]
- Wilson E.B.1925The cell in development and heredity. New York, NY: Macmillan [Google Scholar]