Dynamic Regulation of a Metabolic Multi-enzyme Complex by Protein Kinase CK2 (original) (raw)
Abstract
The reversible association and dissociation of a metabolic multi-enzyme complex participating in de novo purine biosynthesis, the purinosome, was demonstrated in live cells to respond to the levels of purine nucleotides in the culture media. We also took advantage of in vitro proteomic scale studies of cellular substrates of human protein kinases (e.g. casein kinase II (CK2) and Akt), that implicated several de novo purine biosynthetic enzymes as kinase substrates. Here, we successfully identified that purinosome formation in vivo was significantly promoted in HeLa cells by the addition of small-molecule CK2-specific inhibitors (i.e. 4,5,6,7-tetrabromo-1_H_-benzimidazole, 2-dimethylamino-4,5,6,7-tetrabromo-1_H_-benzimidazole, tetrabromocinammic acid, 4,4′,5,5′,6,6′-hexahydroxydiphenic acid 2,2′,6,6′-dilactone (ellagic acid) as well as by silencing the endogenous human CK2α catalytic subunit with small interfering RNA. However, 4,5,6,7-tetrabromobenzotriazole, another CK2-specific inhibitor, triggered the dissociation of purinosome clusters in HeLa cells. Although the mechanism by which 4,5,6,7-tetrabromobenzotriazole affects purinosome clustering is not clear, we were capable of chemically reversing purinosome formation in cells by the sequential addition of two CK2 inhibitors. Collectively, we provide compelling cellular evidence that CK2-mediated pathways reversibly regulate purinosome assembly, and thus the purinosome may be one of the ultimate targets of kinase inhibitors.
Keywords: Metabolism, Metabolic Regulation, Nucleoside Nucleotide Biosynthesis, Protein Assembly, Protein Kinases, Casein Kinase 2, Fluorescent Live Cell Imaging, Purine Biosynthesis
Introduction
Sequential metabolic enzymes have been hypothesized to form into macromolecular complexes in cells to respond to events that require changes in metabolic flux (1–3). However, in vitro experimental evidence for such complexes has been sparse, likely due to the transient nature of the protein-protein interactions, the possible requirement of post-translational modifications in given enzymes, and/or the participation of unknown accessory proteins in macromolecular complex formation. After a prolonged search in vitro and in vivo for such a complex, we successfully identified in human cell lines a subcellular organization, the purinosome (4), involved in de novo purine biosynthesis (see Fig. 1). Dual color colocalization demonstrated that all the six human de novo purine biosynthetic enzymes were clustered in the cytoplasm upon purine depletion. Importantly, the reversible formation of the purinosome was observed in response to the availability of purine sources in the growth media, implying the functional role of the purinosome in live cells. However, the underlying molecular mechanism that ties purine depletion to purinosome function remained to be elucidated. Here, we propose that human protein kinase CK23 (hCK2; previously, casein kinase 2) plays a pivotal role for the assembly and disassembly of the purinosome in vivo.
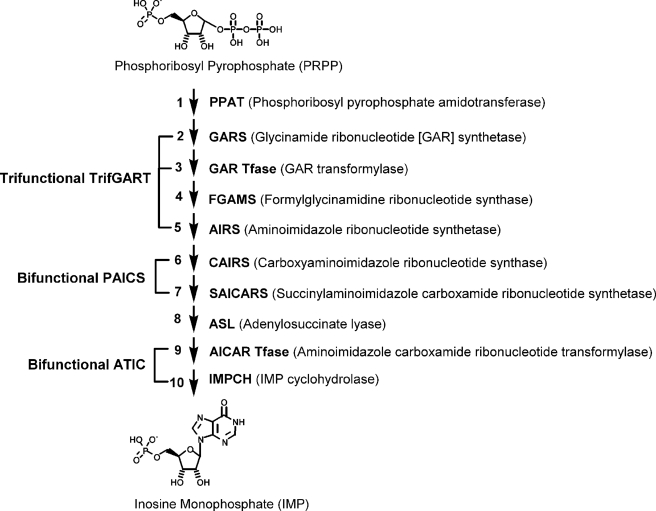
FIGURE 1.
Enzymes in human de novo purine biosynthesis. The de novo pathway transforms phosphoribosyl pyrophosphate (PRPP) to inosine monophosphate (IMP) in ten steps. Steps 2, 3, and 5 are catalyzed by a trifunctional enzyme TrifGART; steps 6 and 7 are catalyzed by a bifunctional enzyme PAICS; and steps 9 and 10 are catalyzed by a bifunctional enzyme ATIC.
CK2 is categorized as a member of the serine/threonine protein kinases, but in some cases the enzyme also phosphorylates tyrosine residues (5, 6). CK2 is a pleiotropic, ubiquitous protein kinase that is active in a myriad of cellular processes, including gene expression and cell growth (7–9). To support the diverse roles of CK2, the enzyme can translocate to loci throughout the cell to interact with a number of its cellular substrates (5, 10). Two catalytic subunits (α and α′) and two regulatory subunit (β2) in three configurations (α2β2, αα′β2, or α′2β2) constitute the heterotetrameric CK2 holoenzyme. However, individual catalytic subunits (CK2α or CK2α′) can also function without regulatory subunits. Of note, hCK2 expression is noticeably up-regulated in a wide variety of tumors (8). Accordingly, a number of small molecules have been developed for cancer chemotherapy to specifically inhibit CK2 (11, 12).
However, it is important to note that two specific kinases, CK2 and Akt (also known as protein kinase B) have been implicated to interact with de novo purine biosynthetic enzymes based on two different in vitro proteomic scale experiments; (i) hPPAT, hTrifGART, and hFGAMS (see Fig. 1) are implicated as substrates for hCK2 (13), and (ii) hFGAMS is implicated as a substrate for Akt (14). We took into account both kinases in this work because several key metabolic enzymes were previously implicated as their substrates; for example, glycogen synthase, acetyl-CoA carboxylase, and ornithine decarboxylase for CK2 (5, 15) and ATP-citrate lyase for Akt (16). Therefore, our in vivo experiments center on whether protein kinase-mediated regulation of these pathway enzymes is linked to purinosome assembly/disassembly in HeLa cells through the use of potent kinase inhibitors and small interfering RNAs (siRNAs) combined with single-cell fluorescence imaging.
EXPERIMENTAL PROCEDURES
Cloning
All plasmids expressing de novo purine biosynthetic enzymes with monomeric enhanced green fluorescent protein (GFP) and/or monomeric orange fluorescent protein (OFP) were prepared as before (4).
Transfection of Mammalian Cells
A human cervical cancer cell line, HeLa (ATCC), and a human carcinoma liver cell line, C3A (ATCC), were maintained and transfected for this study as described before (4). Briefly, we subjected HeLa and C3A cells to both purine-rich medium: minimal essential medium (Mediatech), 10% fetal bovine serum (Atlanta Biological) and 50 μg/ml gentamicin sulfate (Sigma); and purine-depleted medium: Roswell Park Memorial Institute 1640 (RPMI 1640; Mediatech) supplemented with dialyzed 5% fetal bovine serum and 50 μg/ml gentamicin sulfate. Fetal bovine serum was dialyzed against 0.9% NaCl at 4 °C for ∼2 days. Lipofectamine™ 2000 (Invitrogen) as a transfection reagent was used by following the manufacturer's protocol as described previously (4).
Fluorescence Microscopy of Live Cells
All samples were imaged at ambient temperature (∼25 °C) with a 60× 1.49 numerical aperture objective (Nikon Apo TIRF) using a Photometrics CoolSnap ES2 CCD detector mounted onto a Nikon TE-2000E inverted microscope. GFP detection was accomplished using a S484/15× excitation filter (Chroma Technology), S517/30m emission filter (Chroma Technology), and Q505LP/HQ510LP dichroic (Chroma Technology); and OFP detection was carried out using a S555/25× excitation filter (Chroma Technology), S605/40m emission filter (Chroma Technology), and Q575LP/HQ585LP dichroic (Chroma Technology). Nikon imaging software, NIS-Elements (version 3.0) was used for collecting images from samples enlightened with the Nikon mercury fiber illuminator.
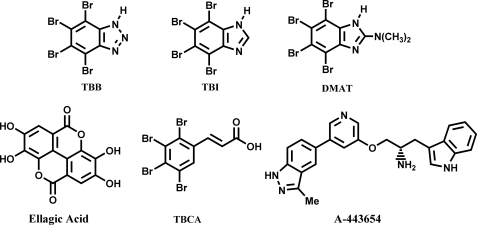
Small molecules were added to cells after washing three times with buffered saline solution (BSS: 20 mm HEPES (pH 7.4), 135 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2, and 5.6 mm glucose). Images were acquired immediately prior to drug addition and after cells had been incubated with drugs at various time points. Various ranges of five CK2 inhibitors (stock: 10 mg/ml in dimethyl sulfoxide or ethanol; see Fig. 2) were tested to give final concentrations of ∼5–150 μg/ml; 4,5,6,7-tetrabromo-1_H_-benzimidazole (TBI; Sigma; 12, 23, 58, or 116 μm), 2-dimethylamino-4,5,6,7-tetrabromo-1_H_-benzimidazole (DMAT; Sigma; 21, 52, 105, or 314 μm), tetrabromocinammic acid (TBCA; Calbiochem; 54 or 108 μm), 4,4′,5,5′,6,6′-hexahydroxydiphenic acid 2,2′,6,6′-dilactone (ellagic acid; Calbiochem; 148 μm) and 4,5,6,7-tetrabromobenzotriazole (TBB; Sigma; 23, 58, 115, or 345 μm). However, in depth investigations using TBI were mostly carried out at a final concentration of 23 μm unless its concentration is specified. Experiments targeting Akt kinases were performed by the addition of 25–100 μm of A-443654. Control experiments were also carried out with 1–30 μl dimethyl sulfoxide or ethanol (i.e. 0.05–1.5% dimethyl sulfoxide or ethanol in final working solution).
FIGURE 2.
Chemical structures of small molecules that inhibit CK2 and Akt kinases.
RNA Interference
A siRNA expression system was prepared using the psiRNAhH1GFPzeo vector (InvivoGen) according to the manufacturer's protocol. To specifically knock down the human CK2α catalytic subunit in HeLa cells, four different DNA sequences generating short hairpin RNAs were inserted into the siRNA expression vector using restriction enzymes Acc65I and HindIII: 5′-GTACCAGACGTTAACAGACTA-3′ (siCK2α-1), 5′-GTGGATTTATAGTAGTTCAGT-3′ (siCK2α-2), 5′-GAAGGGAGGACCCAATCTATA-3′ (siCK2α-3), and 5′-GCTTGCTGGTCGCTTACATCA-3′ (siCK2α-4). Each siRNA transfection was monitored by a GFP marker present in the plasmid under fluorescence microscopy. hFGAMS-OFP was cotransfected to assess the efficiency of purinosome formation affected by siRNAs.
Immunoblotting of HeLa Lysates
Western blot analysis using commercially available anti-hCK2α antibody (C-18; Santa Cruz Biotechnology) was carried out to measure the expression level of hCK2α as decreased by individual siRNA constructs. To be consistent with cellular imaging, HeLa cells grown in purine-depleted medium were transfected with plasmids expressing siRNAs by Lipofectamine™ 2000. Cells were harvested 24 h after transfection and lysed in mammalian protein extraction reagent solution (Pierce) containing both protease inhibitor mixture (Roche Applied Science) and phosphatase inhibitor mixture (Roche Applied Science). Cell lysates then were loaded onto SDS-PAGE gels, and enhanced chemiluminescence signals were captured with the Fluor-S imaging system (Bio-Rad) as described before (4). The CK2α-specific band was normalized for quantitative analysis using the chemiluminescent signal of endogenous hTrifGART enhanced by anti-hTrifGART antibody (4).
RESULTS
Small Molecule Inhibition of CK2 in HeLa Cells Expressing GFP Fusion Proteins of de Novo Purine Biosynthetic Enzymes
Among the potential CK2 substrate entities (13), we were specifically interested in three enzymes that participate in de novo purine biosynthesis: hPPAT, hFGAMS, and hTrifGART (Fig. 1) and thus the effect of CK2 inhibition on their incorporation into a purinosome. We first applied four CK2-specific inhibitors: TBI, DMAT, TBCA, and ellagic acid (Fig. 2). When these drugs were individually introduced into HeLa cells expressing hFGAMS-GFP as a purinosome marker, almost all transfected cells (∼95%) exhibited the formation of the cytoplasmic purinosomes (Fig. 3, a–b, and supplemental Fig. S1). The phenotypic change was detected at ∼15 min and nearly completed across all the cells within an hour after drug addition. These CK2 inhibition studies were carried out using live HeLa cells grown in either purine-rich or purine-depleted medium and consistently provided similar robust signals. The observed changes in localization were also routinely detected at various concentrations of the drugs as indicated in the “Experimental Procedures” (e.g. TBI, 12–116 μm). We also confirmed that HeLa cells incubated with CK2 inhibitors for <2 h were able to proliferate again on Petri dishes when drugs were washed off and growth media restored. Because CK2 was previously identified as a bona fide in vivo target of TBI and DMAT in suspended HeLa cells (17), the phenotypic changes we observed in adherent HeLa cells appear to be a consequence of CK2 inhibition.
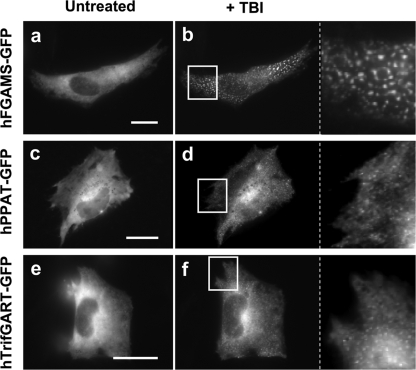
FIGURE 3.
Effects of a CK2-specific inhibitor, TBI, on the purinosome formation in HeLa cells transiently expressing a GFP fusion protein. Representative cells expressing hFGAMS-GFP (a and b), hPPAT-GFP (c and d), or hTrifGART-GFP (e and f) were subjected to cellular imaging prior to addition of TBI (untreated: a, c, and e) and after the cells had been incubated with TBI for a given period (b, 30 min; d, 40 min; f, 60 min). The indicated regions of interest (white boxes) were enlarged for clarification. Scale bars, 10 μm.
With hPPAT-GFP as a purinosome marker in HeLa cells the addition of TBI also resulted in ∼85–90% of the transfected cells exhibiting purinosomes within an hour (Fig. 3, c–d). In cells expressing hTrifGART-GFP, the effect of TBI on the majority of the cells (∼85%) was subtle (supplemental Fig. S2), with a smaller fraction (∼15%) of the cells forming distinct clusters (Fig. 3, e–f). However, with TBCA, >70% HeLa cells transiently expressing hTrifGART-GFP readily formed purinosomes, implying that each drug may possess different in vivo efficacy. Collectively, hCK2 inhibition appears to perturb the balance between purinosome association/disassociation by possibly reducing the ratio of phosphorylated to dephosphorylated species for the three pathway-specific enzymes.
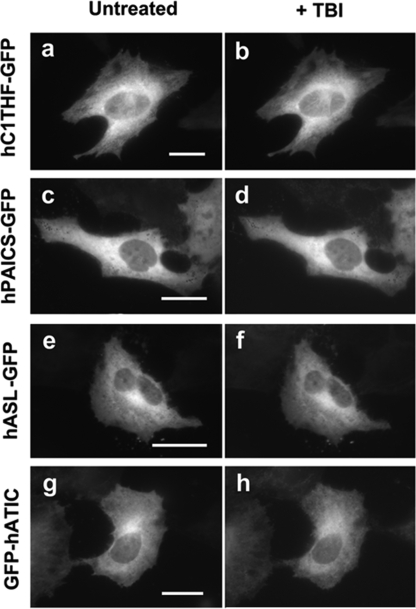
To validate the enzyme-specific responses to the CK2 drugs and to obviate localization artifacts, a series of control experiments were conducted. hC1THF, the enzyme that synthesizes a critical cofactor, 10-formyl-tetrahydrofolate, for the transformylase enzymes in the de novo purine biosynthetic pathway is also identified as a substrate for CK2 (13) but is not a member of the purinosome cluster (4). In cells expressing hC1THF-GFP, TBI did not affect its cellular localization (Fig. 4, a–b). Thus, CK2 inhibition did not introduce a localization artifact of the targeted enzymes. Additional control experiments monitored intracellular hPAICS-GFP, which is not a CK2 substrate and does not possess a hypothetical CK2 phosphorylation site, according to the Scansite database (18), an Internet-based phosphosite prediction algorithm. The addition of TBI to the cells transiently expressing hPAICS-GFP did not induce or dissociate the purinosomes (Fig. 4, c–d). Similarly, the loci in the cytoplasm of human adenylosuccinate lyase-GFP and GFP-hATIC that likewise do not possess a CK2 phosphorylation site were not changed upon the addition of TBI (Fig. 4, e–h). Collectively, it is apparent that CK2 is the principal regulatory kinase for the complexation of the de novo purine biosynthetic enzymes.
FIGURE 4.
No alteration in cellular distribution of a non-purinosome member or non-CK2-substrate enzymes upon addition of TBI into singly transfected HeLa cells. Representative cells expressing hC1THF-GFP (a and b), hPAICS-GFP (c and d), human adenylosuccinate lyase (hASL)-GFP (e and f), or GFP-hATIC (g and h) were subjected to cellular imaging prior to addition of TBI (untreated: a, c, e, and g) and after the cells had been incubated with TBI for a given period (b, 60 min; d, 30 min; f, 30 min; and h, 50 min). Scale bars, 10 μm.
Small Molecule Inhibition of Akt Kinases in HeLa Cells Expressing hFGAMS-GFP
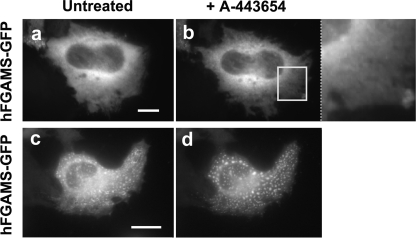
To explore for participation by another kinase in the assembly/disassembly of purinosomes in HeLa cells, we applied a small molecule, A-443654 (Fig. 2), that was validated as a specific inhibitor against Akt kinases (Akt1, Akt2, and Akt3) (19, 20). This experiment using hFGAMS-GFP as a purinosome marker showed that A-443654 did not promote the formation of purinosome clusters (Fig. 5, a–b) nor the dissociation of purinosomes (Fig. 5, c–d). Taking into account that the Akt substrate identification study (14) utilized cytokine-dependent bone marrow-derived Ba/F3 cells, whereas HeLa S3 cells were employed for CK2 substrate identification (13), our cellular investigation supported a functional role of CK2, instead of Akt, for the purinosome assembly at least in HeLa cells.
FIGURE 5.
No effect of an Akt-specific inhibitor, A-443654, added to the HeLa cells expressing hFGAMS-GFP. Representative cells grown in either purine-rich (a) or -depleted (c) medium were imaged prior to addition of A-443654, (untreated: a and c) and after the cells had been incubated with A-443654 for 70 min (b and d). The indicated region of interest (a white box) was enlarged for clarification. Scale bars, 10 μm.
CK2 Inhibition in Dually Transfected HeLa Cells
To directly confirm the observed cluster formation in the presence of CK2 inhibitors, two enzymes in the pathway, hFGAMS-OFP and hTrifGART-GFP, were dually transfected into HeLa cells. Purinosomes were clearly induced in ∼70–80% cells after the addition of TBI (supplemental Fig. S3, a–d). This CK2 inhibition study was also tested on the cells grown in either purine-rich or -depleted medium, consistently reproducing a similar phenotypic transformation. Co-clustering of hFGAMS and hTrifGART was also independent of the position of the fluorescent probes (i.e. hFGAMS-GFP and hTrifGART-OFP). Another combination of the three CK2-substrate pathway enzymes (i.e. hPPAT-GFP and hFGAMS-OFP) also exhibited a similar trend of purinosome formation by TBI-induced CK2 inhibition.
We further tracked the behavior of two proteins, including a non-CK2-substrate enzyme, in dually transfected HeLa cells. ∼85–90% of HeLa cells coexpressing hPAICS-GFP and hFGAMS-OFP clearly induced co-clusters upon TBI addition (supplemental Fig. S3, e–h). We also confirmed co-cluster formation by TBI in HeLa cells coexpressing human adenylosuccinate lyase-GFP or GFP-hATIC with hFGAMS-OFP. Therefore, we conclude that perturbation in post-translational modification (i.e. phosphorylation/dephosphorylation) on the three pathway enzymes, hPPAT, hFGAMS and hTrifGART, would be sufficient to nucleate other purine biosynthetic pathway enzymes into clusters.
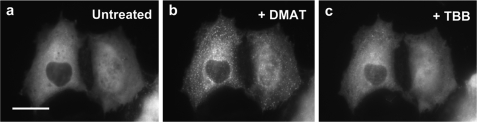
Purinosome Assembly/Disassembly in the Presence of TBB
The addition of another CK2 inhibitor, TBB (Fig. 2), at a high concentration (345 μm) to cells expressing hFGAMS-GFP revealed a two-phase time-dependent transition (supplemental Fig. S4); the first phase was the formation of purinosomes, and the second phase was the dissociation of the purinosome clusters. Although the first phase transition was consistent with observed effect of other CK2 inhibitors, the second phase transition was unexpected. At a lower concentration (23 μm) of TBB, the inhibitor acted to dissociate the purinosome. Although the mechanism by which TBB affects purinosome disassembly is not clear, we were capable of chemically reversing purinosome formation in cells by the sequential addition of two small molecule CK2 inhibitors. For example, DMAT or TBI synchronized ∼90–95% cells to form clusters (tracked by hFGAMS-GFP, Fig. 6, a–b) that dissolved upon the subsequent addition of TBB (Fig. 6, b–c).
FIGURE 6.
Reversibility of purinosome formation in HeLa cells by the sequential addition of DMAT and TBB. Representative cells expressing hFGAMS-GFP were monitored for purinosome formation prior to drug addition (a) and after incubating with DMAT for 25 min (b). DMAT-containing buffer was exchanged for fresh buffer containing TBB, subsequently promoting the dissociation of purinosomes within 20 min in the cells (c). Scale bars, 10 μm.
TBI-promoted Purinosome Formation in C3A Cells
In addition to HeLa cells, a human liver carcinoma cell line, C3A, also utilized purinosomes preferentially under purine depleted conditions relative to purine-rich medium (supplemental Fig. S5, a–b). Subsequently, we confirmed that TBI was able to promote the compartmentalization of the de novo purine biosynthetic enzymes in the cytoplasm of C3A cells (supplemental Fig. S5, c–d). This study was carried out by tracking hFGAMS-GFP as a purinosome marker. However, we noted that clustering in HeLa cells occurred at ∼15 min after the addition of TBI, and C3A cells required ∼60 min to exhibit such phenotypic changes after the same doses (58 or 116 μm) of TBI.
Silencing hCK2α Catalytic Subunit by siRNA
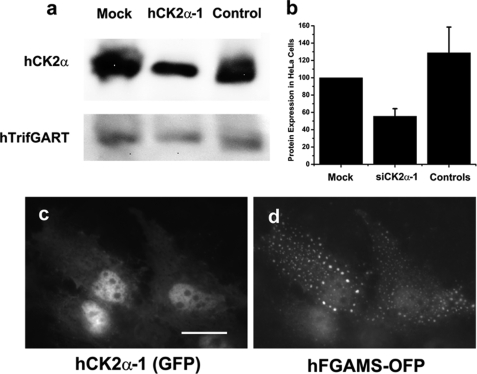
An alternative experimental technique in addition to the multiple CK2 inhibitors was next used to confirm the functional participation of CK2 in purinosome assembly. We constructed four plasmids expressing siRNAs targeting the hCK2α catalytic subunit in HeLa cells. Western blot analysis revealed that one construct (i.e. siCK2α-1) successfully knocked down ∼45% of the endogenous level of the hCK2α catalytic subunit in HeLa cells (Fig. 7, a–b). However, the other three constructs (i.e. siCK2α-2, -3, and -4) did not noticeably reduce the expression level of hCK2α, thus serving as negative controls. In addition, cellular imaging experiments revealed that nearly all HeLa cells coexpressing hFGAMS-OFP and siCK2α-1 (the latter carries a GFP marker in the expression plasmid) exhibited purinosomes (Fig. 7, c–d), whereas co-transfection of the other siRNAs did not effect purinosome formation. Imaging experiments with siCK2α-1 were again conducted for HeLa cells maintained in either purine-rich or -depleted medium. Under both conditions, ∼95% of the co-transfected HeLa cells demonstrated the formation of cytoplasmic clusters upon RNA interference. Consequently, the siRNA studies corroborate those involving imaging-based CK2 drug inhibition that together strongly implicate CK2 as the primary kinase regulating purinosome appearance.
FIGURE 7.
Silencing the endogenous hCK2α catalytic subunit by RNA interference. Western blot analysis revealed that siCK2α-1 knocked down ∼45% of the expression level of the endogenous hCK2α catalytic subunit in HeLa cells (a and b). hCK2α expression was quantitatively normalized relative to the expression level of the endogenous hTrifGART (a and b). S.D. were indicated by error bars (b). In addition, cellular imaging experiments revealed that purinosomes were clearly detected in >95% of dually transfected HeLa cells coexpressing siCK2α-1 (c) and hFGAMS-OFP (d). Scale bars, 10 μm.
DISCUSSION
Previously, we demonstrated the reversibility of the purinosome in cells, consequently proposing a regulatory mechanism for the purinosome assembly in vivo (4). Aware of growing in vitro evidence (13, 14, 21) obtained from proteomic scale experiments for de novo purine biosynthetic enzymes as substrates for kinases, we investigated the hypothesis that, in vivo, a kinase-mediated signal transduction would control the purinosome assembly/disassembly in cells. Excitingly, an in vitro kinase substrate identification method (13) revealed significant numbers of metabolic enzymes as new CK2 substrates; including three of six de novo purine biosynthetic enzymes (i.e. hPPAT, hTrifGART, and hFGAMS) as well as hC1THF, which synthesizes a key cofactor for the de novo purine biosynthesis. Our subsequent investigation of this finding was carried out by adding multiple CK2 inhibitors into cells transiently expressing a GFP fusion of the CK2-substrate pathway enzymes (i.e. hPPAT-GFP, hTrifGART-GFP, or hFGAMS-GFP). All three targeted enzymes were incorporated into the purinosome upon CK2 inhibition. Additionally, their nucleation in the cytoplasm appeared to recruit the other pathway enzymes to form functional purinosomes. The drugs we used (i.e. TBI, DMAT, TBCA, and ellagic acid) are fairly specific and potent enough to inhibit CK2 in vitro and in vivo (11, 12, 17). Taken together with a series of control experiments, CK2 inhibition influences the assembly of the purinosome only when the substrates of the CK2 enzyme were used as imaging markers.
Subsequently, we genetically knocked down the expression level of the hCK2α catalytic subunits in HeLa cells by RNA interference. Since CK2 was previously identified as an essential gene product (22), significant silencing of hCK2α expression might be fatal for the cells. However, from Western blot analysis (∼45% silencing from lawns of HeLa cells; Fig. 7) and given the transfection efficiency of the siCK2α-1 expression plasmid (∼70–80%), we estimate a level of ∼55–65% silencing of hCK2α in single cells. Such moderate silencing of hCK2α was apparently sufficient to stimulate purinosome clustering without cell death. Collectively, the subtle fluctuation of cellular CK2 activity appears to be sufficient to perturb cellular processes, including the de novo purine biosynthetic pathway.
Our experimental evidence raises the possibility that a reversible phosphorylation and dephosphorylation process may be mediated by a kinase and phosphatase pair. Unlike the other CK2 inhibitors, the addition of TBB unexpectedly promoted the dissociation of purinosomes. Although TBB, TBI, and DMAT were previously shown to inhibit CK2α catalytic subunits in cells (17), the TBB-specific cellular response tracked by a purinosome marker clearly implies that potential off target(s) of TBB and/or downstream of CK2-mediated signal transductions may have a profound influence on a hypothetical phosphatase-activating pathway. This hypothesis gains support from the study of the sequential addition of two CK2 drugs, DMAT and TBB (Fig. 6). When DMAT followed by TBB was supplied to the cells, purinosome formation was first observed followed by a second phase transition to dissociation. Although an inactive conformation of human CK2 was recently reported in a structural study (23), CK2 is generally believed to be a constitutively active kinase. Therefore, the switching on and off of an unknown phosphatase activity in conjunction with CK2 kinase activity may also constitute a part of a key signal transduction for reversible purinosome assembly.
At present, we cannot rule out additional kinase participation in the de novo purine biosynthetic pathway. A recent report suggested that a phosphatidylinositol 3-kinase/Akt signaling pathway (24) seems to regulate both upstream and downstream of de novo purine biosynthesis through phosphoribosyl pyrophosphate synthetase and ATIC (24). Nucleophosmin-anaplastic lymphoma kinase also appears to phosphorylate ATIC, consequently increasing its enzymatic activity (25). Moreover, hFGAMS and hTrifGART were postulated to be phosphorylated on their 14-3-3 protein binding motifs (21). Considering that several metabolic pathways are interconnected with de novo purine biosynthesis, it is certainly possible for cells to possess a much more complicated regulatory mechanism for the purinosome. Nevertheless, we have provided compelling evidence to support our contention that protein kinase CK2 dynamically regulates the purinosome in cells through three de novo purine biosynthetic enzymes.
Investigation of anti-metabolites targeting de novo purine biosynthesis has led to a number of drugs for the treatment of human diseases; for example, methotrexate, 6-mercaptopurine, thioguanine, hydroxyurea, and deoxycoformycin for acute leukemia (26, 27). In addition, two folate-dependent enzymatic activities in this pathway (i.e. the activities of GAR transformylase (step 3) in TrifGART and aminoimidazolecarboxamide ribonucleotide transformylase (step 9) in ATIC) are considered as key targets for the development of antineoplastic agents, for example Lometrexol, LY309887, and Pemetrexed (Alimta) now in the clinic (28). Many of these drugs were designed primarily to perturb enzymatic activities on the basis of chemical structures resembling purines or precursors and may have additional effects on the context of the purinosome. Additionally, elevated CK2 activity has been associated with the malignant transformation of several tissues and with aggressive tumor behavior (11), leading to the development of CK2 inhibitors now in clinical trials (29). As presented here, the direct correlation between purinosome assembly and hCK2 activity would tie inhibition of de novo purine biosynthesis to anti-cancer drug discovery. In conclusion, the purinosome (4) and other metabolic organizations (30, 31) provides a novel class of drug targets whose manipulation by pharmacophores might provide a means of controlling flux through those pathways.
Supplementary Material
Supplemental Data
Acknowledgment
We thank Dr. Ye Fang (Corning, Inc.) for sharing a human C3A cell line.
*
This work was supported, in whole or in part, by National Institutes of Health Grants GM24129 (to S. J. B.) and RO1 EB0001987 (to K. M. S.).
3
The abbreviations used are:
CK2
casein kinase 2
hCK2
human protein kinase CK2
GFP
green fluorescent protein
OFP
orange fluorescent protein
IMP
inosine monophosphate
PPAT
phosphoribosyl pyrophosphate amidotransferase
GARS
glycinamide ribonucleotide (GAR) synthetase
FGAMS
formylglycinamidine ribonucleotide synthase
CAIRS
carboxyaminoimidazole ribonucleotide synthase
SICARS
succinylaminoimidazolecarboxamide ribonucleotide synthetase
IMPCH
IMP cyclohydrolase
TrifGART
trifunctional GARS-GAR transformylase-aminoimidazole ribonucleotide synthetase
PAICS
bifunctional CAIRS-SICARS
ATIC
bifunctional aminoimidazolecarboxamide ribonucleotide transformylase-IMPCH
siRNA
small interfering RNA
TBI
4,5,6,7-tetrabromo-1_H_-benzimidazole
DMAT
2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole
TBCA
tetrabromocinammic acid
ellagic acid
4,4′,5,5′,6,6′-hexahydroxydiphenic acid 2,2′,6,6′-dilactone
TBB
4,5,6,7-tetrabromobenzotriazole.
REFERENCES
- 1.Rowe P. B., McCairns E., Madsen G., Sauer D., Elliott H. (1978) J. Biol. Chem. 253, 7711–7721 [PubMed] [Google Scholar]
- 2.Smith G. K., Mueller W. T., Wasserman G. F., Taylor W. D., Benkovic S. J. (1980) Biochemistry 19, 4313–4321 [DOI] [PubMed] [Google Scholar]
- 3.Srere P. A. (1987) Annu. Rev. Biochem. 56, 89–124 [DOI] [PubMed] [Google Scholar]
- 4.An S., Kumar R., Sheets E. D., Benkovic S. J. (2008) Science 320, 103–106 [DOI] [PubMed] [Google Scholar]
- 5.Meggio F., Pinna L. A. (2003) FASEB J. 17, 349–368 [DOI] [PubMed] [Google Scholar]
- 6.Wilson L. K., Dhillon N., Thorner J., Martin G. S. (1997) J. Biol. Chem. 272, 12961–12967 [DOI] [PubMed] [Google Scholar]
- 7.Filhol O., Cochet C. (2009) Cell. Mol. Life Sci. 66, 1830–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trembley J. H., Wang G., Unger G., Slaton J., Ahmed K. (2009) Cell. Mol. Life Sci. 66, 1858–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominguez I., Sonenshein G. E., Seldin D. C. (2009) Cell. Mol. Life Sci. 66, 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faust M., Montenarh M. (2000) Cell Tissue Res. 301, 329–340 [DOI] [PubMed] [Google Scholar]
- 11.Duncan J. S., Litchfield D. W. (2008) Biochim. Biophys. Acta 1784, 33–47 [DOI] [PubMed] [Google Scholar]
- 12.Pagano M. A., Bain J., Kazimierczuk Z., Sarno S., Ruzzene M., Di Maira G., Elliott M., Orzeszko A., Cozza G., Meggio F., Pinna L. A. (2008) Biochem. J. 415, 353–365 [DOI] [PubMed] [Google Scholar]
- 13.Allen J. J. (2008) Development and Application of Technologies to Study Individual Kinase Substrate Relationships. Ph.D. thesis, University of California, San Francisco [Google Scholar]
- 14.van Gorp A. G. (2007) The Identification and Functional Characterization of Novel Targets of Protein Kinase B (PKB/c-akt) Action. Ph.D. thesis, Proefschrift Universiteit, Utrecht, Netherlands [Google Scholar]
- 15.Pinna L. A. (1990) Biochim. Biophys. Acta 1054, 267–284 [DOI] [PubMed] [Google Scholar]
- 16.Berwick D. C., Hers I., Heesom K. J., Moule S. K., Tavare J. M. (2002) J. Biol. Chem. 277, 33895–33900 [DOI] [PubMed] [Google Scholar]
- 17.Duncan J. S., Gyenis L., Lenehan J., Bretner M., Graves L. M., Haystead T. A., Litchfield D. W. (2008) Mol. Cell. Proteomics 7, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 18.Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuzumi T., Fiedler D., Zhang C., Gray D. C., Aizenstein B., Hoffman R., Shokat K. M. (2009) Nat. Chem. Biol. 5, 484–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y., Shoemaker A. R., Liu X., Woods K. W., Thomas S. A., de Jong R., Han E. K., Li T., Stoll V. S., Powlas J. A., Oleksijew A., Mitten M. J., Shi Y., Guan R., McGonigal T. P., Klinghofer V., Johnson E. F., Leverson J. D., Bouska J. J., Mamo M., Smith R. A., Gramling-Evans E. E., Zinker B. A., Mika A. K., Nguyen P. T., Oltersdorf T., Rosenberg S. H., Li Q., Giranda V. L. (2005) Mol. Cancer Ther. 4, 977–986 [DOI] [PubMed] [Google Scholar]
- 21.Pozuelo Rubio M., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., Mackintosh C. (2004) Biochem. J. 379, 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lou D. Y., Dominguez I., Toselli P., Landesman-Bollag E., O'Brien C., Seldin D. C. (2008) Mol. Cell. Biol. 28, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raaf J., Issinger O. G., Niefind K. (2009) J. Mol. Biol. 386, 1212–1221 [DOI] [PubMed] [Google Scholar]
- 24.Wang W., Fridman A., Blackledge W., Connelly S., Wilson I. A., Pilz R. B., Boss G. R. (2009) J. Biol. Chem. 284, 3521–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boccalatte F. E., Voena C., Riganti C., Bosia A., D'Amico L., Riera L., Cheng M., Ruggeri B., Jensen O. N., Goss V. L., Lee K., Nardone J., Rush J., Polakiewicz R. D., Comb M. J., Chiarle R., Inghirami G. (2009) Blood 113, 2776–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elion G. B. (1989) Science 244, 41–47 [DOI] [PubMed] [Google Scholar]
- 27.Riscoe M. K., Brouns M. C., Fitchen J. H. (1989) Blood Rev. 3, 162–173 [DOI] [PubMed] [Google Scholar]
- 28.Field M. S., Anguera M. C., Page R., Stover P. J. (2009) Arch. Biochem. Biophys. 481, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinna L. A., Allende J. E. (2009) Cell. Mol. Life Sci. 66, 1795–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campanella M. E., Chu H., Low P. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narayanaswamy R., Levy M., Tsechansky M., Stovall G. M., O'Connell J. D., Mirrielees J., Ellington A. D., Marcotte E. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data