Noninvasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells (original) (raw)
Abstract
We have developed a novel molecular methodology that utilizes stool samples containing intact sloughed epithelial cells to quantify intestinal gene expression profiles in the developing human neonate. Since nutrition exerts a major role in regulating neonatal intestinal development and function, our goal was to identify gene sets (combinations) that are differentially regulated in response to infant feeding. For this purpose, fecal mRNA was isolated from exclusively breast-fed (n = 12) and formula-fed (n = 10) infants at 3 mo of age. Linear discriminant analysis was successfully used to identify the single genes and the two- to three-gene combinations that best distinguish the feeding groups. In addition, putative “master” regulatory genes were identified using coefficient of determination analysis. These results support our premise that mRNA isolated from stool has value in terms of characterizing the epigenetic mechanisms underlying the developmentally regulated transcriptional activation/repression of genes known to modulate gastrointestinal function. As larger data sets become available, this methodology can be extended to validation and, ultimately, identification of the main nutritional components that modulate intestinal maturation and function.
Keywords: systems biology, breast feeding, biomarkers, microarray
translational highlights Given the need to better understand gastrointestinal health and development, the use of noninvasive tests is expected to become increasingly relevant tools in tailoring diet to meet the nutritional needs of the growing infant. Chapkin and colleagues developed a novel molecular methodology that utilizes stool samples containing intact sloughed epithelial cells, to noninvasively quantify intestinal gene expression profiles in the developing human neonate. Linear discriminant analysis was successfully used to identify the best single genes and two- to three-gene combinations for distinguishing the feeding groups.
Gastrointestinal (GI) maturation involves a continuous cascade of growth, differentiation, and renewal of epithelial cells. In humans, the intestinal tract is functionally immature and immunologically naïve at birth (18). Evidence from animal models (48) and human infants (7, 12) demonstrates that the intestinal tract of the newborn undergoes marked structural and functional adaptation in response to feeding. The trophic response to human milk exceeds that to formula, suggesting that bioactive components in human milk are important in this response (19, 20). Nutrition during early postnatal development can modulate intestinal development in at least three ways: 1) directly through stimulation of transcription and/or translation by milk-derived nutrients (5), 2) indirectly through modulation of the intestinal microbiota (9) or the mucosal immune system (29), and/or 3) through induction of maturation of gene-specific methylation, thereby permanently altering the expression of developmental genes in the intestine (46, 47). Therefore, it is essential to determine the effects of postnatal nutrition on intestinal global gene expression profiles in the infant. However, because of the ethical constraints associated with obtaining tissue biopsies from healthy infants, no investigators have comprehensively profiled intestinal gene expression during early postnatal development.
Recent interest has focused on exfoliated cells as a tool to investigate the impact of therapeutic and nutritional regimens on the maturation of GI functions (17, 30). Approximately one-sixth to one-third of normal adult colonic epithelial cells are shed daily (37). This number corresponds to the daily exfoliation of ∼1010 cells. Because the number of intact cells that can be isolated from fecal material is low (1, 14), we have developed a patented noninvasive mRNA-based method as a highly sensitive technique for detecting molecular markers of intestinal development and function. This methodology has the advantage of using exfoliated cell mRNA directly isolated from feces, which contain sloughed small intestinal and colon cells, and, therefore, does not result in any discomfort to the subject. The method is capable of isolating and quantifying specific mRNAs under various intestinal conditions, including mode of feeding, and has been tested in a rat experimental colon carcinogenesis model (8, 13) and in adult humans (15, 51). We propose that the ability to use exfoliated cell mRNA, instead of biopsy or autopsy material, would be highly advantageous to document the impact of nutrition on the continuum of intestinal development and maturation in the healthy newborn. Therefore, in this study, we noninvasively examined intestinal transcriptional profiles in infants exclusively fed formula or breast milk 1) to validate that this technique can be applied to infant fecal samples and 2) to identify gene sets (combinations) in response to mode of feeding, i.e., human milk or infant formula. We demonstrate for the first time that two- and three-gene combinations provide classifiers with potential to noninvasively identify discriminative signatures for the development of molecular markers to explore nutritional effects on maturation of intestinal function. In addition, we utilized complementary systems biology approaches, e.g., multivariate measurement of gene expression relationships, to identify “master” genes, which regulate the transcriptional states of other “slave” genes in the infant intestine.
METHODS
Subject Recruitment and Inclusion and Exclusion Criteria
Mothers of infants were recruited into the study between the third trimester of pregnancy and 1 mo postpartum. Healthy, full-term infants, who were exclusively breast-fed (BF) or fed commercially available infant formula (FF; Enfamil LIPIL, Mead Johnson Nutrition, Evansville, IN) and medically certified as healthy, i.e., asymptomatic and with no clinical indication of disease, were eligible for enrollment into the study. Infants who were diagnosed with intolerance to cow's milk or who were receiving both breast milk and formula, soy or casein hydrolysate formula, juice, or solid foods were excluded from the study. Enrolled infants who became clinically ill (fever, contagious diseases, or active diarrhea) or had received antibiotic treatment within 2 wk of the sample collection were excluded from the study. Mothers of the FF infants were provided with Enfamil LIPIL formula for the duration of the study, and mothers of the BF infants received a $25.00 gift card for each stool sample collection. Fecal samples were coded with a unique numerical identification to maintain confidentiality of the participants. The experimental protocol was approved by the University of Illinois and Texas A & M Institutional Review Boards, and informed consent was obtained from parents prior to participation in the study.
Sample Collection
Stool specimens were collected by the parent from exclusively BF and FF infants at 3 mo postpartum. Instructions on sample collection were provided to the parents. A sterile spoon was used to place ∼10 g of freshly voided fecal material into a sterile 50-ml conical tube containing 20 ml of guanidinium denaturation solution (Ambion, Austin, TX). Samples were mixed by hand to create a homogenous sample, which was immediately frozen at −20°C until it was transported on ice to the laboratory by the research staff. Samples were held at −80°C until they were shipped on dry ice to Texas A & M University for analysis.
Breast Milk and Formula Intake
For a full 24-h period preceding the stool sample collection, parents weighed the infant before and after each feeding on an electronic scale (Medela, McHenry, IL) without a change of clothing or diaper. The change in body weight was determined as an estimate of breast milk or formula intake. All data are presented as means ± SD. Maternal age and infant birth weight and length were compared using a Student's _t_-test (SAS version 6.0, SAS, Cary, NC). Differences in breast milk and formula intake, Z scores, and body weight at 1, 2, and 3 mo of age were determined using a repeated-measures ANOVA (SAS). Statistical significance was set at P ≤ 0.05.
mRNA Expression Microarray Analysis
From each subject, polyA+ RNA was isolated from feces as previously described (15). Because of the high level of bacterial RNA in fecal samples, polyA+ RNA was isolated to obtain a highly enriched mammalian polyA+ RNA population (14). In addition, an Agilent 2100 Bioanalyzer was used to assess integrity of fecal polyA+ RNA, and quantification was performed by spectrophotometer (NanoDrop, Wilmington, DE). Samples were processed in strict accordance to the CodeLink Gene Expression Assay manual (Applied Microarray, Tempe, AZ) and analyzed using the Human Whole Genome Expression Bioarray, as we previously described (16, 51). Each array contained the entire human genome derived from publicly available, well-annotated mRNA sequences. Arrays were inspected for spot morphology. Marginal spots were flagged as background contaminated, irregularly shaped, or saturated in the output of the scanning software. Spots that passed the quality-control standards were categorized as good (G). In addition, a reading of L indicated “near background.” The low-L measurements reflect true low gene expression levels or may have been caused by degradation of the mRNA, resulting in a low signal. Typically, samples collected from colonic mucosa (16) exhibit a relatively low proportion (30–45%) of L spots. In comparison, we previously reported that the proportion of L spots obtained from adult fecal samples is significantly higher (65–83%) (51). In the present study, the proportion of L spots was 45–77%; therefore, we performed statistical and classification analyses using only the common G spots (4,250) for all 22 samples.
Microarray Data Normalization
For the purpose of interarray normalization, a set of housekeeping genes was used. These were determined as follows.
Housekeeping gene preparation.
Common G probes (4,250) across all 22 microarrays were identified. Using a list of 575 housekeeping genes (24), we identified 33 housekeeping genes from the 4,250 common G probes found in the previous step (see supplemental methods, supplemental Fig. 1, and supplemental Table 1 in the online version of this article).
Additive normalization procedure.
Arrays were grouped across the type of feeding, and the average values of the 33 housekeeping genes were calculated (see supplemental Fig. 1). Median values of the averages were also calculated. Subsequently, a robust piecewise linear regression was performed, and the corresponding regression value for each array was calculated. Then the difference between the median and regression values for each array was determined, and the raw expression values of the common 4,250 genes on each array were shifted by the corresponding discrepancies.
Identifying Multivariate Discriminators (Feature Gene Sets) for Diet Classification
We used a previously described algorithm for feature set identification (51; also see supplemental methods). Estimation of the classification error is of critical importance when the number of potential feature sets is large. When sample size is limited, an error estimator may have a large variance and, therefore, may often be low, even if it is approximately unbiased. This can produce many feature sets and classifiers with low error estimates. We mitigate this problem by applying bolstered error estimation (3). This procedure places a kernel (density) at each data point and computes the error by integrating the kernels over their misclassification regions, rather than simply by counting incorrectly classified points, as is done in resubstitution error estimation, thereby giving more weight to points near the classification boundary (see supplemental material for details on bolstering). Bolstered error estimation performs especially well compared with other error estimation methods in ranking feature sets, which was important in this analysis (41). The bolstered error estimated can be computed analytically in some cases, such as linear discriminant analysis (LDA; see supplemental material). Using prior knowledge consisting of a set of 529 biomarkers (genes) known to be involved in intestinal biology (51), we identified 146 potential features (genes) that were biomarkers and exhibited a G-flagged expression value for all the arrays. The relatively small size of this gene set allows for the comparison of the errors of all the possible single-, two-, and three-gene feature sets, thereby avoiding feature selection, which can be highly unreliable in small sample settings (42). The result of the overall approach is a list of “best” feature sets among all possible feature sets, i.e., those possessing minimum classification error.
Identifying Potential Master and Slave Genes
Mathematical modeling of “master-slave” relationships in a gene regulatory network was originally described by Dougherty et al. (22). While examining a portion of a gene regulatory network, one can view genes at various positions in that cascade as regulators/masters or regulated/slaves. This is a relative characterization: in certain situations, a gene might act as a master; in other situations, it might act as a slave. The intuitive notion of regulatory cascade can be formalized using the coefficient of determination (CoD) (22, 31). The CoD measures the relative increase in prediction accuracy by using the optimal predictor for the target based on the predictors compared with the best estimate of the target in the absence of predictors, i.e., the increase in prediction accuracy vs. only knowledge of the target distribution. The CoD has previously been used to measure multivariate nonlinear gene interaction (22, 31) and is defined as follows: (ε0 − ε·)/ε0, where ε· and ε0 are the errors of the optimal predictor and the predictor for the target using only statistics relating to the target itself, respectively. The key concept in defining master genes is the expectation that this type of regulator, when expressed, will be responsible for activating one or more cascades of subsequent changes in the expression of other genes. Thus one can achieve a more accurate prediction of the state of a master gene in a specific microarray sample if inferences are based on the observation of many of its slaves, rather than only on the behavior of a single slave. This modeling approach postulates probabilistic relations between masters and slaves and derives the CoD for the master relative to the slaves from the postulated relations (21). The validation of the model was performed using expression profiles from a series of 40 cell lines from a study of the response of benchmark cancer cell lines, the National Cancer Institute 60 Anti-Cancer Drug Screen cell line (27). The results suggest that the shapes of the histograms of the CoD values, when all the possible triples of genes are used as predictors of a given gene, can identify both master and slave genes in a particular cell context. In general, CoD histograms for master genes are skewed toward larger (closer to 1) CoD values, while the CoD histograms for the slaves tend to exhibit the opposite behavior (skewed toward 0) (21, 32).
RESULTS
Subject Demographics, Birth Weight, and Length
A total of 12 mothers of BF infants and 10 mothers of FF infants were recruited for the study. There were no differences in mean age or parity between mothers of BF and FF infants (Table 1). The sex distribution of the infants was similar in both groups, and most infants enrolled in the study were Caucasian. Birth length and weight were similar in both groups (Table 1).
Table 1.
Demographics and growth of study subjects
| Breast-Fed | Formula-Fed | |
|---|---|---|
| Sample size | 12 | 10 |
| Maternal age, yr | 28.0 ± 5.4 | 30.5 ± 5.0 |
| Parity | 2.0 ± 0.0 | 2.1 ± 0.6 |
| Infant sex | 8 male, 4 female | 7 male, 3 female |
| Infant ethnicity | 10 Caucasian, | 9 Caucasian, |
| 1 Hispanic, | 1 African-American | |
| 1 African-American | ||
| Length at birth, cm | 52.7 ± 3.6 | 51.0 ± 3.1 |
| Body weight, kg | ||
| Birth | 3.8 ± 0.6* | 3.5 ± 0.6* |
| 1 mo | 4.9 ± 0.7† | 4.6 ± 0.7† |
| 2 mo | 5.9 ± 0.7‡ | 5.7 ± 0.8‡ |
| 3 mo | 6.5 ± 0.7§ | 6.4 ± 0.9§ |
| Breast milk or formula intake, ml·kg−1·day−1 | ||
| 1 mo | 168.8 ± 36.4* | 175.4 ± 30.0* |
| 2 mo | 128.0 ± 20.6* | 162.9 ± 43.2*† |
| 3 mo | 123.0 ± 35.7† | 141.4 ± 14.2† |
Breast Milk and Formula Intake
Breast milk and formula intake (ml·kg−1·day−1) were unaffected by sex but differed over time (P = 0.001) and between diets (P = 0.038). Intake was affected by an interaction of feeding mode and time (P = 0.004). BF infant intake decreased between 1 and 2 mo of age and then remained stable (Table 1). FF infant intake was similar at 1, 2, and 3 mo of age. Intake differed between BF and FF infants at 2 mo of age, with FF infants having significantly higher intake per kilogram of body weight than BF infants (P = 0.007).
Postnatal Growth
Body weights (kg) of BF and FF infants at birth and 1, 2, and 3 mo of age are shown in Table 1. Body weight was significantly different across all time points, but no differences were detected between males and females or by feeding method. To compare the growth of infants in the study with reference standards, weight-for-age Z scores for individual infants were calculated in reference to the 2000 Centers for Disease Control growth standards for male and female infants (www.cdc.gov/GrowthCharts/). Z scores describe how far a child's weight is from the average weight of a child of the same age in the reference data. It is expressed in multiples of the standard deviation, with 0 reflecting the 50th percentile. Weight-for-age of all infants was within +2 to −1 Z scores, supporting normal growth and good nutritional status of the study population.
Classification of Diet
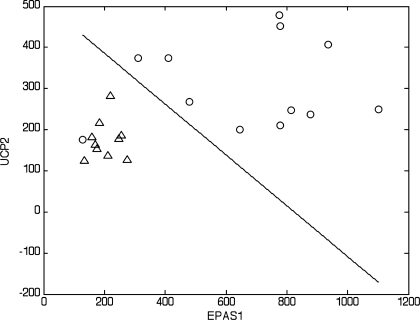
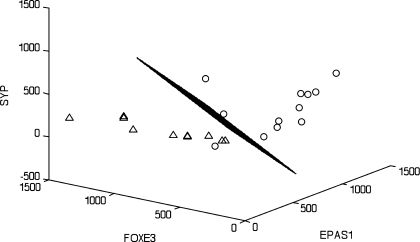
Although our primary goal was to validate this methodology for use in infants, we also wanted to identify if mode of feeding differentially influenced mRNA expression patterns. We applied a linear discriminator algorithm initially described by Zhao et al. (51). The number of genes (features) for each linear classifier was limited to 3, which allowed for an exhaustive search and, thus, avoided errors associated with small sample setting feature extraction. We identified the 10 best single-, two-, and three-gene classifiers to distinguish between the two groups of infants at 3 mo of age (Table 2). The results show several cases where single genes can provide good (in terms of the error estimate) classification (Table 2; see supplemental Table 2). However, when considering these features as part of two- or three-gene classifiers, we observed a significant decrease in the classification error. A similar phenomenon was recently documented in the context of gene network modeling (35). Specifically, the target (a gene or a phenotype) was predicted with greater accuracy by the expression profiles of a group of genes than by any proper subset of these genes (Table 2; see supplemental Tables 2–4). For example, the best single-gene classifier (1-feature) based on endothelial PAS domain-containing protein (EPAS1; ranked 1 in the list of single-gene classifiers) had an estimated classification error of 0.1214. Interestingly, conbination of this gene with a poorly performing single-gene classifier, uncoupling protein 2 (UCP2; ranked 33 in the list of single-gene classifiers), resulted in a two-gene classifier with a much lower estimated error of 0.0869 (Table 2). Moreover, UCP2 appeared frequently in the 10 best three-gene classifiers. These data clearly illustrate why complex phenotypes can be explained better by multivariate feature sets. To identify sets of genes that perform in a multivariate manner to provide strong classification, we specifically looked for pairs of genes that performed better than either of the genes individually and triplets of genes that performed well and substantially better than the best-performing pair among the three, and so on. To estimate the improvements of the classification performance, we introduced two quantities for each feature set: εbolstered and Δεbolstered. εbolstered denotes the bolstered resubstitution error for the LDA classifier for the respective feature set of size n, and Δεbolstered denotes the decrease in error with respect to the highest ranked of its subsets of features (in the list of features of size n − 1, n = 2,3). The feature sets were ranked on the basis of the value of εbolstered. Figures 1 and 2 show LDA classifiers for the best two- and three-gene feature sets listed in Table 2. The segregation of the two groups of infants can be readily seen in Figs. 1 and 2.
Table 2.
Classification of breast-fed vs. formula-fed infants
| Gene | εbolstered | Δεbolstered | ||
|---|---|---|---|---|
| EPAS1 | 0.1214 | |||
| NR5A2 | 0.1356 | |||
| NR3C1 | 0.1364 | |||
| PCDH7 | 0.1367 | |||
| ITGB2 | 0.1374 | |||
| FGF5 | 0.1385 | |||
| TJP1 | 0.1400 | |||
| MYB | 0.1456 | |||
| EPIM | 0.1478 | |||
| BAD | 0.1496 | |||
| EPAS1 | UCP2 | 0.0869 | 0.0345 | |
| CTDSPL | NR3C1 | 0.0975 | 0.0381 | |
| NR3C1 | TNFRSF10B | 0.0987 | 0.0377 | |
| FOXP4 | NR3C1 | 0.1026 | 0.0341 | |
| CDK4 | EPAS1 | 0.1039 | 0.0335 | |
| EPAS1 | SYP | 0.1045 | 0.0340 | |
| NR3C1 | SLC26A2 | 0.1057 | 0.0343 | |
| GPR41 | TJP1 | 0.1060 | 0.0396 | |
| FOXP1 | NR3C1 | 0.1064 | 0.0414 | |
| HSPA1A | NR3C1 | 0.1064 | 0.0432 | |
| EPAS1 | FOXE3 | SYP | 0.0778 | 0.0091 |
| CTDSPL | FOXE3 | NR3C1 | 0.0785 | 0.0190 |
| EPAS1 | TLR5 | UCP2 | 0.0785 | 0.0202 |
| EPAS1 | REG4 | UCP2 | 0.0802 | 0.0224 |
| EPAS1 | LIFR | UCP2 | 0.0808 | 0.0231 |
| EPAS1 | NODAL | UCP2 | 0.0821 | 0.0224 |
| EPAS1 | HIF3A | UCP2 | 0.0823 | 0.0234 |
| EPAS1 | HOXD10 | UCP2 | 0.0823 | 0.0237 |
| EPAS1 | KIT | UCP2 | 0.0824 | 0.0240 |
| ALOX5 | EPAS1 | UCP2 | 0.0827 | 0.0237 |
Fig. 1.
Linear discriminant analysis (LDA) phenotype classification using the genes endothelial PAS domain-containing protein 1 (hypoxia-inducible factor 2α, EPAS1) and uncoupling protein 2 (UCP2) provides the best-performing 2-gene feature set. Classification is between breast-fed (○) and formula-fed (△) infants (see Table 2 for additional details). Axes represent normalized intensity values of the indicated genes. Note clear separation between the groups, except for 1 outlier.
Fig. 2.
LDA phenotype classification using the genes EPAS1, forkhead box protein E3 (FOXE3), and synaptophysin (SYP) provides the best-performing 3-gene feature set. The 3-dimensional LDA hyperplane discriminates between breast-fed (○) and formula-fed (▵) infants (see Table 2 for additional details). Axes represent normalized intensity values of the corresponding genes. Note clear separation between the groups, except for 1 outlier (the same infant outlier in Fig. 1).
To evaluate how this classification approach relates to differential expression, we compared the 10 best one-feature sets and the genes showing differential expression between the two groups at P < 0.05 (Table 3), where _t_-tests are performed using normalized and logarithmically transformed gene intensity values. The comparison revealed that 9 of the 10 best one-feature (gene) sets identified by the linear (LDA) classifier also have P < 0.05. This is not surprising, because individual differentially expressed genes have been traditionally used to discriminate between phenotypes (38). We emphasize that these P values are only for comparison purposes; therefore, the cutoff of 0.05 has no bearing on classification in this study.
Table 3.
Relative exfoliated cell gene expression levels in breast-fed vs. formula-fed infants following a 3-mo feeding period
Identification of Master Regulatory Genes
Table 2 lists the 10 best single-gene classifiers among the 146 genes considered. We subsequently posed the following question: Do these genes provide good discrimination between the classes because they regulate large numbers of other genes? Although this question cannot be answered directly using the CoD method, a master gene that controls many slaves will possess a CoD histogram, the mass of which is concentrated to the right (toward 1). Thus we examined whether the best discriminators have master-like CoD histograms (see supplemental methods). After binarization, genes exhibiting less than 4 changes in their binary profiles were excluded from further CoD analyses, leaving only 55 of the original 146 biomarker genes. Of the 10 best single-gene classifiers in Table 2, all but tight junction protein 1 (TJP1) are among the 55 genes. TJP1 is missing because its binary profile exhibited less than 4 changes over the 22 infant samples. Rather than performing the CoD analysis with this small set, which includes only genes selected on the basis of putative biological relevance, we randomly selected 100 of the remaining 764 genes to provide a total of 155 genes to construct CoD histograms. To compute the CoD distributions, each gene was treated as the target in turn, and triples of the remaining 154 genes were queried to predict the target. This approach was used to examine a total of C(154,3) = 596,904 different combinations of triple predictors for each target gene and its CoD distribution histograms.
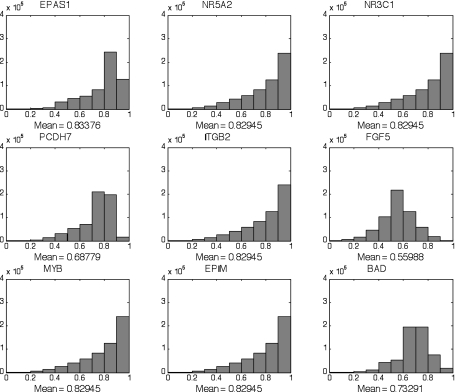
The CoD histograms for the single-classifier genes in Table 2, excluding TJP1, are shown in Fig. 3. CoD-histogram means >0.7 are considered strong, and those >0.8 are considered extremely strong. CoD means are >0.8 for six and very close to 0.7 for two of the nine genes from Table 2 (see supplemental Fig. 2 for CoD histograms of the 9 genes with the lowest CoD means).
Fig. 3.
Association between expression patterns of genes (triple predictors) was determined using coefficient of determination (CoD). CoD measures the degree to which the transcriptional levels of an observed gene set can be used to improve the prediction of the transcriptional state of a target gene relative to the best-possible prediction in the absence of observations. Examples for the strong single-gene classifiers in Table 2, excluding tight junction protein 1 (TJP1), are shown. NR5A2, nuclear receptor subfamily 5, group A, member 2; NR3C1, nuclear receptor subfamily 3, group C, member 1; PCDH7, protocadherin 7; ITGB2, integrin-β2; EPIM, epimorphin; BAD, Bcl2 antagonist of cell death.
DISCUSSION
The interaction between the major components of the intestinal ecosystem, including nutrients, microflora, epithelium, and aspects of nonspecific and specific immunity, has important implications for GI development and function, as well as overall health (6). Nutrition, in particular, is believed to play a major role in the modulation of the evolving infant gut. Recent evidence suggests that host factors in amniotic fluid and breast milk contribute to gut maturation (19, 44). The regulatory role of nutrition is particularly crucial during the early postnatal period, impacting GI barrier function, gut motility, mucosal immunity, digestive and absorptive capacity, and microbial colonization (10, 44). With respect to underlying mechanisms, data indicate that epigenetic regulation plays an important role in intestinal development and pathology (18, 46, 47). Unfortunately, because of the lack of tissue biopsies, no investigators have performed a global transcriptional analysis of the developing human intestine in the early postnatal period. Therefore, the overall objectives of this proof-of-principle study were 1) to validate the use of methodology to extract mRNA from infant fecal samples and 2) to noninvasively fingerprint and compare intestinal transcriptional profiles in exclusively BF or FF infants. As part of this effort, gene sets or combinations were identified in response to human milk or infant formula feeding. As opposed to using expression levels of significantly up- or downregulated genes, we applied novel mRNA-based noninvasive methods to identify the best single-gene and two- to three-gene combinations for distinguishing treatment effects. Similar to previous studies, we demonstrate that three-gene combinations provide classifiers with potential to noninvasively identify discriminative signatures for diagnostic purposes (32, 36). Collectively, these data indicate for the first time that genes that are modulated during neonatal gut epithelial differentiation are differentially represented in BF and FF infants.
It is likely that some of the variability among the BF infants results from potential differences in milk composition, namely, oligosaccharides and, potentially, lipids, as well as differences in the intestinal microbiota. Human milk contains >200 different oligosaccharides, which vary by maternal genetics (25). Human milk oligosaccharides have direct interactions with intestinal epithelial cells (33), as well as the microbiota (45), and, thus, may directly or indirectly affect host gene expression. Lipids are the most variable components of human milk and are directly impacted by maternal diet (4). This may be relevant, because several long-chain polyunsaturated fatty acids can directly regulate transcriptional activation of gene expression via activation of nuclear transcription factors (43). Lastly, the microbiota of BF and FF infants are reported to differ (2), and it is known that the microbiota and soluble metabolites of the microbiota can directly influence host gene expression (49).
From a biological context, the best-performing gene, on the basis of its ranking of single-, two-, and three-gene classifiers, was EPAS1. The EPAS1 protein is a basic helix-loop-helix PAS transcription factor involved in cellular response to hypoxia, whereby its activation is stimulated by hypoxia, with subsequent induction of vascular development (26, 40). Interestingly, EPAS1 was significantly (3-fold) upregulated in BF infants. This is noteworthy, because hypoxia-triggered angiogenesis may affect predisposition to proinflammatory states, including necrotizing enterocolitis (50). Although the role of EPAS1 in the evolving human neonatal intestine is not well defined, CoD analyses indicated its role as a master regulator of signaling cascades. This novel method finds associations between the expression patterns of individual genes by determining whether knowledge of the transcriptional levels of a small gene set can be used to predict the associated transcriptional state of another gene (21, 31). The power of this approach lies in its ability to find unexpected links between processes not previously known to be coordinated. Interestingly, the majority of master genes identified in this study are transcription factors associated with angiogenesis and wound repair, e.g., EPAS1, nuclear receptor subfamily 5, group A, member 2 (NR5A2), MYB, and nuclear receptor subfamily 3, group C, member 1 NR3C1 (11, 23, 26, 28, 34, 39). Other genes have also been implicated in GI morphogenesis: integrin-β2 (ITGB2), Bcl2 antagonist of cell death (BAD), protocadherin 7 (PCDH7), and fibroblast growth factor 5 (FGF5). It is noteworthy that several genes exhibited the same binary profile; i.e., their CoD distributions were identical (Fig. 3). This represents the interplay of three factors: 1) the sample size is small; 2) binarization has removed small differences, which indeed is a salient purpose of binarization; and 3) we conjecture that these genes are reacting concordantly in response to cellular conditions, which is supported by their strong CoD values and discriminatory power. We also must keep in mind that identical profiles might indicate that the genes are part of a tightly controlled regulated pathway in which one of them is actually the master over the pathway and that this master, or the pathway as a whole, controls a large family of genes. The validity and reliability of these molecular master-slave gene relationships may be better defined at the protein expression level and, therefore, warrant further studies to confirm physiological relevance.
Although the precise origin of exfoliated cells is not known, results from this study indicate that a number of genes associated with discrete epithelial cell types are detectable. Examples include absorptive enterocytes (lactase, sucrase-isomaltase), goblet cell (mucin-2), enteroendocrine cells (chromogranin A, intestinal mucin 3a), and paneth cell (lysozyme; data not shown). Thus it is likely that transcriptome signatures of the small and large intestine can be monitored over time. Clearly, additional studies are needed to elucidate the contribution of small and large intestine exfoliated cells to the pool of mRNA used in our screening test. We previously tested the feasibility of the noninvasive mRNA procedure in 1) a rat experimental colon carcinogenesis model (13) and 2) human subjects at high risk for colonic adenoma recurrence (15, 51). In a significant advancement, we reported that two- and three-gene combinations provide classifiers with potential to noninvasively identify discriminative molecular signatures. These findings support our hypothesis that it is possible to noninvasively detect molecular markers (combination gene sets) in exfoliated cells, which may provide an alternative approach to evaluating intestinal development and function in infants.
In conclusion, we demonstrate that fecal samples containing exfoliated cells hold potential for evaluating intestinal exposure to food and possibly other nondietary components in infants. Response to type of infant feeding based on one-, two-, and three-gene feature sets was accomplished using mRNA directly isolated from infant feces. In addition, on the basis of these preliminary data, the concordance between classification and CoD analyses emphasizes the potential usefulness of exfoliated cells in the identification of master controlling genes, many of which may be associated with mucosal morphogenesis. Given the need to better understand the role of nutrition in promoting GI health and development, noninvasive tests are expected to become increasingly relevant tools in tailoring diet to meet the nutritional needs of the growing infant. By continuing to build on this data set, this methodology may be used as a molecular tool to noninvasively evaluate the performance of infants, at the intestinal level, in response to different dietary regimens.
GRANTS
This work was supported by Mead Johnson and by National Institutes of Health Grants CA-59034, CA-129444, DK-71707, and P30 ES-09106.
DISCLOSURES
No conflicts of interest are declared by the authors.
Supplementary Material
[Supplemental Figures and Tables]
REFERENCES
- 1.Albaugh GP, Iyengar V, Lohani A, Malayeri M, Bala S, Nair P. Isolation of exfoliated colonic epithelial cells, a novel, non-invasive approach to the study of cellular markers. Int J Cancer 52: 347–350, 1992 [DOI] [PubMed] [Google Scholar]
- 2.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program 62: 13–29, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Braga-Neto UM, Dougherty ER. Bolstered error estimation. Pattern Recognition 37: 1267–1281, 2004 [Google Scholar]
- 4.Brenna JT, Salem N, Jr, Sinclair AJ, Cunnane SC. α-Linolenic acid supplementation and conversion to n-3 long-chain polyunsaturated fatty acids in humans. Prostaglandins Leukot Essent Fatty Acids 80: 85–91, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Burrin DB, Shulman RG, Reeds PJ, Davis TA, Gravitt KR. Porcine colostrum and milk stimulate visceral organ and skeletal muscle protein synthesis in neonatal piglets. J Nutr 122: 1205–1213, 1992 [DOI] [PubMed] [Google Scholar]
- 6.Canani RB, Passareillo A, Buccigrossi V, Terrin G, Guarino A. The nutritional modulation of the evolving intestine. J Clin Gastroenterol 42: S197–S200, 2008 [DOI] [PubMed] [Google Scholar]
- 7.Catassi C, Bonucci A, Coppa GV, Carlucci A, Giorgi PL. Intestinal permeability changes during the first month: the effect of natural versus artificial feeding. J Pediatr Gastroenterol Nutr 21: 383–386, 1995 [DOI] [PubMed] [Google Scholar]
- 8.Chapkin RS, Clark AE, Davidson LA, Schroeder F, Zoran DL, Lupton JR. Dietary fiber differentially alters cellular fatty acid binding protein expression in exfoliated colonocytes during tumor development. Nutr Cancer 32: 107–112, 1998 [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury SR, King DE, Willing BP, Band MR, Beever JE, Lane AB, Loor JJ, Marini JC, Rund LA, Schook LB, Van Kessel AG, Gaskins HR. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics 8: 215, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conroy ME, Shi HN, Walker WA. The long-term effects of neonatal microbial flora. Curr Opin Allergy Clin Immunol 9: 197–201, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Coste A, Dubuquoy L, Barnouin R, Annicotte JS, Magnier B, Notti M, Corazza N, Antal MC, Metzger D, Desreumaux P, Brunner T, Auwerx J, Schoonjans K. LRH-1-mediated glucocorticoid synthesis in enterocytes protects against inflammatory bowel disease. Proc Natl Acad Sci USA 104: 13098–13103, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cummings AG, Thompson FM. Postnatal changes in mucosal immune response: a physiological perspective of breast feeding and weaning. Immunol Cell Biol 75: 419–429, 1997 [DOI] [PubMed] [Google Scholar]
- 13.Davidson LA, Aymond CM, Jiang YH, Turner ND, Lupton JR, Chapkin RS. Non-invasive detection of fecal protein kinase C βII and ζmessenger RNA: putative biomarkers for colon cancer. Carcinogenesis 19: 253–257, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Davidson LA, Jiang YH, Lupton JR, Chapkin RS. Non-invasive detection of putative biomarkers for colon cancer using fecal mRNA. Cancer Epidemiol Biomarkers Prev 4: 643–647, 1995 [PubMed] [Google Scholar]
- 15.Davidson LA, Lupton JR, Miskovsky E, Fields AP, Chapkin RS. Quantification of human intestinal gene expression profiles using exfoliated colonocytes: a pilot study. Biomarkers 8: 51–61, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Davidson LA, Nguyen DV, Hokanson RM, Callaway ES, Isett RB, Turner ND, Dougherty ER, Wang N, Lupton JR, Carroll RJ, Chapkin RS. Chemopreventive n-3 polyunsaturated fatty acids reprogram genetic signatures during colon cancer initiation and progression in the rat. Cancer Res 64: 6797–6804, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis CD. Use of exfoliated cells from target tissues to predict responses to bioactive food components. J Nutr 133: 1769–1772, 2003 [DOI] [PubMed] [Google Scholar]
- 18.De Santa Barbara P, van den Brink GR, Roberts DJ. Development and differentiation of the intestinal epithelium. Cell Mol Life Sci 60: 1322–1332, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donovan SM. Role of human milk components in gastrointestinal development: current knowledge and future needs. J Pediatr 149: S49–S61, 2006 [Google Scholar]
- 20.Donovan SM, Odle J. Growth factors in milk as mediators of infant development. Annu Rev Nutr 14: 147–167, 1994 [DOI] [PubMed] [Google Scholar]
- 21.Dougherty ER, Brun M, Trent JM, Bittner ML. Conditioning-based modeling of contextual genomic regulation. IEEE/ACM Trans Comp Biol Bioinf 6: 310–320, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Dougherty ER, Kim S, Chen Y. Coefficient of determination in nonlinear signal processing. Signal Processing 80: 2219–2235, 2000 [Google Scholar]
- 23.Dusing MR, Wiginton DA. Epithelial lineages of the small intestine have unique patterns of GATA expression. J Mol Histol 36: 15–24, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet 19: 362–365, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Erney RM, Malone WT, Skelding MB, Marcon AA, Kleman-Leyer KM, O'Ryan ML, Ruiz-Palacios G, Hilty MD, Pickering LK, Prieto PA. Variability of human milk neutral oligosaccharides in a diverse population. J Pediatr Gastroenterol Nutr 30: 181–192, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Favier J, Kempf H, Corvol P, Gasc JM. Coexpression of endothelial PAS protein 1 with essential angiogenic factors suggests its involvement in human vascular development. Dev Dyn 222: 377–388, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Fornace AJ, Jr, Alamo I, Jr, Hollander M. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci USA 85: 8800–8804, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gartner H, Graul MC, Oesterreicher TJ, Finegold MJ, Henning SJ. Development of the fetal intestine in mice lacking the glucocorticoid receptor (GR). J Cell Physiol 194: 80–87, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Jenkins SL, Wang J, Vazir M, Vela J, Sahagun O, Gabbay P, Hoang L, Diaz RL, Aranda R, Martin MG. Role of passive and adaptive immunity in influencing enterocyte-specific gene expression. Am J Physiol Gastrointest Liver Physiol 285: G714–G725, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Kaeffer B, Des Robert C, Anlexandre-Gouabau MC, Pagniez A, Legrand A, Amarger V, Kuster A, Piloquet H, Champ M, Le Huerou-Luron I, Roze JC. Recovery of exfoliated cells from gastrointestinal tract of premature infants: a new tool to perform “noninvasive biopsies”? Pediatr Res 62: 564–569, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Dougherty ER, Bittner ML, Chen Y, Sivakumar K, Meltzer P, Trent JM. General nonlinear framework for the analysis of gene interaction via multivariate expression arrays. J Biomed Optics 5: 411–424, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Kim S, Dougherty ER, Chen Y, Sivakumar K, Meltzer P, Trent JM, Bittner M. Multivariate measurement of gene expression relationships. Genomics 67: 201–209, 2000 [DOI] [PubMed] [Google Scholar]
- 33.Kuntz S, Kunz C, Rudloff S. Oligosaccharides from human milk induce growth arrest via G2/M by influencing growth-related cell cycle genes in intestinal epithelial cells. Br J Nutr 101: 1306–1315, 2009 [DOI] [PubMed] [Google Scholar]
- 34.Mace KA, Hansen SL, Myers C, Young DM, Boudreau N. HOXA3 induces cell migration in endothelial and epithelial cells promoting angiogenesis and wound repair. J Cell Sci 118: 2567–2577, 2005 [DOI] [PubMed] [Google Scholar]
- 35.Martins D, Braga-Neto U, Hashimoto R, Bittner ML, Dougherty ER. Intrinsically multivariate predictive genes. IEEE J Selected Topics Signal Processing 2: 424–439, 2008 [Google Scholar]
- 36.Morikawa J, Li H, Kim S, Nishii K, Ueno S, Suh E, Dougherty ER, Shmulevich I, Shiku H, Zhang W, Kobayashi T. Identification of signature genes by microarray for acute myeloid leukemia without maturation (FAB-M1) and AML with t(15;17)(q22;q12)(PML/RARalpha). Int J Oncol 23: 617–625, 2003 [PubMed] [Google Scholar]
- 37.Potten CS, Schofield R, Lajtha LG. A comparison of cell replacement in bone marrow, testis and three regions of epithelium. Biochim Biophys Acta 560: 281–299, 1979 [DOI] [PubMed] [Google Scholar]
- 38.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R, Harpole D, Marks J, Berchuck A, Ginsburg GS, Febbo P, Lancaster J, Nevins JR. Genomic signatures to guide the use of chemotherapeutics. Nat Med 11: 1294–1300, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer 8: 523–534, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Scortegagna M, Morris MA, Oktay Y, Bennett M, Garcia JA. The HIF family member EPAS1/HIF-2a is required for normal hematopoiesis in mice. Blood 102: 1634–1640, 2003 [DOI] [PubMed] [Google Scholar]
- 41.Sima C, Attoor S, Braga-Neto U, Lowey J, Suh E, Dougherty ER. Impact of error estimation on feature-selection algorithms. Pattern Recognition 38: 2472–2482, 2005 [Google Scholar]
- 42.Sima C, Dougherty ER. What should be expected from feature selection in small-sample settings. Bioinformatics 22: 2430–2436, 2006 [DOI] [PubMed] [Google Scholar]
- 43.Uauy R, Mena P, Rojas C. Essential fatty acids in early life: structural and functional role. Proc Nutr Soc 59: 3–15, 2000 [DOI] [PubMed] [Google Scholar]
- 44.Wagner CL, Taylor SN, Johnson D. Host factors in amniotic fluid and breast milk that contribute to gut maturation. Clin Rev Allergy Immunol 34: 191–204, 2008 [DOI] [PubMed] [Google Scholar]
- 45.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol 72: 4497–4499, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterland RA. Epigenetic mechanisms and gastrointestinal development. J Pediatr 149: S137–S142, 2006 [DOI] [PubMed] [Google Scholar]
- 47.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet 15: 705–716, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Widdowson EM, Colombo VE, Artavanis CA. Changes in the organs of pigs in response to feeding for the first 24 hours after birth. II. The digestive tract. Biol Neonate 23: 272–281, 1976 [DOI] [PubMed] [Google Scholar]
- 49.Wiling BP, Van Kessel AG. Intestinal microbiota differentially affect brush border enzyme activity and gene expression in the neonatal gnotobiotic pig. J Anim Physiol Anim Nutr 93: 586–595, 2009 [DOI] [PubMed] [Google Scholar]
- 50.Zamora R, Vodovotz Y, Betten B, Wong C, Zuckerbraun B, Gibson KF, Ford HR. Intestinal and hepatic expression of BNIP3 in necrotizing enterocolitis: regulation by nitric oxide peroxynitrite. Am J Physiol Gastrointest Liver Physiol 289: G822–G830, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Zhao C, Ivanov I, Dougherty ER, Hartman TJ, Lanza E, Colburn NH, Lupton JR, Davidson LA, Chapkin RS. Non-invasive detection of candidate molecular biomarkers in subjects with a history of insulin resistance and colorectal adenomas. Cancer Prev Res 2: 590–597, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Figures and Tables]