Salivary Proteomics for Oral Cancer Biomarker Discovery (original) (raw)
. Author manuscript; available in PMC: 2010 May 26.
Abstract
Purpose
This study aims to explore the presence of informative protein biomarkers in the human saliva proteome and to evaluate their potential for detection of oral squamous cell carcinoma (OSCC).
Experimental Design
Whole saliva samples were collected from patients (n = 64) with OSCC and matched healthy subjects (n = 64). The proteins in pooled whole saliva samples of patients with OSCC (n = 16) and matched healthy subjects (n = 16) were profiled using shotgun proteomics based on C4 reversed-phase liquid chromatography for prefractionation, capillary reversed-phase liquid chromatography with quadruple time-of-flight mass spectrometry, and Mascot sequence database searching. Immunoassays were used for validation of the candidate biomarkers on a new group of OSCC (n = 48) and matched healthy subjects (n = 48). Receiver operating characteristic analysis was exploited to evaluate the diagnostic value of discovered candidate biomarkers for OSCC.
Results
Subtractive proteomics revealed several salivary proteins at differential levels between the OSCC patients and matched control subjects. Five candidate biomarkers were successfully validated using immunoassays on an independent set of OSCC patients and matched healthy subjects. The combination of these candidate biomarkers yielded a receiver operating characteristic value of 93%, sensitivity of 90%, and specificity of 83% in detecting OSCC.
Conclusion
Patient-based saliva proteomics is a promising approach to searching for OSCC biomarkers. The discovery of these new targets may lead to a simple clinical tool for the noninvasive diagnosis of oral cancer. Long-term longitudinal studies with large populations of individuals with oral cancer and those who are at high risk of developing oral cancer are needed to validate these potential biomarkers.
Oral cancer, predominantly oral squamous cell carcinoma (OSCC), is a high-effect local disease in the oral cavity affecting over 300,000 people worldwide annually (1, 2). The American Cancer Society estimated that more than 30,000 new cases of oral cancer were diagnosed in 2006, representing ~3% of all malignancies in men and 2% of all malignancies in women (3). Patients with OSCC often present with symptoms at a late stage, and there is a high recurrence rate after treatment, especially in those with neck lymph node metastasis. The overall 5-year survival rates for oral cancer have remained low and are essentially unchanged during the past few decades (4, 5). Delayed detection is likely to be a primary reason for the high morbidity rate of oral cancer patients, and this supports the imperative need for sensitive biomarkers to improve early detection of oral cancers.
Currently, oral cancer diagnosis depends on a thorough oral examination, usually by a dentist or other qualified health care provider, for possible signs and symptoms of the disease. If an exam shows an abnormal area in the oral cavity, a small tissue biopsy may be removed for a pathologist to check for cancer cells under a microscope. Scientists are searching for biomarkers in saliva, an easy-to-obtain body fluid, for noninvasive detection of oral cancer. For instance, mitochondrial DNA mutation and aberrant promoter hypermethylation of cancer-related genes are common in head and neck/oral cancer. Detection of these genetic alterations in saliva may be useful for diagnosis and monitoring of the disease (6–9). We have identified seven salivary mRNAs that can discriminate OSCCs from matched control subjects (10). Validation of this set of signature on a large patient population is currently being done in a multicenter trial. Last, several studies have investigated the use of salivary proteins as potential diagnostic markers for oral cancer (11–14). Elevated levels of salivary soluble CD44 were shown in the majority of OSCC and could distinguish cancer from benign disease with high specificity (11). Three tumor markers, cytokeratin 19 fragment Cyfra21-1, tissue polypeptide antigen, and cancer antigen 125, were found significantly elevated in the saliva of OSCC patients, and combined use of these markers resulted in similar diagnostic value to those obtained when measuring them in the sera of OSCC patients (12). The level of p53 autoantibody in saliva was also found correlated with its serum levels in OSCC and analysis of p53 antibody in saliva may offer a specific method for detection of a subset of OSCC with p53 aberrations (13). Considering that these candidate biomarkers were discovered at an individual basis, their predicting power for OSCC detection is limited. Our study is aimed for the identification of a panel of candidate biomarkers, which may collectively improve the sensitivity and specificity for detecting OSCC.
The purpose of this present study is to discover and validate differentially expressed proteins in saliva from patients with OSCC that could serve as potential biomarkers for OSCC detection. By using a subtractive proteomics approach to profile salivary proteins from oral cancer and matched healthy subjects followed by immunoassay validation, we have revealed a panel of candidate protein biomarkers for potential detection of OSCC.
Materials and Methods
Patients and saliva samples
In total, 128 participants, including 64 OSCC patients and 64 control subjects, were recruited for this study. The cancer and control groups were completely matched in terms of gender (P = 1) and ethnicity (P = 1). The mean ages for OSCC patients and healthy controls were 54.0 and 46.9 y (P = 0.006, Student’s t test), respectively. The sex distribution in OSCC or control groups was 20:44 (female: male), and the smoking history was matched by determining the pack per year history (P = 0.65). All of the OSCC patients were recently diagnosed and had not received any prior treatment in the form of chemotherapy, radiotherapy, surgery, or alternative remedies before sample collection. All the subjects involved in this study signed the institutional review board–approved consent form. None of these subjects had a history of prior malignancy, immunodeficiency, autoimmune disorders, hepatitis, or HIV infection.
A well-defined and standardized protocol was used for collection, storage, and processing of all the samples under the exactly same conditions. Unstimulated whole saliva (WS) samples were collected between 9 a.m. and 10 a.m. with prior mouth rinsing with water (15). The donors were asked to abstain from eating, drinking, smoking, or using oral hygiene products for at least 1 h before collection. The samples, once collected, were centrifuged at 2,600 × g for 15 min at 4°C to remove debris and cells. The supernatant was then removed and protease inhibitors were included in the collected samples to ensure preservation of the protein integrity. The samples were immediately aliquoted into smaller volumes and stored at −80°C. To avoid the effects of protein degradation, the samples that had been thawed were not reused.
Shotgun proteome analysis
Shotgun proteomics based on reversed-phase liquid chromatography (LC) off-line prefractionation of intact proteins and subsequent LC-tandem mass spectrometry (LC-MS/MS) analysis of proteolyzed peptides was used for profiling of saliva proteins from OSCC or matched control subjects. The total protein concentration in each sample was measured using a 2D Quant kit (GE Healthcare). We then pooled equal amount of WS proteins from 16 OSCC patients or 16 control subjects, respectively, for the comparative analysis.
Both pooled OSCC and control samples (100 μg proteins in total each) were separated by LC (HP1100, Agilent Technologies) using a Vydac C4 RP column (particle size, 5 μm; 250 × 2.1 mm inner diameter; The Nest Group, Inc.) at a flow rate of 250 μL/min. This allowed for 35 fractions collected from each pooled sample (1 fraction per minute). The proteins in each fraction were reduced with DTT (10 mmol/L, 1 h), alkylated with iodoacetamide (55 mmol/L, 1 h), and digested by trypsin (60 ng trypsin for each fraction) at 37°C for overnight. The resulting peptide digests were dried, reconstituted in 0.1% formic acid, and then analyzed by online capillary LC-quadruple time-of-flight (QqTOF) MS.
LC-MS/MS analysis was done using a LC Packings nano-LC system (Dionex) with a nanoelectrospray interface (Protana) and QqTOF mass spectrometer (QSTAR XL, Applied Biosystems). A New Objective PicoTip tip (inner diameter, 8 mm) was used for electrospraying with the voltage at 1,850 V for online MS and MS/MS analyses. The samples were first loaded onto a home-packed C18 precolumn (300 μm × 1 mm; particle size, 5 μm) and then injected onto a LC Packings PepMap C18 column (75 μm × 150 mm; particle size, 5 μm) for nano-LC separation at a flow rate of 250 nL/min.
Identification of the peptides and represented proteins was realized by using the Mascot database search engine (version 1.9; Matrix Science). All searches were done against the EBI human IPI database (version 2.0.1). In all searches, one missed tryptic cleavage was allowed, and a mass tolerance of 0.3 Da was set for the precursor and product ions. A Mascot score of >25 with P < 0.05 was considered a significant match. False-positive rates were determined using the method described in the Mascot manual. Briefly, false-positive rate was calculated by multiplying the number of false-positive identifications (hits to the decoy database constructed from randomized sequences) and dividing by the number of total identifications. The estimated false-positive rate was determined to be 2% or less.
Two-dimensional gel electrophoresis
Two-dimensional gel electrophoresis (2-DE) with Sypro-Ruby staining was used to map out the proteins in the same pooled saliva samples from either OSCC (n = 16) or matched control subjects (n = 16). The proteins in both samples were precipitated by cold ethanol at −20°C for overnight. Following centrifugation at 14,000 × g for 20 min, the supernatants were removed and the pellets were washed with cold ethanol and then collected for total protein assay using a 2D Quant kit (Amersham). Briefly, 100 μg of total proteins from each pooled samples were used for comparative 2-DE analysis. Isoelectric focusing was done using immobilized pH gradient strips (11-cm long, p_I_ 3–10 NL; Bio-Rad) on a Protean isoelectric focusing cell (Bio-Rad), and SDS-PAGE was done in 8% to 16% precast gels on a Criterion Dodeca Cell (Bio-Rad). Fluorescent Sypro-Ruby stain (Molecular Probes-Invitrogen) was used to visualize protein spots.
Gel images were acquired and analyzed using PDQuest software (Bio-Rad). Initially, the images were filtered to remove small noise features without affecting the protein spot. After normalization of spot signals based on the total gel density, a matched set was created between the two-dimensional gel image of the OSCC and control samples. Protein spots were automatically detected and matched and subsequently manually reviewed. Finally, the relative levels of protein spots between the disease and control patients were determined.
Immunoassays
ELISAs were done to determine the levels of s90K/Mac-2 binding protein (M2BP; IBL-Hamburg), histone H1 (ALPCO Diagnostics), and S100A9 and S100P (CycLex Co.) in saliva samples from OSCC and matched control subjects. Samples were analyzed, in duplicate, and the protein levels were determined according to calibration curves established from standards.
Immunoblotting was also done to compare the protein levels between OSCC and matched control subjects. The antibodies used included MRP14 (S100A9), profilin (Cell Signaling), CD59 (Abcam), catalase (Abcam), Ras-related protein Rab-7 (Lab Vision), hematopoietic lineage cell–specific protein (Novus Biologicals, Polyclonal) and moesin (Abcam). Saliva samples from equal numbers of OSCC and control subjects were separated on a 4% to 12% NuPAGE gel (Invitrogen) at 150 V and then transferred to polyvinylidene difluoride membranes using an Invitrogen iBLOT transfer module. After saturating with 5% milk in TBS-Tween 20 buffer overnight at 4°C, the blots were sequentially incubated with primary antibody and horseradish peroxidase–conjugated IgG secondary antibody (Amersham). Finally, the bands were detected by enhanced chemiluminescence (Amersham) and quantified using the Scion Imaging software. Immunoblotting of actin in OSCC and control subjects was also done for normalization of densitometric signals.
Statistical analysis
The Western blotting data for MRP14, CD59, profilin, and catalase were normalized against the corresponding actin levels from the same samples. The normalized values for MRP14, CD59, profilin, and catalase as well as the ELISA data for M2BP were used for further analysis. The reported P values were based on a nonparametric test using the Wilcoxon rank sum test.
We built a logistic regression model and conducted receiver operating characteristic (ROC) curve analyses to evaluate overall predictive power of the combined five candidate protein biomarkers (M2BP, MRP14, CD59, profilin, and catalase). The optimal cut point was determined for each biomarker by identifying the value that yielded the maximum corresponding sensitivity and specificity. ROC curves were then plotted based on the set of optimal sensitivity and specificity values. Area under the curve was computed via numerical integration of the ROC curves. All statistical data analysis was done using the statistical software package R 2.5.0.
We used leave-one-out cross-validation to validate the logistic regression model. The cross-validation strategy first removes one observation and then fits a logistic regression model from the remaining cases with all of the markers. Stepwise model selection is used for each of these models to remove variables that do not improve the model. Subsequently, we used the marker values for the case that was left out to compute a predicted class for that observation. The cross-validation error rate is then the number of samples predicted incorrectly divided by the number of samples.
Results
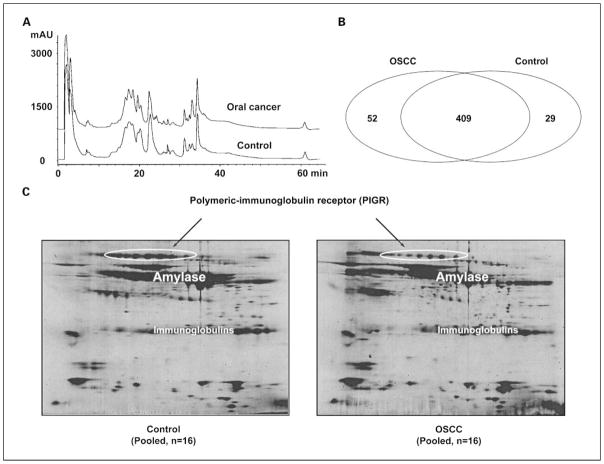
To discover target proteins that can serve as potential markers for OSCC detection, we used a subtractive proteomics approach to profile proteins in pooled saliva samples from 16 OSCC and 16 healthy subjects. These cancer patients and healthy control subjects were very well matched in terms of gender (P = 1), ethnicity (P = 1), and age (P = 0.94) to minimize potential bias from these factors during the discovery phase. The total protein concentrations for each group were determined as 1.01 ± 0.43 mg/mL (OSCC, n = 16) and 1.07 ± 0.59 mg/mL (control, n = 16; P = 0.74). Both pooled samples were initially prefractionated using reversed-phase LC, and subsequent in-solution tryptic digestion with LC-MS/MS of the resulting peptides allowed the identification of proteins in each collected fraction. As shown in Fig. 1A, the C4-LC profiles of intact saliva proteins from OSCC and healthy subjects are remarkably similar. We repeated the analysis of collected protein fractions using the same method and combined the searched results. In total, we identified 461 and 438 nonredundant proteins from both pooled samples, respectively, for OSCC and control samples and as shown in Fig. 1B. Most proteins (n = 409) overlap between the disease and control samples. However, 52 proteins were found to be present in OSCC but are absent in the healthy control subjects (Supplementary Table S1), whereas 29 proteins were found only in the healthy subjects but absent in OSCC patients (Supplementary Table S2). The proteins listed in both tables were all verified manually by examining the raw MS/MS spectra. Because our shotgun proteome analysis was done on the pooled cancer and control samples instead of the samples from individual subjects, we did not have the statistics for the spectral counting data. Some of the listed proteins were identified based on a single-peptide assignment. However, this study was based on accurate QqTOF MS analysis. Tandem mass spectra of peptides were obtained with better than 50 ppm mass accuracy and resolution routinely in excess of 5,000, providing very confident identification of proteins. Nevertheless, these protein identifications need to be verified. We tested four proteins (CD59, involucrin, Ras-related protein Rab-7, and moesin) with single-peptide assignment. Their presence in OSCC saliva was all confirmed by immunoassays. As a comparison, the same pooled samples from OSCCs and matched controls were also profiled with 2-DE and remarkably similar patterns were observed (Fig. 1C). It has been challenging to produce decent two-dimensional gel patterns of WS proteins because WS contains amylase and immunoglobulins at extremely high abundance, which are very difficult to deplete.
Fig. 1.
Subtractive proteomics and 2-DE profiling of proteins in pooled WS samples from 16 OSCC or 16 matched control subjects. A, the intact proteins in each pooled sample were initially prefractionated using C4 reversed-phase LC. In total, 35 fractions were collected from each pooled sample and proteins in each fraction were reduced, alkylated, and digested using trypsin. The resulting peptides were analyzed using capillary LC-QqTOF MS/MS. B, database searching using Mascot was carried out to identify a total of 461proteins from OSCC subjects and 438 proteins from the matched control subjects. Most proteins (n = 409) overlap between the disease and control samples. However, 52 proteins were only found in OSCC, whereas 29 proteins were only identified in the healthy subjects. C, 2-DE patterns of WS proteins from the same pooled cancer and control subjects. The circled spots were identified to be PIGR, which is a down-regulated glycoprotein in OSCC.
Among the large number of proteins that we found present in both OSCC and healthy subjects, many of them are differentially expressed as reflected by the differential number of MS/MS spectra (spectrum counting) observed from the shotgun proteomic analysis (Supplementary Table S3). For instance, MRP14 is a calcium-binding protein that has been implicated in different types of human cancers. From the two repeated analyses, 66 MS/MS spectra were observed for this specific protein in OSCCs, whereas only 18 MS/MS spectra were observed in healthy subjects. Further validation of MRP14 in a new patient cohort (n = 48 OSCCs; n = 48 controls) by immunoblotting confirmed that this protein is significantly overexpressed in saliva of patients with OSCC (Table 1). Conversely, polymeric-immunoglobulin receptor (PIGR; also known as hepatocellular carcinoma–associated protein TB6) is down-regulated in OSCC as reflected by the 24 MS/MS spectra observed in OSCC but 49 MS/MS spectra observed in healthy subjects. We were unable to validate this protein with immunoassay due to unavailability of a commercial antibody. However, down-regulation of PIGR in OSCC patients compared with healthy control subjects was verified by two-dimensional gel analysis. As shown in Fig. 1C, salivary PIGR was heavily glycosylated and exhibited a 2.1-fold decrease in OSCC patients.
Table 1.
Immunoassay validation of salivary proteins differentially abundant in OSCC and healthy control subjects
| Protein | Fold change (mean level) | P | Validation method |
|---|---|---|---|
| M2BP | 1.99 | 0.006 | ELISA |
| Profilin | 1.93 | 0.0003 | Immunoblotting |
| CD59 | 2.45 | 0.00001 | Immunoblotting |
| MRP14 | 2.15 | 0.000002 | Immunoblotting |
| Catalase | 2.07 | 0.0000005 | Immunoblotting |
| Histone H1 | 1.10 | 0.92 | ELISA |
| S100A12 | 1.01 | 0.82 | ELISA |
| Ras-related protein Rab-7 | 1.12 | 0.61 | Immunoblotting |
| Moesin | 1.19 | 0.32 | Immunoblotting |
| Involucrin | 1.67 | 0.11 | ELISA |
| S100P | 1.24 | 0.05 | ELISA |
| Hematopoietic lineage cell-specific protein | 1.43 | 0.002 | Immunoblotting |
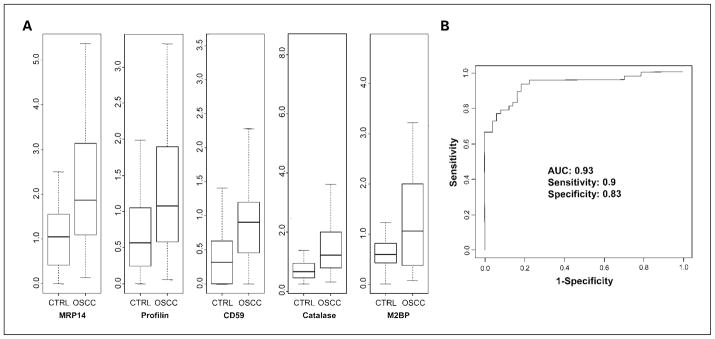
Twelve candidates showing up-regulated levels in OSCC (Supplementary Tables S1 and S2) were chosen for further validation because antibodies and ELISA assays for these proteins are commercially available. Table 1 summarizes the validation results between OSCC and matched controls for these proteins. ELISA was used for validation of M2BP, involucrin, histone H1, S100A12, and S100P, whereas immunoblotting was used for validation of eight other proteins, including MRP14, CD59, catalase, profilin, moesin, hematopoietic lineage cell-specific protein, and Ras-related protein Rab-7. Actin was used for normalization of densitometric signals in immunoblotting. Among these proteins, five potential biomarkers showed significant differences (P ≤ 0.006) between OSCC (n = 48) and matched healthy cohorts (n = 48). The normalized signals for these five potential biomarkers (M2BP, MRP14, CD59, catalase, and profilin) between the OSCC and matched healthy groups are presented in Fig. 2A.
Fig. 2.
A, validation of five candidate biomarkers (M2BP, MRP14, CD59, catalase, and profilin) between OSCC (n = 48) and matched control (n = 48) subjects. MRP14, CD59, catalase, and profilin were measured by Western blotting and normalized against corresponding actin levels, whereas M2BP was measured with ELISA (μg/mL). B, ROC analysis based on the validation results of these five candidate markers. The area under the curve (AUC) was determined as 0.93. The sensitivity and specificity were 90% and 83%, respectively.
Discussion
Oral carcinogenesis arises through a series of histopathologic stages from benign hyperplasia to dysplasia to carcinoma in situ followed by invasive squamous cell carcinoma. At the molecular level, the development of OSCC is a multistep process accompanied by genetic mutations and expression changes of many genes that lead to uncontrolled cellular growth (16, 17). The cellular and molecular heterogeneity of OSCC and the large number of genes potentially involved in oral carcinogenesis emphasize the importance of studying gene expression changes in a global scale by proteomics. This also suggested that multiple proteins (pathways) should be simultaneously targeted as an effective strategy to diagnose or treat the disease.
Because oral cancer cells are immersed in the salivary milieu, analysis of the salivary proteomes from OSCC patients represents a potentially promising approach to finding potential biomarkers for the disease. Saliva is an easily accessible fluid compared with tissue biopsies. Therefore, a large number of saliva samples can be collected and analyzed, and this should allow for a robust study design with sufficient statistical power to reveal true signatures characteristic of the disease. Furthermore, the identified protein biomarkers can be translated into simple clinical assays, allowing the disease to be detected or monitored in a noninvasive body fluid. We have done in-depth analysis of the human salivary proteome using a combination of different separation techniques combined with QqTOF MS (18–20), and the data from WS have been deposited into the Saliva Proteome Knowledge Base for public access. We have also identified 84 _N_-glycosylated peptides representing 45 unique _N_-glycoproteins in WS using hydrazide chemistry and LC-MS/MS analysis of _N_-glycosylated peptides selectively released from the captured glycoproteins (21). In addition, a 3-year effort by the Human Salivary Proteome Project, a consortium of three research groups including the Scripps Research Institute/University of Rochester, the University of California-San Francisco, and the University of California-Los Angeles/University of Southern California, has led to the identification of over 1,100 nonredundant proteins in human parotid and submandibular/sublingual secretions (22). With the successful compilation of the saliva proteome, a next step would be to identify potential diagnostic and/or prognostic biomarkers that could be used in a clinical context for disease detection and monitoring in saliva. For example, saliva proteome analysis of patients with primary Sjögren’s syndrome (pSS) has suggested that pSS is associated with increased inflammatory proteins and decreased acinar proteins in saliva when compared with non-SS (23, 24). Current diagnosis of pSS requires salivary gland biopsy. However, if these newly discovered targets are successfully validated, a noninvasive diagnosis of pSS based on salivary biomarkers will be possible in the future.
Subtractive proteomics refers to direct profiling of proteins expressed in samples from two cellular or pathologic states using multidimensional LC separation and data-dependent MS/MS analysis (25, 26). This method does not use gels or stable isotopes but simply quantifies proteins by counting the number of MS/MS spectra for the tryptic peptides identified for each proteins (27, 28). The quantitative measurement is because the number of MS/MS spectra of tryptic peptides identified for a given protein is correlated to its relative abundance in the sample. Although subtractive proteomics is imprecise compared with other quantitative MS such as stable isotope labeling methods, this approach is attractive for biomarker discovery mainly because of its inherent simplicity. Very little sample preparation before MS analysis is required, with no covalent labeling involved (29). Our study suggests that subtractive proteomics is a more feasible approach than 2-DE/MS in terms of revealing the OSCC-associated saliva proteins at differential levels. As shown in Fig. 1C, remarkably similar patterns between OSCC and control subjects were observed, although some faint protein spots showed differential levels. The high abundance of salivary amylase and immunoglobulins undermined the analysis of low-abundance proteins in WS. Both glycogen precipitation and antibody binding were tested in our study to deplete amylase; however, the efficiency was low and depletion of many other proteins occurred as well (data not shown). On the contrary, the subtractive proteomics approach allowed the identification of many low-abundance proteins at differential levels and seems to be a more rational approach than 2-DE/MS for discovery of salivary protein biomarkers of human diseases.
Many proteins at elevated levels in OSCC patients’ saliva (Supplementary Tables S1 and S3) have been previously associated with human cancers (e.g., squamous cell carcinoma antigen 2, calcyclin, Rho GDP dissociation inhibitor, heat shock 70-kDa protein 1, Annexin I, cathepsin G, peroxiredoxin II, thioredoxin, short palate, and lung and nasal epithelium carcinoma-associated protein; refs. 30–34). Apart from potential clinical applications, these target proteins may contribute to an understanding of the molecular mechanism of the disease. Meanwhile, some of the salivary proteins were underexpressed in OSCC (Supplementary Tables S2 and S3). For instance, clusterin was found present in control but absent in OSCC by subtractive proteomic analysis. This secretory protein is involved in programmed cell death (apoptosis; ref. 35), and down-regulation of clusterin in esophageal squamous cell carcinoma and prostate cancer has been reported in previous studies (36, 37).
Obviously, the candidates discovered by subtractive proteomics need to be further validated. From the overexpressed proteins in OSCC, we chose 12 candidates that have commercially available antibodies or ELISA assays for further validation (Table 1). Six candidates (50%) were validatable based on the immunoassays, including M2BP, a tumor antigen (38–40). In a previous study using cancer cell–secreted proteomes as a basis for searching potential tumor markers, M2BP was found significantly up-regulated in nasopharyngeal carcinoma. The serum levels of M2BP were also significantly higher in both nasopharyngeal carcinoma patients and nasopharyngeal carcinoma nude mice model compared with healthy people or tumor-free mice (41). Other successfully validated proteins included MRP14, CD59, profilin 1, and catalase. An increased level of MRP14 has been previously reported in tissue cells of oral tongue cancer (42). CD59 (protectin) is one of the complement restriction factors that are overexpressed on tumor cells, and they enable tumor cells to escape from complement-dependent and antibody-mediated killing (43). Profilin 1 is a regulator of the microfilament system and is involved in various signaling pathways via interactions with cytoplasmic and nuclear ligands. It may be secreted into tumor microenvironments during the early progressive stage of tumor formation (44). Finally, catalase protects the cell against oxidative stress, and altered levels of catalase (as well as other antioxidative enzymes such as superoxide dismutase and glutathione peroxidase) are evident in many human tumors and are fundamentally involved in carcinogenesis and tumor progression (45).
OSCC is a complex disease resulting from an interdependent series of genetic alterations rather than a single decisive event. Therefore, combination of these candidate protein markers can improve the sensitivity and specificity for OSCC detection. ROC analysis indicates that the five candidate markers (M2BP, MRP14, CD59, catalase, and profilin), collectively, provide a sensitivity of 90% and a specificity of 83% for OSCC detection, with a ROC value (area under the curve) of 0.93 (Fig. 2B). The total accuracy was determined as 85% through a leave-one-out cross-validation approach.
In summary, our study has shown that patient-based saliva proteomics is a promising approach to discovery of biomarkers for oral cancer detection. However, at this stage, we have only verified that the results obtained by shotgun proteomics are true findings. The discovered candidate biomarkers need to be extensively validated considering that the sampling efficiency for LC-MS/MS might vary from one experiment to another and some of the targets were identified based on single-peptide assignment. Clearly, it is challenging to translate candidate biomarkers from proteomic investigations into real-world diagnostic or prognostic applications. Approval of use of a biomarker or set of biomarkers for a given clinical decision relies on the results of large-scale multicenter clinical trials, and approval of the use of the detection technology for that purpose. The successful completion of biomarker development requires adherence to guidelines set forth by the Early Detection Research Network, which was established by the National Cancer Institute in response to the recent development of emerging technologies (e.g., proteomics and microarrays) for cancer screening (46, 47). We are currently working with the National Cancer Institute/Early Detection Research Network–designated Biomarker Reference Lab laboratory at the University of California at Los Angeles to validate the identified protein biomarkers and test if the combination of these biomarkers with our previously discovered mRNA biomarkers can further improve the detection of OSCC. The appropriate application of a biomarker in clinics can also be aided by novel diagnostic devices (e.g., microfluidics-based chips) for simple and high-throughput measurement of the biomarker in patients’ saliva (48, 49). If appropriately validated on larger patient cohorts, testing of the discovered candidate markers coupled with microfluidic devices may become a powerful tool for oral cancer diagnosis in the future. The biology behind these promising targets should also be well studied to understand the mechanism in a clinical setting. Lastly, OSCC is usually detected at late stages when the cancer has advanced and therefore results in poor prognosis and survival. It is therefore critical to detect oral cancer as early as possible, when it can be treated more successfully, and thus enhancing the rate of survival. Considering that ~10% of the general population have oral mucosal abnormalities, and precancerous and early cancerous lesions rarely show distinct clinical characteristics, there is a growing realization that some premalignant and early cancerous lesions are not readily detectable by visual inspection (50). Therefore, the integration of early detection and screening based on protein biomarkers, in conjunction with a conventional oral examination, is very important. This warrants retrospective proteomic analyses of oral precancer and cancer to achieve discriminatory biomarkers for true early detection.
Translational Relevance
By using patient-based saliva proteomics, this study has shown that informative protein biomarkers are present in oral cancer patients’ saliva that can be used for potential detection of the disease. Because it is very simple to collect and process saliva fluids, the discovery of these biomarkers may lead to a useful clinical tool for noninvasive diagnosis of oral cancer in the future. Large-scale clinical trials are needed for further testing those potential biomarkers on new populations of individuals with oral cancer and those who are at high risk for developing oral cancer. Novel diagnostic devices will also be developed for fast and high-throughput measurement of these biomarkers in cancer patients’ saliva.
Supplementary Material
Supplemental Table 3
Supplemental table 1
Supplemental table 2
Acknowledgments
Grant support: USPHS grants R03-DE017144 (S. Hu), R21-CA122806 (S. Hu), R01DE15970 (D.T. Wong), and R01DE17170 (D.T. Wong).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Greenlee RT, Murray T, Bolden S, et al. Cancer tatistics, 2000. CA Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Cancer facts and figures 2006. Atlanta: American Cancer Society; 2006. [Google Scholar]
- 4.Lippman SM, Hong WK. Molecular markers of the risk of oral cancer. N Engl J Med. 2001;344:1323–6. doi: 10.1056/NEJM200104263441710. [DOI] [PubMed] [Google Scholar]
- 5.Vokes EE, Weichselbaum RR, Lippman SM, et al. Head and neck cancer. N Engl J Med. 1993;328:184–94. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 6.Fliss MS, Usadel H, Caballero OL, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–9. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 7.Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278:1054–9. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- 8.Rosas SL, Koch W, da Costa Carvalho MG, et al. Promoter hypermethylation patterns of p16, O6-methyl-guanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–42. [PubMed] [Google Scholar]
- 9.Righini CA, de Fraipont F, Timsit JF, et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13:1179–85. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, St John MAR, Zhou X, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–50. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 11.Franzmann EJ, Reategui EP, Pedroso F, et al. Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1348–55. doi: 10.1158/1055-9965.EPI-06-0011. [DOI] [PubMed] [Google Scholar]
- 12.Nagler R, Bahar G, Shpitzer T, et al. Concomitant analysis of salivary tumor markers—a new diagnostic tool for oral cancer. Clin Cancer Res. 2006;12:3979–84. doi: 10.1158/1078-0432.CCR-05-2412. [DOI] [PubMed] [Google Scholar]
- 13.Tavassoli M, Brunel N, Maher R, et al. p53 antibodies in the saliva of patients with squamous cell carcinoma of the oral cavity. Int J Cancer. 1998;78:390–1. doi: 10.1002/(SICI)1097-0215(19981029)78:3<390::AID-IJC23>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Onsongo G, Popko J, et al. Proteomics analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol Cell Proteomics. 2008;7:486–98. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Mavazesh M. Methods for collecting saliva. Ann N Y Acad Sci. 1993;694:72–7. doi: 10.1111/j.1749-6632.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- 16.Lippman SM, Sudbo J, Hong WK. Oral cancer prevention and the evolution of molecular-targeted drug development. J Clin Oncol. 2005;23:346–56. doi: 10.1200/JCO.2005.09.128. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, He QY, Yuen AP, Chiu JF. Proteomics of buccal squamous cell carcinoma: the involvement of multiple pathways in tumorigenesis. Proteomics. 2004;4:2465–75. doi: 10.1002/pmic.200300762. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, Li Y, Wang J, et al. Human saliva proteome and transcriptome. J Dent Res. 2006;85:1129–33. doi: 10.1177/154405910608501212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Loo JA, Wong DT. Human saliva proteome analysis and disease biomarker discovery. Expert Rev Proteomics. 2007;4:531–8. doi: 10.1586/14789450.4.4.531. [DOI] [PubMed] [Google Scholar]
- 20.Hu S, Xie Y, Ramachandran P, et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–28. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 21.Ramachandran P, Boontheung P, Xie Y, et al. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5:1493–503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 22.Denny P, Hagen FK, Hardt M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu OH, Atkinson JC, Hoehn GT, et al. Identification of parotid salivary biomarkers in Sjogren’s syndrome by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry and two-dimensional difference gel electrophoresis. Rheumatology. 2006;45:1077–86. doi: 10.1093/rheumatology/kei212. [DOI] [PubMed] [Google Scholar]
- 24.Hu S, Wang J, Meijer J, et al. Salivary proteomic and genomic biomarkers for primary Sjögren’s Syndrome. Arthritis Rheum. 2007;56:3588–600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schirmer EC, Florens L, Guan T, et al. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–2. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 27.Dieguez-Acuna FJ, Gerber SA, Kodama S, et al. Characterization of mouse spleen cells by subtractive proteomics. Mol Cell Proteomics. 2005;4:1459–70. doi: 10.1074/mcp.M500137-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Zybailov B, Coleman MK, Florens L, et al. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–24. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 29.Veenstra TD. Global and targeted quantitative proteomics for biomarker discovery. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:3–11. doi: 10.1016/j.jchromb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Kaur J, Kaur J, Ralhan R. Induction of apoptosis by abrogation of HSP70 expression in human oral cancer cells. Int J Cancer. 2000;85:1–5. doi: 10.1002/(sici)1097-0215(20000101)85:1<1::aid-ijc1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 31.Vimalachandran D, Greenhalf W, Thompson C, et al. High nuclear S100A6 (Calcyclin) is significantly associated with poor survival in pancreatic cancer patients. Cancer Res. 2005;65:3218–25. doi: 10.1158/0008-5472.CAN-04-4311. [DOI] [PubMed] [Google Scholar]
- 32.Chen CH, Tsai TL, Yang YS, et al. Studies of the serum HER-2/neu and squamous cell carcinoma-related antigen expression in patients with oral squamous cell carcinoma. J Oral Pathol Med. 2007;36:83–7. doi: 10.1111/j.1600-0714.2007.00502.x. [DOI] [PubMed] [Google Scholar]
- 33.Park S-H, Chung YM, Lee Y-S, et al. Antisense of human peroxiredoxin II enhances radiation-induced cell death. Clin Cancer Res. 2000;6:4915–20. [PubMed] [Google Scholar]
- 34.Hu S, Yu T, Xie Y, et al. Discovery of oral fluid biomarkers for human oral cancer by mass spectrometry. Cancer Genomics Proteomics. 2007;4:55–64. [PubMed] [Google Scholar]
- 35.Wong P, Taillefer D, Lakins J, et al. Molecular characterization of human TRPM-2/clusterin, a gene associated with sperm maturation, apoptosis and neurodegeneration. Eur J Biochem. 1994;221:917–25. doi: 10.1111/j.1432-1033.1994.tb18807.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang LY, Ying WT, Mao YS, et al. Loss of cluster in both in serum and tissue correlates with the tumorigenesis of esophageal squamous cell carcinoma via proteomics approaches. World J Gastroenterol. 2003;9:650–4. doi: 10.3748/wjg.v9.i4.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scaltriti M, Brausi M, Amorosi A, et al. Clusterin (SGP-2, Apo J) expression is downregulated in low-and high-grade human prostate cancer. Int J Cancer. 2004;108:23–30. doi: 10.1002/ijc.11496. [DOI] [PubMed] [Google Scholar]
- 38.Park YP, Choi SC, Kim JH, et al. Up-regulation of Mac-2 binding protein by hTERT in gastric cancer. Int J Cancer. 2007;120:813–20. doi: 10.1002/ijc.22369. [DOI] [PubMed] [Google Scholar]
- 39.Ulmer TA, Keeler V, Loh L, et al. Tumor-associated antigen 90K/Mac-2-binding protein: possible role in colon cancer. J Cell Biochem. 2006;98:1351–66. doi: 10.1002/jcb.20784. [DOI] [PubMed] [Google Scholar]
- 40.Marchetti A, Tinari N, Buttitta F, et al. Expression of 90K (Mac-2 BP) correlates with distant metastasis and predicts survival in stage Inon-small cell lung cancer patients. Cancer Res. 2002;62:2535–9. [PubMed] [Google Scholar]
- 41.Wu CC, Chien KY, Tsang NM, et al. Cancer cell-secreted proteomes as a basis for searching potential tumor markers: nasopharyngeal carcinoma as a model. Proteomics. 2005;5:3173–82. doi: 10.1002/pmic.200401133. [DOI] [PubMed] [Google Scholar]
- 42.He QY, Chen J, Kung HF, et al. Identification of tumor-associated proteins in oral tongue squamous cell carcinoma by proteomics. Proteomics. 2004;4:271–8. doi: 10.1002/pmic.200300550. [DOI] [PubMed] [Google Scholar]
- 43.Ravindranath NM, Shuler C. Cell-surface density of complement restriction factors (CD46, CD55, and CD59): oral squamous cell carcinoma versus other solid tumors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;103:231–9. doi: 10.1016/j.tripleo.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 44.Huang CM, Ananthaswamy HN, Barnes S, et al. Mass spectrometric proteomics profiles of in vivo tumor secretomes: capillary ultrafiltration sampling of regressive tumor masses. Proteomics. 2006;6:6107–16. doi: 10.1002/pmic.200600287. [DOI] [PubMed] [Google Scholar]
- 45.McEligot AJ, Yang S, Meyskens JFL. Redox regulation by intrinsic species and extrinsic nutrients in normal and cancer cells. Annu Rev Nutr. 2005;25:261–95. doi: 10.1146/annurev.nutr.25.050304.092633. [DOI] [PubMed] [Google Scholar]
- 46.Henson DE, Srivastava S, Kramer BS. Molecular and genetic targets in early detection. Curr Opin Oncol. 1999;11:419–25. doi: 10.1097/00001622-199909000-00018. [DOI] [PubMed] [Google Scholar]
- 47.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–61. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Denny P, Ho CM, et al. The Oral Fluid MEMS/NEMS Chip (OFMNC): diagnostic and translational applications. Adv Dent Res. 2005;18:3–5. doi: 10.1177/154407370501800102. [DOI] [PubMed] [Google Scholar]
- 49.Wong DT. Salivary diagnostics powered by nano-technologies, proteomics and genomics. J Am Dent Assoc. 2006;137:313–21. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 50.Lingen MW. Oral cancer screening aids: where is the science? Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;103:153–4. doi: 10.1016/j.tripleo.2006.11.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 3
Supplemental table 1
Supplemental table 2