Lysine Propionylation and Butyrylation Are Novel Post-translational Modifications in Histones (original) (raw)
. Author manuscript; available in PMC: 2010 Jul 29.
Published in final edited form as: Mol Cell Proteomics. 2007 Jan 30;6(5):812–819. doi: 10.1074/mcp.M700021-MCP200
Abstract
The positively charged lysine residue plays an important role in protein folding and functions. Neutralization of the charge often has a profound impact on the substrate proteins. Accordingly all the known post-translational modifications at lysine have pivotal roles in cell physiology and pathology. Here we report the discovery of two novel, in vivo lysine modifications in histones, lysine propionylation and butyrylation. We confirmed, by in vitro labeling and peptide mapping by mass spectrometry, that two previously known acetyltransferases, p300 and CREB-binding protein, could catalyze lysine propionylation and lysine butyrylation in histones. Finally p300 and CREB-binding protein could carry out autopropionylation and autobutyrylation in vitro. Taken together, our results conclusively establish that lysine propionylation and lysine butyrylation are novel post-translational modifications. Given the unique roles of propionyl-CoA and butyryl-CoA in energy metabolism and the significant structural changes induced by the modifications, the two modifications are likely to have important but distinct functions in the regulation of biological processes.
Molecular anatomy of post-translational modifications that regulate cellular processes and disease progression stands as one of the major goals of postgenomics biological research. To date, more than 200 post-translational modifications have been described, providing an efficient way to diversify the primary structure of a protein and possibly its functions (1–3). The remarkable complexity of these molecular networks is exemplified by modifications at the side chain of lysine, one of the 15 ribosomally coded amino acid residues known to be modified (1). The electron-rich and nucleophilic nature of the lysine side chain makes it suitable for undergoing covalent post-translational modification reactions with diverse substrates. The residue can be potentially modulated by several post-translational modifications including methylation, acetylation, biotinylation, ubiquitination, and sumoylation, which have pivotal roles in cell physiology and pathology.
Lysine acetylation is an abundant, reversible, and highly regulated post-translational modification. Although initially discovered in histones (4), the modification was later identified in non-histone proteins, such as p53 (5). A recent proteomics screening showed that acetyllysine is abundant and present in substrates that are affiliated with multiple organelles and have diverse functions (6). Interestingly the modification is enriched in mitochondrial proteins and metabolic enzymes, implying its roles in fine tuning the functions of the organelle and energy metabolism (6). The modification plays important roles in diverse cellular processes, such as apoptosis, metabolism, transcription, and the stress response (7–10). In addition to their roles in fundamental biology, lysine acetylation and its regulatory enzymes (acetyltransferases and deacetylases) are intimately linked to aging (11) and several major diseases such as cancer, neurodegenerative disorders, and cardiovascular diseases (12–14).
Acetyl-CoA, a member of the high energy CoA compounds, is the substrate used by acetyltransferases to catalyze the lysine acetylation reaction. It remains unknown, however, whether cells can use other short-chain CoAs, such as propionyl- and butyryl-CoA (which are structurally close to acetyl-CoA), to carry out similar post-translational modifications at lysine. Nevertheless several lines of evidence suggest such a possibility. First, like acetyl-CoA, propionyl-CoA and butyryl-CoA are high energy molecules, making it thermodynamically feasible to carry out a reaction with a lysine side chain. Second, propionyl-CoA and butyryl-CoA are structurally similar to acetyl-CoA with a difference of only one or two CH2. Third, propionyl-CoA and butyryl-CoA are present at high concentrations in cells. In the case of starved mouse liver, the concentrations of the two CoAs are only 1–3 times less than acetyl-CoA (15). Finally it appears, from structural studies on some HATs1 (such as Hat1), that the enzyme has ample space within the cofactor binding pocket to accept propionyl-CoA without steric interference (16). Despite such evidence, the short-chain CoAs with the exception of acetyl-CoA have not been described as a substrate for protein modification.
Here we report the identification and validation of two novel post-translational protein modifications, propionylation and butyrylation at lysine residues, by a proteomics study. The unbiased global screening involved exhaustive peptide identification by nano-HPLC/MS/MS analysis, protein sequence database search, and manual verification. The resulting propionylated and butyrylated peptides were verified by MS/MS of their corresponding synthetic peptides. Using in vitro labeling with isotopic propionyl-CoA and butyryl-CoA as well as mass spectrometry, we identified two acetyltransferases, p300 and CBP, that could perform robust lysine modifications at histones in vitro. Furthermore we demonstrated that p300 and CBP could carry out autopropionylation and autobutyrylation at lysine residues in a fashion similar to autoacetylation. Taken together, these results reveal that lysine propionylation and butyrylation are novel lysine modifications that can be catalyzed by acetyltransferases. Given the unique roles of propionyl-CoA and butyryl-CoA in energy metabolism (17), their distinct structure, and significant structural changes induced by the modifications, it is anticipated that lysine propionylation and butyrylation will have important but likely distinct functions in the regulation of biological processes. The identification of lysine-propionylated and lysine-butyrylated substrates described here provides an entry point for future functional studies of the two modifications in cellular physiology and pathology.
EXPERIMENTAL PROCEDURES
In-gel Digestion
Protein in-gel digestion, peptide extraction, and peptide cleaning using a μ-C18 ZipTip were carried out as reported previously (18).
HPLC/MS/MS Analysis
HPLC/MS/MS analysis for mapping propionylation and butyrylation sites in histone H4, p53, p300, and CBP were carried out by nano-HPLC/LTQ mass spectrometry as described previously (6). Briefly each tryptic digest was dissolved in 10 µl of HPLC buffer A (0.1% formic acid in water (v/v)), and 2 µl were injected into an Agilent HPLC system (Agilent, Palo Alto, CA). Peptides were separated on a home-made capillary HPLC column (50-mm length × 75-µm inner diameter, 4-µm particle size, 90-Å pore diameter) with Jupiter C12 resin (Phenomenex, St. Torrance, CA) and directly electrosprayed into the mass spectrometer using a nanospray source. The LTQ mass spectrometer was operated in the data-dependant mode acquiring fragmentation spectra of the 10 strongest ions.
Protein Sequence Database Search and Manual Verification
All MS/MS spectra were searched against the National Center for Biotechnology Information non-redundant (NCBI-nr) protein sequence database (updated November 28, 2006 with 4,125,643 entries) specifying lysine modifications using the MASCOT database search engine (version 2.1). A low cutoff peptide score of 20.0 was selected to maximize the identification of lysine-modified peptides. For each Mascot search, the peptide mass error was set to ±4 Da, fragment ion mass error was set to ±0.6 Da, and six missing cleavages were allowed. All lysine-propionylated or -butyrylated peptides identified with MASCOT score >20.0 were manually examined with the rules described previously (19), and all lysine propionylation or butyrylation sites had to be identified by consecutive b- or y-ions so that the possibility that propionylation (+56 Da) or butyrylation (+70 Da) occurred on adjacent residues was eliminated.
Synthesis of Lysine-propionylated and Lysine-butyrylated Peptides
The synthesis procedure can be found in the supplemental information.
In Vitro Propionylation and Butyrylation Assay
In vitro propionylation and butyrylation assays were carried out essentially as described previously (20) with some modifications. Tagged human FLAG-p300, hemagglutinin-CBP, FLAG-MOF, and FLAG-PCAF proteins from transfected 293 cells and tagged human GST-Tip60 and GST-p53 expressed in bacteria were purified to homogeneity under stringent conditions (500 mm NaCl + 1% Triton X-100). Ten-microliter reactions contained 50 mm Tris, pH 7.9, 10% glycerol, 1 mm DTT, 10 mm sodium butyrate, 1 µl of [14C]acyl-CoA (55 mCi/mmol; acetyl-CoA from Amersham Biosciences and propionyl-CoA and butyryl-CoA from American Radiolabeled Chemicals, Inc.). Two and a half micrograms of substrates (human core histones and recombinant human histone H4 (Upstate Biotechnology, Lake Placid, NY) or GST-p53) and 20–100 ng of the enzyme protein, as indicated, were incubated at 30 °C for 1 h. The reaction mixture was then subjected to electrophoresis by SDS-PAGE followed by either autoradiography or Coomassie Blue staining.
RESULTS
Initial Identification of Lysine-propionylated and -butyrylated Peptides in Histone H4 Protein
We hypothesized that propionyl-CoA could be used by acetyltransferases for lysine propionylation. In addition, we further assumed that some propionylated, tryptic peptides could be affinity-purified by anti-acetyllysine antibody due to the close structural similarity between the acetyllysine residue and the propionyllysine residue (Fig. 1). To identify lysine-propionylated peptides, we searched the MS/MS datasets of affinity-enriched acetyllysine-containing tryptic peptides acquired in nano-HPLC/LTQ mass spectrometry (the study on lysine acetylation proteomics was published previously (6)). During the protein sequence database search, the lysine was considered as unmodified, acetylated, or propionylated. The database search and manual verification of peptide hits led to the identification of 11 lysine-propionylated histone H4 peptides (Table I; see Supplemental Fig. F4 for relevant MS/MS spectra). The identification of lysine-propionylated peptides motivated us to hunt for the lysine-butyrylated peptides. The same datasets were searched again with the lysine considered as unmodified, acetylated, or butyrylated. The analysis identified two additional histone H4 peptides with lysine butyrylation sites (Table I; see Supplemental Fig. F4 for MS/MS spectra).
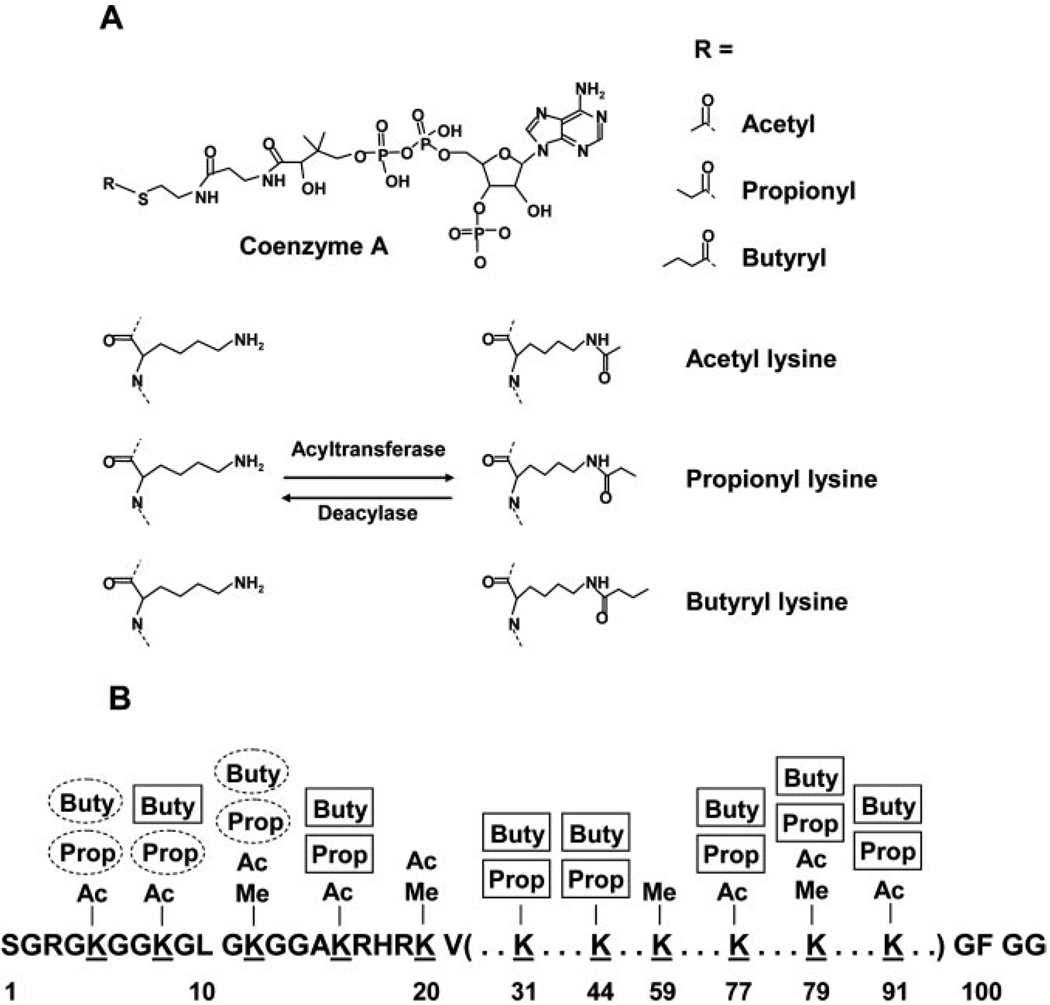
FIG. 1.
A, structures of three short-chain CoAs, the acetyl-CoA, propionyl-CoA, and butyryl-CoA, as well as the three modified lysines, acetyllysine, propionyllysine, and butyryllysine. B, an illustration of novel lysine propionylation and butyrylation sites in histone H4 (unmarked labels, lysine acetylation and methylation sites identified previously; circled labels, novel, in vivo lysine modification sites identified in this study; boxed labels, additional novel, in vitro lysine modification sites identified in this study). The known sites of lysine acetylation and methylation were obtained from Millipore/Upstate. Ac, acetyl; Me, methyl; Buty, butyryl; Prop, propionyl.
TABLE I.
The list of lysine-propionylated and -butyrylated peptides identified in vivo and the number of modification sites identified in three proteins in vitro
| No. | Protein name | gif no. | Sequence | No. of propionyl-Lys sites |
|---|---|---|---|---|
| 1 | Histone 4, H4 | 28173560 | GK^GGK*GLGK^GGAK*R | 2 |
| 2 | Histone 4, H4 | 28173560 | GGK^GLGK*GGAK*R | 1 |
| 3 | Histone 4, H4 | 28173560 | GGK*GLGK^GGAK*R | 1 |
| 4 | Histone 4, H4 | 28173560 | GK^GGK*GLGK*GGAK*R | 1 |
| 5 | Histone 4, H4 | 28173560 | GK*GGK^GLGK*GGAK*R | 1 |
| 6 | Histone 4, H4 | 28173560 | GK*GGK*GLGK^GGAK*R | 1 |
| 7 | Histone 4, H4 | 28173560 | GK^GGK*GLGK*GGAK | 1 |
| 8 | Histone 4, H4 | 28173560 | GK*GGK^GLGK*GGAK | 1 |
| 9 | Histone 4, H4 | 28173560 | GK*GGK*GLGK^GGAK | 1 |
| 10 | Histone 4, H4 | 28173560 | GGK*GLGK^GAK | 1 |
| 11 | Histone 4, H4 | 28173560 | GGK^GLGK*GGAK | 1 |
| No. | Protein name | gif no. | Sequence | No. of butyryl-Lys sites |
| 12 | Histone 4, H4 | 28173560 | GK″GGK*GLGK*GGAK*R | 1 |
| 13 | Histone 4, H4 | 28173560 | GK*GGK*GLGK″GGAK*R | 1 |
| In vitro analysis | ||||
| p53 | p300 | Histone H4 | CBP | |
| Propionyl-Lys | 2 | 21 | 9 | 12 |
| Butyryl-Lys | 2 | 11 | 9 | 7 |
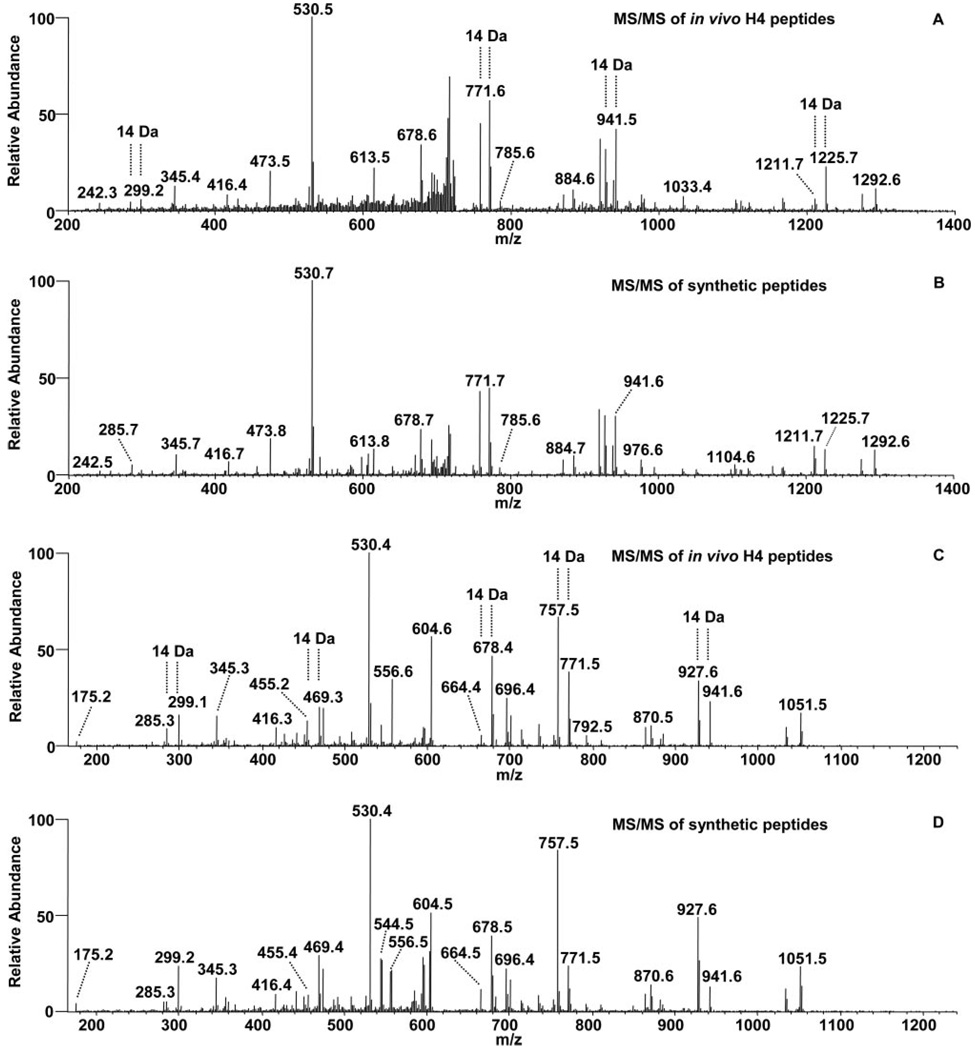
Two examples of MS/MS spectra are presented to show the identification of lysine-propionylated and lysine-butyrylated peptides (Fig. 2 and Supplemental Fig. 4). In the initial peptide identification by protein sequence database search, the spectrum in Fig. 2_A_ led to the identification of lysine-propionylated Peptide 1 (Table I), whereas that in Fig. 2_C_ led to the identification of lysine-propionylated Peptide 2 (Table I). A careful inspection of the spectra revealed that there were a number of fragment ions in both spectra with strong intensities that could not be assigned to the fragment ions derived from Peptide 1 and Peptide 2, respectively, suggesting the presence of additional peptide(s). Three lines of evidence suggest that the spectra were from peptide isomers. (i) Multiple peak pairs with mass difference of 14 Da were observed in both spectra, (ii) the peptides have the same molecular weights, and (iii) the peptides co-eluted. Indeed the remaining peaks in spectrum Fig. 2_A_ could be accounted for by two additional lysine-butyrylated peptides: Peptide 12 and Peptide 13 (Table I and Supplemental Fig. F4, L and M). Likewise an additional lysine-propionylated peptide isomer, Peptide 3 (Table I and Supplemental Fig. F4C), could also be identified from the spectrum in Fig. 2_C_.
FIG. 2. Identification and verification of lysine-propionylated and lysine-butyrylated histone H4 peptides.
A, tandem mass spectrum of a tryptic peptide ion from a peptide mixture that was affinity-enriched with an anti-acetyllysine antibody from tryptic peptides of HeLa nuclear extracts. The spectrum was used to identify one lysine-propionylated and two lysine-butyrylated peptides, Peptides 1, 12, and 13 (Table I) from histone H4. The fragmentation ions for each peptide are available in the supplemental information. B, tandem mass spectrum of a peptide mixture from the three synthetic peptides corresponding to the sequences identified in A, showing similar ion intensity. C, MS/MS of a tryptic peptide ion led to the identification of two modified peptides, Peptides 2 and 3 (Table I) from histone H4. D, tandem mass spectrum of a peptide mixture from the two synthetic peptides corresponding to the sequences identified in C, showing similar ion intensity.
The chemical nature of an identified peptide can be confirmed by MS/MS of their corresponding synthetic peptides, a gold standard for verification of peptide identification and chemical identity. To validate the identities of the propionylated and butyrylated peptides in Fig. 2_A_, MS/MS of three synthetic peptides with the sequences and modification patterns corresponding to Peptides 1, 12, and 13 in Table I were analyzed (Supplemental Fig. F1, A, B, and C). To validate the in vivo fragmentation spectrum by the peptide mixture observed in Fig. 2_A_, three synthetic peptides were mixed at a molar ratio of 4:2:1 (Peptide 1:Peptide 12:Peptide 13). MS/MS analysis of the peptide mixture resulted in a fragmentation pattern (Fig. 2_B_) that matched perfectly with the in vivo spectrum in Fig. 2_A_, verifying the identification of one peptide with lysine propionylation and two peptides with lysine butyrylation. Likewise, to validate Peptides 2 and 3 (Table I) identified from the in vivo spectrum in Fig. 2_C_, two corresponding synthetic peptides were analyzed (Supplemental Fig. F1, D and E). The fragmentation pattern of a peptide mixture at a 2:1 molar ratio (Peptide 2:Peptide 3; Fig. 2_D_) matched perfectly with the in vivo spectrum in Fig. 2_C_, confirming the identification of two lysine-propionylated peptides.
Together we identified in vivo lysine propionylation at Lys-5, Lys-8, and Lys-12, as well as in vivo lysine butyrylation at Lys-5 and Lys-12 of histone H4 (Table I). The Lys-5, Lys-8, and Lys-12 of histone H4 are known to be acetylated, whereas Lys-12 is the subject of lysine methylation (Millipore/Upstate). Lysine acetylation at the four H4 lysine residues is associated with transcriptional activation, transcriptional silencing, chromatin high order structure, and DNA repair (21, 22). Some of the acetyllysine residues (e.g. Lys-8 of histone H4) provide a docking site to recruit a bromodomain-containing chromatin-remodeling enzyme SWI/SNF (23). Although biological functions of lysine propionylation and butyrylation in histones remain unknown, it is intriguing to speculate that propionyllysine or butyryllysine would be involved in the interaction/recruiting of a distinct set of proteins/enzymes to control the structure of chromatin and transcriptional activities.
Propionylation and Butyrylation of Core Histones Can Be Catalyzed by p300/CBP
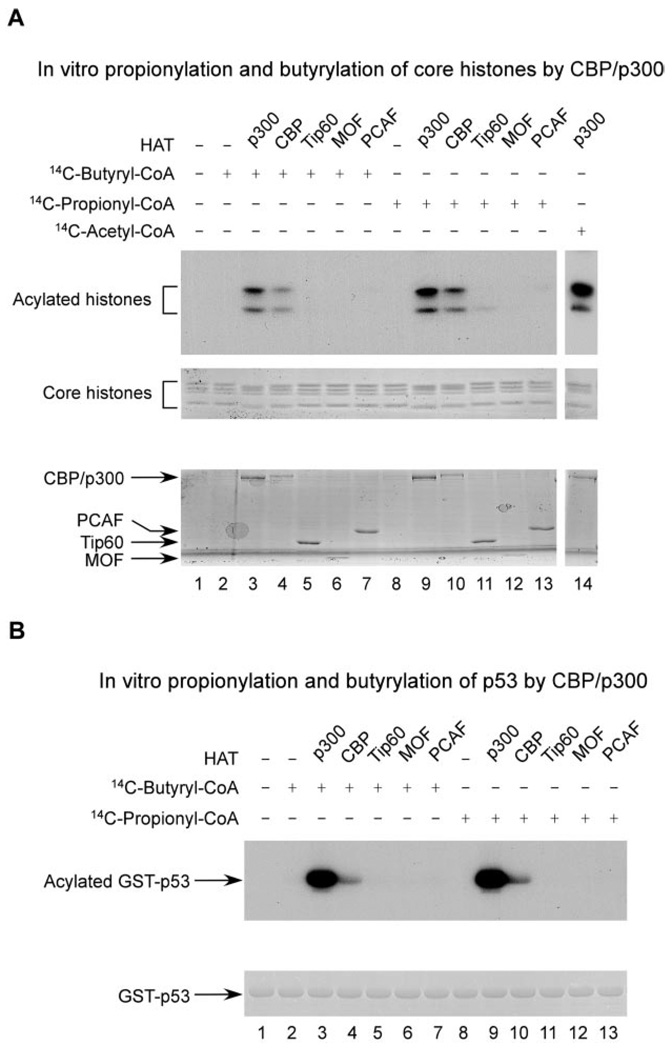
Because histone H4 can be propionylated and butyrylated in vivo, we next tested whether core histones can be propionylated and butyrylated in vitro by acetyltransferases using either [14C]propionyl-CoA or [14C]butyryl-CoA. Five acetyltransferases were tested, CBP, p300, Tip60, MOF, and PCAF. CBP and p300 are known acetyltransferases for Lys-5, Lys-8, Lys-12, and Lys-16 of histone H4.
The core histones were incubated with one of the CoAs and the acetyltransferase of interest. The protein mixture was then resolved by SDS-PAGE and visualized by autoradiography. CBP and p300 showed significant activities in catalyzing modifications on histone H4 (Fig. 3_A_). On the other hand, no significant propionylation or butyrylation products were detected for the other three acetyltransferases, Tip60, MOF, and PCAF.
FIG. 3. In vitro propionylation and butyrylation of core histones and p53 by acetyltransferases.
A, in vitro propionylation and butyrylation of core histones. The core histone proteins were incubated with the purified acetyltransferase in the presence of either [14C]propionyl-CoA or [14C]butyryl-CoA as indicated. The reaction products were resolved by SDS-PAGE and visualized by autoradiography (top panel). The amounts of the core histone substrates and the acetyltransferases are shown in the middle and bottom panels, respectively, by Coomassie Blue staining. B, in vitro propionylation and butyrylation of p53. The GST-p53 recombinant protein was incubated with the purified acetyltransferase in the presence of [14C]acyl-CoA as indicated. The reaction mixtures were resolved by SDS-PAGE and visualized by autoradiography (upper panel). The amount of the GST-p53 substrate is shown in the lower panel stained by Coomassie Blue.
To corroborate evidence of an in vitro modification reaction at lysine residues, we used nano-HPLC/mass spectrometric analysis to map the CBP-catalyzed, lysine-modified residues in histone H4. Lys-5, Lys-8, Lys-12, Lys-16, Lys-31, Lys-44, Lys-77, Lys-79, and Lys-91 were found to be both propionylated and butyrylated by CBP (supplemental information). Together these data establish that histone H4 can be lysine-propionylated and -butyrylated directly by CBP and p300 in vitro.
p53 Can Be Propionylated and Butyrylated by p300/CBP in Vitro
To examine whether acetyltransferases can catalyze lysine propionylation and butyrylation reactions in non-histone proteins, we evaluated in vitro propionylation and butyrylation reactions in p53. CBP/p300, co-activators of p53, augment its transcriptional activity and modulate its biological functions (5, 24). Multiple lysine residues in p53 can be acetylated, some of which are known to be modified by CBP/p300 (5, 25, 26). Given the fact that CBP/p300 are the acetyltransferases for p53 and that they have enzymatic activities for lysine propionylation and butyrylation in histones, we tested whether the HATs could catalyze similar reactions in p53. Toward this aim, we repeated the in vitro enzymatic reactions for p53 using the procedure described above. Again only two of the five acetyltransferases, CBP and p300, could carry out propionylation and butyrylation reactions at p53 at a significant reaction rate under our experimental conditions (Fig. 3_B_). Interestingly p300 showed higher catalytic activity than CBP for p53. In contrast, the two enzymes had comparable activities toward histones.
CBP and p300 are acetyltransferases that can catalyze autoacetylation reactions. To test whether the proteins can carry out autopropionylation and autobutyrylation reactions, we mapped the modification sites at p300 and CBP by mass spectrometry. Twenty-one lysine propionylation sites and 11 lysine butyrylation sites were localized in p300, whereas 12 lysine propionylation sites and seven lysine butyrylation sites were mapped in CBP (Table I and supplemental information). Identification of propionylated and butyrylated peptides in non-histone proteins, p53, CBP, and p300, suggests the possibility that the two modifications are not restricted to histones.
DISCUSSION
In summary, we report the identification of in vivo lysine propionylation and lysine butyrylation in histones. The modifications were validated by synthetic peptides and in vitro enzymatic reactions. After testing five acetyltransferases, we showed that two previously known acetyltransferases, CBP and p300, could catalyze lysine modification reactions using either propionyl-CoA or butyryl-CoA, in histones and p53, at a significant reaction rate in vitro.
Lysine is one of the two major ribosomally coded amino acid residues with a positive charged side chain at physiological pH, playing an important role in protein folding and functions. We do not know yet what will be the functional consequences of the two modifications. Nevertheless given the regulatory functions of every known lysine modification and significant structural changes induced by lysine propionylation and butyrylation, it is anticipated that the two modifications are likely to have biological functions in the same fashion as other lysine modifications.
Histones are known to be modified by an array of post-translational modifications, including methylation, acetylation, ubiquitination, small ubiquitin-like modification, and ribosylation (27). A combinatorial array of post-translational modifications in histones, termed the “histone code,” dictates the functions of the proteins in gene expression and chromatin dynamics (28). Post-translational modifications of histones have been elegantly studied by both biochemistry (28) and mass spectrometry (29–31). Nevertheless lysine propionylation and butyrylation at histones have not been reported before. Our results suggest that the complexity of histone modifications remains to be explored.
Acetyl-CoA can arise during the catabolism of sugars, fatty acids, and amino acids. Propionyl-CoA is derived only from odd-chain fatty acid and amino acid catabolism, whereas butyryl-CoA is a metabolic intermediate formed during the β-oxidation of fatty acids as well as a substrate for fatty acid elongation. The concentration of the short-chain CoAs fluctuates depending on diet and cellular physiological conditions (15). If the rate of the modifications depends on the concentration of the short-chain CoAs, directly or indirectly, it would be appealing to speculate that lysine propionylation and butyrylation may regulate cellular metabolic pathways in response to cellular physiological conditions. Such a scenario then opens up the potential for the biochemical intermediates thus produced to lead to tissue-specific and environmentally responsive regulatory programs.
The activities and specificities of regulatory enzymes responsible for propionylation and butyrylation might be influenced by other factors. For some acyltransferases (e.g. acetyltransferases), the dimensions of the CoA binding pocket might determine the binding affinity to the short-chain CoA. Alternatively it is also possible that the binding affinity might be modulated by other regulatory molecules, such as association of cofactors or interaction proteins. Finally identification of lysine propionylation and lysine butyrylation further suggests the possible existence of novel enzymes specific for the modifications (other than the known acetyltransferases).
Discovery of lysine propionylation and butyrylation raises many interesting questions. What are the enzymes responsible for controlling the status of lysine propionylation and butyrylation? Given the close structural similarity, some of the HATs and histone deacetylases might be able to catalyze the reactions. What determines the specificities of a transferase among three types of lysine modifications: acetylation, propionylation, and butyrylation? What are the biological pathways regulated by lysine propionylation and butyrylation? The study described here is really the beginning of future research into addressing the fundamental questions of the novel lysine modifications.
Supplementary Material
S1
Acknowledgment
We thank Kai Zhang for technical help for this work.
Footnotes
1
The abbreviations used are: HAT, histone acetyltransferase; CBP, CREB (cAMP-response element-binding protein)-binding protein; PCAF, p300/CBP-associated factor.
*
This work was supported by National Institutes of Health Grants CA107943 (to Y. Z.), CA098821 and CA104843 (to W. G.), GM31278 (to J. F.), and AG025688 (to J. P.) and Robert A. Welch Foundation Grant I1550 (to Y. Z. and to J. F.).
REFERENCES
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT. Posttranslational Modifications of Proteins: Expanding Nature’s Inventory. Greenwood Village, CO: Roberts and Co. Publishers; 2006. [Google Scholar]
- 3.Schweppe RE, Haydon CE, Lewis TS, Resing KA, Ahn NG. The characterization of protein post-translational modifications by mass spectrometry. Acc. Chem. Res. 2003;36:453–461. doi: 10.1021/ar020143l. [DOI] [PubMed] [Google Scholar]
- 4.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 6.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 8.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 9.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 10.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigis MC, Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 12.McKinsey TA, Olson EN. Cardiac histone acetylation—therapeutic opportunities abound. Trends Genet. 2004;20:206–213. doi: 10.1016/j.tig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br. J. Cancer. 2004;90:761–769. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King MT, Reiss PD. Separation and measurement of short-chain coenzyme-A compounds in rat liver by reversed-phase high-performance liquid chromatography. Anal. Biochem. 1985;146:173–179. doi: 10.1016/0003-2697(85)90412-9. [DOI] [PubMed] [Google Scholar]
- 16.Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Structure of the yeast histone acetyltransferase Hat1: insights into substrate specificity and implications for the Gcn5-related N-acetyltransferase superfamily. Cold Spring Harb. Symp. Quant. Biol. 1998;63:501–507. doi: 10.1101/sqb.1998.63.501. [DOI] [PubMed] [Google Scholar]
- 17.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York: W. H. Freeman and Co.; 2005. pp. 631–652. [Google Scholar]
- 18.Zhao Y, Zhang W, Kho Y, Zhao Y. Proteomic analysis of integral plasma membrane proteins. Anal. Chem. 2004;76:1817–1823. doi: 10.1021/ac0354037. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 21.Peterson CL, Laniel MA. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Shia WJ, Pattenden SG, Workman JL. Histone H4 lysine 16 acetylation breaks the genome’s silence. Genome Biol. 2006;7:217. doi: 10.1186/gb-2006-7-5-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 24.Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 25.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 29.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 30.Boyne MT, II, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 31.Medzihradszky KF, Zhang X, Chalkley RJ, Guan S, McFarland MA, Chalmers MJ, Marshall AG, Diaz RL, Allis CD, Burlingame AL. Characterization of Tetrahymena histone H2B variants and posttranslational populations by electron capture dissociation (ECD) Fourier transform ion cyclotron mass spectrometry (FT-ICR MS) Mol. Cell. Proteomics. 2004;3:872–886. doi: 10.1074/mcp.M400041-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1