Redox signaling and protein phosphorylation in mitochondria: progress and prospects (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 16.
Published in final edited form as: J Bioenerg Biomembr. 2009 Apr;41(2):159–168. doi: 10.1007/s10863-009-9217-7
Abstract
As we learn more about the factors that govern cardiac mitochondrial bioenergetics, fission and fusion, as well as the triggers of apoptotic and necrotic cell death, there is growing appreciation that these dynamic processes are finely-tuned by equally dynamic post-translational modification of proteins in and around the mitochondrion. In this minireview, we discuss the evidence that S-nitrosylation, glutathionylation and phosphorylation of mitochondrial proteins have important bioenergetic consequences. A full accounting of these targets, and the functional impact of their modifications, will be necessary to determine the extent to which these processes underlie ischemia/reperfusion injury, cardioprotection by pre/post-conditioning, and the pathogenesis of heart failure.
Keywords: Mitochondria, Heart, Signaling, Redox, Phosphorylation, Ischemia, Preconditioning, Bioenergetics
Introduction
Mitochondria are the sites of aerobic metabolism where energy in the form of nutrition is converted to a more usable energy currency, ATP. A hallmark of mitochondrial function in the heart is its ability to step-up ATP production promptly in response to increased workloads. Failure to do so has disastrous consequences. And yet mitochondria are also capable of much more. Work in diverse fields has led to the emergence of mitochondria as loci of control within a cell, at times acting as integrators of intracellular signals, while at others, they initiate or amplify signals, in turn. Specifically, mitochondria figure prominently in shaping two prominent signaling modalities within the cell, redox balance and protein phosphorylation cascades. In this minireview, we briefly consider intramitochondrial redox signaling with emphasis on nitrosylation and glutathionylation, as well as the growing evidence for the importance of protein phosphorylation by resident kinases. Finally, inspired by cell signaling literature outside the mitochondrion, we consider specific examples of cross-talk that may hint at a more extensive intramitochondrial redox-phosphorylation network.
Redox signaling in mitochondria
Reactive oxygen and nitrogen species
The redox balance within the cell results from the balance of pro-oxidant and anti-oxidant species. Mitochondrial superoxide (O2−•) arises from the incomplete reduction of O2 as a low-level byproduct of aerobic respiration; yields range from up to 1–2% of oxygen consumed by isolated mitochondria in vitro, though the yield is likely lower in vivo (St-Pierre et al. 2002; Turrens 2003). Superoxide undergoes spontaneous dismutation to yield hydrogen peroxide (H2O2), which can, in turn, decompose to generate reactive a hydroxyl radical (OH•) via the Fenton reaction. Collectively, O2−•, H2O2, and OH• are commonly termed reactive oxygen species (ROS). Respiratory complexes I and III have been implicated as prominent sites of mitochondrial ROS production. In the heart, a high proportion of superoxide is produced at the Q0 site of complex III as a result of oxidation of the ubisemiquinone radical during the Q cycle (Brookes et al. 2004). Excessive ROS production is often deleterious to mitochondrial and cell function. In particular, unchecked OH• accumulation leads to its indiscriminate reaction with proteins, lipids and DNA. ROS are, therefore, tightly monitored by scavenging enzymes. Superoxide from complex III is released into both the mitochondrial matrix, where it is converted to H2O2 by manganese-dependent superoxide dismutase (MnSOD), and into the intermembrane space (IMS) where it is similarly reduced by copper/zinc-dependent superoxide dismutase (CuZnSOD). Hydrogen peroxide, in turn is quickly cleared (reduced to water) by the enzymes glutathione peroxidase, peroxiredoxins, and catalase.
In addition to ROS-scavenging enzymes, the cell is equipped with low molecular weight antioxidant molecules, the most abundant of which is the tripeptide, glutathione (γ-Glu-Cys-Gly). It is synthesized in the cytosol and transported to the mitochondria (Griffith and Meister 1985; Jocelyn 1975; Jocelyn and Kamminga 1974), where its concentration has been estimated between 5 and 10 mM (Wahllander et al. 1979). Upon oxidation, glutathione forms a cystine-linked dimer (GSSG). It is efficiently recycled to the reduced form through the action of NADPH-dependent glutathione reductase (GR), of which there are both cytosolic and mitochondrial isoforms. GR helps maintain the cellular GSH/GSSG ratio in the range of 30–100:1 (Hwang et al. 1992). GSH participates as an antioxidant both by acting as a thiol buffer, directly reacting with protein thiols and ROS/RNS, and also by serving as a substrate for the glutathione peroxidases (Hurd et al. 2005a) that reduce ROS.
The term, reactive nitrogen species (RNS) refers to the family of compounds derived from nitric oxide (NO•), the pleiotropic signaling molecule synthesized from L-arginine by the nitric oxide synthases, nNOS (NOS1), eNOS (NOS3) and iNOS (NOS2). In the heart, eNOS is found in the coronary vascular endothelium, while cardiac myocytes constitutively express eNOS and nNOS; iNOS expression can be triggered by inflammation, ischemia and heart failure (Brown and Borutaite 2007; Jones and Bolli 2006). NO• exerts many of its physiological effects, including classical vasodilation, through a canonical pathway wherein it binds to guanylyl cyclase, thereby stimulating the formation of cyclic GMP from GTP, which subsequently activates PKG (Costa et al. 2008; Schlossmann et al. 2003). Yet NO• can also be converted, by oxidation or reduction, into many chemically reactive forms including nitrite (NO2−), peroxynitrite (ONOO−), and nitroxyl (HNO), each of which yields distinct functional consequences (Brown and Borutaite 2007; Burwell and Brookes 2008; Poderoso 2009; Sinha et al. 2008; Wink et al. 2003). Under pathological conditions, peroxynitrite, formed by the reaction of NO• with O2−•, can react directly and irreversibly with proteins (on tyrosine), lipids, and DNA (Burwell and Brookes 2008), a result that is frequently deleterious. It is now clear, however, that when properly regulated under physiological conditions, ROS/RNS, are not purely destructive, but rather, engage in important signaling functions both in and outside of mitochondria (Finkel 2003; Jones 2006).
Redox signaling
The term, “redox signaling” describes a process by which milder physiological levels of ROS/RNS induce modifications to proteins that are discrete, site-specific and reversible. Redox signals may abrogate or enhance activity of the target protein, and have been implicated in physiological signaling processes that include kinase signaling, channel function, apoptotic proteolysis, and regulation of transcription (Hool and Corry 2007; Janssen-Heininger et al. 2008; Ueda et al. 2002). The tenet of redox-sensing is that certain proteins undergo reversible chemical changes in response to changes in local redox potential. The sulfhydryl chemistry of cysteine residues within proteins is uniquely suited to the task, as they can adopt multiple oxidation states (Forman et al. 2004; Winterbourn and Hampton 2008). Moreover, cysteines that are adjacent to basic amino acids or lie in a local hydrophobic pocket within a protein’s tertiary structure are rendered particularly reactive at physiological pH, since these conditions lower the pKa of the thiol sidechain from 8 to between 5 and 7 (Greco et al. 2006; Hess et al. 2001). These reactive cysteines can react with ROS/RNS to yield a number of species including S-nitrosylated proteins (PSNO), intra- and intermolecular disulfide bonds (PSSP), or the formation of mixed disulfides with glutathione (glutathionylation, PSSG) (Hurd et al. 2005a, b; Janssen-Heininger et al. 2008).
S-nitrosylation
S-nitrosylation, the addition of NO• to reactive cysteines, has emerged as an important signaling modality, independently of the induction of cGMP production, effectively acting as a reversible molecular switch analogous to phosphorylation (Hess et al. 2005). Owing to the complexity of NO• and thiol chemistry, there are many possible reaction mechanisms that can lead to S-nitrosylation, though which prevails in vivo is still debated (Costa et al. 2003; Zhang and Hogg 2005). Removal of the nitrosyl moiety can result from transnitrosation with GSH, which yields GSNO, whose accumulation is cleared by glutathione reductase. Alternately, nucleophilic attack by a vicinal protein thiol will yield a disulfide bond and concomitantly release NO−(Costa et al. 2003; Janssen-Heininger et al. 2008). Finally, the search for a bona fide denitrosylase, whose actions would be analogous to the phosphatases in kinase signaling, has implicated the cytosolic and mitochondrial isoforms of thioredoxin (Trx1 and Trx2 respectively) and the reductases that recharge them TR1 and TR2, which in turn, depend directly upon the redox potential of NADPH (Benhar et al. 2008; Sengupta et al. 2007).
Prior to considering S-nitrosylation further however, we digress briefly to consider the best characterized of the interactions of NO• with mitochondria, namely its interaction with respiratory complex IV, cytochrome C oxidase. The function of the complex is to accept electrons from cytochrome C and use them to reduce O2 to H2O, while harnessing the energy to extrude protons from the matrix to the intermembrane space. NO• binds the complex at its oxygen-binding site, the heme a3-CuB center, and inhibits respiration. This interaction fulfills many criteria of a redox signal. NO• binding is fast, reversible and happens under physiological conditions of low oxygen tension (Brookes et al. 2002; Brown and Borutaite 2007; Erusalimsky and Moncada 2007). That binding of NO can compete with O2, has led to the hypothesis that a gradient in the tissue concentration of NO•, as a function of distance from blood vessels, effectively extends the diffusibility of O2 to distal tissues (Thomas et al. 2001). Another consequence of NO• interaction with complex IV is that, depending on the O2 concentration, ROS production may be favored (Brookes et al. 2002; Brown and Borutaite 2007; Erusalimsky and Moncada 2007).
A secondary consequence of prolonged exposure of cells or isolated mitochondria to NO• is inhibition of Complex I activity and oxidation of the mitochondrial glutathione pool. Since inhibition could be reversed by exposure to light and exogenous application of thiols, it was suspected that NO• exerted its effects by S-nitrosylation (Clementi et al. 1998). The inferred nitrosylation could be induced by NO• and peroxynitrite, though perhaps more rapidly by S-nitrosothiols like S-nitrosoglutathione (GSNO) (Borutaite et al. 2000). Direct confirmation of complex I S-nitrosylation has since been verified using chemiluminescence (Burwell et al. 2006; Dahm et al. 2006) and “biotin switch” methods (Burwell et al. 2006). In fact, complex I accounts for approximately 20% of mitochondrial S-nitrosylation induced by exogenous treatment with GSNO. The nitrosylation, up to a stoichiometry of 7 mol/mol appears confined predominantly to the 75-kDa subunit (Burwell et al. 2006) and causes a 20–30% reduction in activity (Burwell et al. 2006; Dahm et al. 2006), though mitochondrial respiration could be inhibited by 80% within an hour of exposure to S-nitrosopenicillamine (SNAP) (Dahm et al. 2006). Inhibition of respiration was accompanied by increased ROS production (Dahm et al. 2006).
Intriguingly, whether nitrosylation-mediated inhibition of complex I is ultimately harmful or beneficial may depend on the source of the mitochondria. In the brain for instance, inhibition of complex I, and the high levels of ROS it engenders, would mimic the oxidative stress that is a hallmark of neurodegenerative diseases including Huntington’s, Alzheimer’s and Parkinson’s disease (Lin and Beal 2006; Przedborski and Ischiropoulos 2005). Yet as noted by Burwell et al. (Burwell et al. 2006), in the heart, complex III accounts for the bulk of ROS production. Therefore complex I inhibition, would likely yield lower ROS levels and serve a protective function. To underscore the point, they found elevated levels of mitochondrial SNO in ischemically-preconditioned hearts (Burwell and Brookes 2008; Burwell et al. 2006).
Indeed, the role of NO• in the cardioprotection afforded by ischemic preconditioning has been well documented (Burwell and Brookes 2008; Jones and Bolli 2006). Until recently however, many studies have centered on elucidating the role of canonical NO• signaling through cGMP to the mitochondria (Costa et al. 2008). Recent work by Sun et al., however, showed that ischemic preconditioning (IPC) of rat hearts caused nitrosylation of proteins involved in Ca2+ handling and energetics (Sun et al. 2006, 2007). Moreover, perfusion of hearts with GSNO improved left ventricular function and protected against ischemia. Adapting the biotin switch assay, by using maleimide-conjugated fluorescent dyes in lieu of HPDP-biotin, enabled researchers to resolve nitrosylated proteins by 2D-gel electrophoresis, and subsequently identify them by mass spectrometry. The results reaffirmed the 75-kDa subunit of complex I as a nitrosoprotein, but also identified F1FoATPase α subunit, α-ketoglutarate dehydrogenase, among others. S-nitrosylation inhibited F1FoATPase activity yet activated α-ketoglutarate dehydrogenase activity in vitro. These results dove-tail nicely with early physiological studies indicating that IPC spares ischemic ATP consumption, and protects against the loss of α-ketoglutarate activity. Clearly, a full account of the role of NO• in cardioprotection must now include both cGMP-dependent and cGMP-independent, redox-dependent mechanisms.
S-glutathionylation
S-glutathionylation refers to modification of a protein’s cysteine residues by covalent modification with glutathione, to form a mixed disulfide species (PS-SG). Typically, under reducing conditions of the cell where ratios of reduced to oxidized glutathione are high, glutathionylation is not favored. Several factors, however, make mitochondria fertile ground for this modification. Firstly, the protein thiols lie in proximity of mitochondrial proteins to ROS derived from the electron transport chain. Secondly mitochondrial proteins are particularly cysteine rich. Heart mitochondrial membranes contain about 90 nmol thiol/mg proteins, of which it is estimated that 40% are potentially reactive (Beer et al. 2004). Finally, mitochondrial proteins within the matrix and inner aspect of the mitochondrial inner membrane are susceptible to glutathionylation owing to the alkaline pH of the mitochondrial matrix (~8) that favors deprotonation of protein thiols.
Glutathionylation can occur by several mechanisms (Hurd et al. 2005a). The first, is simple reaction of oxidized glutathione (GSSG) with protein thiolate ion (S−) in a process called disulfide exchange. This mechanism may occur when a burst of ROS causes accumulation of GSSG. Secondly, glutathiolate anion (GS−) can scavenge protein thiyl radicals that arise in response from one-electron oxidation. Thirdly, GS− on can react with protein thiols that have undergone two-electron oxidation to sulfenic acid (PSOH). Finally, glutathiolate can react with protein nitrosothiols (PSNO). Glutathionylation, like S-nitrosylation, is reversible, and by similar chemistry. For instance, as with the S-nitrosylated proteins, glutathionylated proteins with vicinal thiols can form disulfides, and thereby release GS− (Hurd et al. 2005a). In fact, Beer et al. showed that most mitochondrial proteins are glutathionylated only transiently before they become disulfide bonded. A much smaller fraction is persistently glutathionylated. Glutaredoxin is responsible for deglutathionylation but also reduction of disulfides, provided the GSH pool is reduced (Beer et al. 2004). Thioredoxin also fulfills this function albeit less effectively (Jung and Thomas 1996).
S-glutathionylation likely serves two purposes in mitochondria, defense against oxidant stress and bona fide redox signaling. When the mitochondrial glutathione pool is oxidized, the 75-kDa subunit of complex I, with its bounty of cysteines, is a prominent target (Beer et al. 2004; Hurd et al. 2008). Under these conditions it has been advanced that glutathionylation/oxidation buffer against ROS, protecting protein thiols from progressive oxidation to sulfinic acid (PSO2H) and sulfonic acid (PSO3H) are steps that could lead to irreversible protein dysfunction (Hurd et al. 2005b). Intriguingly, glutathionylation of Complex I, in response to GSH depletion by diamide, inhibits complex I activity, but unlike nitrosylation on the same subunit, does not increase ROS (Hurd et al. 2008).
Mitochondrial targets for nitrosylation and glutathionylation continue to accumulate. Purified NADP+-dependent isocitrate dehydrogenase is glutathionylated at Cys269, in vitro, which renders it less susceptible to proteolysis. Glutathionylation was also observed in oxidant-stressed mitochondria and HEK293 cells (Kil and Park 2005). Complex II is an interesting example of a protein that is persistently glutathionylated. After 24 h of perfusion following 30-min ischemia induced by coronary ligation, glutathionylation of its 70-kDa subunit was markedly reduced, an observation that correlated with a loss of electron transfer activity. The authors suggest that physiological glutathionylation is required for optimal complex II function (Chen et al. 2007). Finally West et al. provide evidence for NO•-induced protein glutathionylation. Treating COS-7 and rat aortic smooth muscle cells with NO• donors increased protein glutathionylation, as assessed with an anti-GSH antibody. Similar results were observed in acetylcholine-treated aortic rings and in transgenic mouse hearts that overexpressed iNOS. Among the targets identified by mass spectrometry were the adenine nucleotide translocator (ANT) and the a subunit of the F1FoATPase (West et al. 2006).
Evidence that nitrosylation/glutathionylation/oxidation also represents a legitimate intramitochondrial redox signal, distinct from simple buffering function, comes from experiments showing that low concentrations of H2O2 and SNAP cause redox modification of a select group of proteins, despite little change in the redox status of the global mitochondrial thiol pool (Hurd et al. 2007). Moreover, several of these proteins were identified as redox-modified in response to endogenous mitochondrial ROS formation caused by reverse electron transport (succinate/antimycin/FCCP; RET). Targets include pyruvate dehydrogenase kinase 2 and propionyl-CoA carboxylase among others. Both enzymes were inhibited by RET-induced oxidation and could be reactivated by reduction with dithiothreitol. No doubt one of the take-home messages from the work of Hurd et al. is that though many mitochondrial proteins can be thiol-modified under oxidant stress, only a much smaller subset of these modifications will meet the criteria of a bona fide physiological redox signal. Since both sets can be reduced glutaredoxin or thioredoxin, the primary distinction between them would appear to be site-specificity.
Kinase signaling to, and within, the mitochondrion
It has long been established that phosphorylation plays an important role in control of mitochondrial metabolism (Linn et al. 1969); the pyruvate dehydrogenase kinases regulate the pyruvate dehydrogenase complex, while branched chain amino acid dehydrogenase kinase regulates a critical step in metabolism of valine, leucine and isoleucine (Harris et al. 1997). Yet apart from these functions, more extensive characterization of the precise role of mitochondrial phosphorylation has lagged other avenues of signaling research. Indeed, conventional wisdom holds that given the prokaryotic ancestry of mitochondria and that phosphorylation in prokaryotes is comparatively primitive, then mitochondrial phosphorylation networks are likely to be similarly primitive. Within the last decade, however, kinase signaling to, and within, the mitochondria has emerged as a prominent research theme as it becomes understood that processes such as fusion, fission, apoptosis and metabolism must respond to environmental cues within the cell (Horbinski and Chu 2005; McBride et al. 2006; Pagliarini and Dixon 2006). In the cardiac realm, mitochondria are the endpoints of cell survival cascades, triggered by cardioprotective preconditioning, that ultimately forestall the mitochondrial permeability transition that would otherwise lead to cellular necrosis (Murphy and Steenbergen 2008). Here, we briefly consider converging lines of cell biological and proteomic evidence that mitochondrial phosphorylation has an impact on diverse mitochondrial functions.
Phosphorylation at the mitochondrial periphery
The external face of the outer mitochondrial membrane (OMM) is adorned with intracellular receptors (scaffolds), and serves as site for the integration of signals for processes ranging from fission, apoptosis, cytoprotection, and energetic demand, to steroidogenesis (Pawson and Scott 1997; Smith et al. 2006). Prominent examples of this type of scaffold would be the PKA anchoring proteins (AKAPs) (Wong and Scott 2004), and the PKC-interacting proteins (Poole et al. 2004), RICKs and RACKs, the receptors for inactive/active kinases respectively (Mochly-Rosen and Gordon 1998).
PKA signaling to the mitochondria is mediated by mitochondrial AKAPs, among them D-AKAP1, Wave1/Scar, PAP7 and RAB32. PKA, tethered to the mitochondrial outer membrane through D-AKAP1, confers the prosurvival effect of IL-3 by phosphorylating the pro-apoptotic protein BAD, thereby mitigating apoptosis (Harada et al. 1999). Mitochondrially-tethered PKA also appears to acutely accelerate the transport of cholesterol into mitochondria, a rate limiting step in steroidogenesis, either through the dual specificity AKAP1 (AKAP121) (Dyson et al. 2008) or the R1-binding AKAP known as PAP7 bound to the peripheral benzodiazepine receptor (Li et al. 2001; Liu et al. 2003; Papadopoulos et al. 2007). Rab32, a Ras-related small molecular weight GTPase, was identified as an AKAP in a yeast two-hybrid screen using the AKAP-binding domain of the RII regulatory subunit of PKA as bait. The dual-function G-protein is localized to mitochondria where it sequesters a portion of the cellular PKA pool (Alto et al. 2002). Moreover, dynamin-related protein 1 (Drp1), a high molecular weight GTPase that participates in mitochondrial fission and cell death (Chang and Blackstone 2007; Cribbs and Strack 2007), is also a substrate for what is presumed to be mitochondrially-tethered PKA.
Aside from PKA, other kinases that have been shown to undergo translocation to the mitochondria include Raf kinases, ERKs and several isoforms of PKC. The Raf kinases A-Raf B-Raf and C-Raf are best known for their role in a growth factor signaling cascade downstream of the small molecular weight GTPase, Ras, and upstream of the MAP kinase kinase, MEK. Yet Rafs, C-Raf in particular, also undergo translocation to the mitochondria (Galmiche and Fueller 2007). It was first shown that raf-1 (C-Raf), is recruited to the mitochondria by its interaction with Bcl-2 (Wang et al. 1996a), inviting the analogy that Bcl-2 may serve a similar role to the AKAPs and RACKs in PKA and PKC signaling. Another potential Raf scaffold, Bag-1, was identified as a BCL-2-binding protein in a yeast two-hybrid screen (Wang et al. 1996b). Bag1, like BCl-2, localizes a population of C-Raf to the mitochondrial membrane but also acts as a co-activator in vitro (Wang et al. 1996b). There, its pro-survival actions appear to be mediated by its ability to elicit phosphorylation of BAD (Jin et al. 2005; Wang et al. 1996a, b).
Several isoforms of PKC have been observed at the OMM. PKCα is targeted to mitochondria via its interaction with PICK1 (Wang et al. 2003). The PKCδ and PKCε isoforms of PKC, members of the novel, or Ca2+-independent, class of PKC isoforms, have received considerable scrutiny as they have emerged as important players in ischemia reperfusion injury and ischemic preconditioning respectively (Churchill and Mochly-rosen 2007; Mackay and Mochly-Rosen 2001). Though the mitochondrial RACK for PKCε is not known, its existence has been inferred by experiments in cardiomyocytes showing that peptide inhibitors of the PKC-RACK interaction abolish cardioprotection from ischemia (Chen et al. 2001) as well as the associated phosphorylation of mitochondrial targets (Ogbi and Johnson 2006). PKCε appears to undergo translocation to the mitochondria where it can interact with, and phosphorylate VDAC, a putative component of the permeability transition pore (Baines et al. 2003); phosphorylation correlated with inhibition of the mitochondrial permeability transition. Moreover, mitochondrial PKCε was found in complexes with MAPK. Formation of these complexes correlates with phosphorylation and inactivation of the aforementioned pro-apoptotic protein BAD (Baines et al. 2002).
MAP Kinases have also been observed at the mitochondrial outer membrane. Steroidogenesis, having already been shown to require mitochondrial PKA activity, has recently been shown to involve mitochondrial ERK1/2. Specifically, ERK1/2 phosphorylates StAR protein, on the outer mitochondrial membrane, at Ser232, in a cAMP-dependent manner. Subsequent mutation of Ser232 to Ala inhibited progesterone production in MA10 cells (Poderoso et al. 2008). In cardiac myocytes, there is evidence that the ERK1/2 and p38 SAPK associate with the mitochondrial outer membrane in a complex with PKCε, though their substrates have not been defined (Baines et al. 2002).
Phosphorylation in the mitochondrial intermembrane space
If phosphorylation at the mitochondrial surface appears to involve regulation of mitochondrial dynamics, control of steroid import and/or apoptotic protein egress, below the surface, in the intermembrane space, prime targets for phosphorylation would be involved in bioenergetics and apoptosis.
To date, the bulk of phosphorylation characterized in the IMS appears to be on tyrosine residues. In fact, mitochondrially localized tyrosine kinase activity was observed over 20 years ago (Piedimonte et al. 1986). Specifically, brain mitochondria undergo extensive tyrosine phosphorylation, in response to tyrosine phosphatase inhibition by sodium peroxovanadate, in the presence of exogenousATP (Arachiche et al. 2008; Lewandrowski et al. 2008; Salvi et al. 2002). Several labs have also observed the tyrosine phosphatases Shp-2 and PTP1B (Arachiche et al. 2008; Augereau et al. 2005; Salvi et al. 2004), in either the IMS (Salvi et al. 2004) or in both the matrix and the inner membrane (Arachiche et al. 2008). Salvi et al. also found members of the Src family tyrosine kinases, Src, Fyn (and to a lesser extent Lyn and Csk) (Salvi et al. 2002). The observation of c-Src in brain has since been confirmed by others (Arachiche et al. 2008).
In heart and liver, evidence for tyrosine phosphorylation in the intermembrane space has come from the detailed study of phosphorylated substrates. Lee et al. showed that cytochrome C oxidase (complex IV) is phosphorylated on tyrosine 304 of the IMS-facing subunit I in a cAMP-dependent manner, and that phosphorylation both increased its Km for cytochrome c and decreased Vmax (Lee et al. 2005). Tyrosine phosphorylation on this subunit was subsequently shown to be responsible for TNFα-induced inhibition of oxidative phosphorylation in hepatocytes (Samavati et al. 2008). Subunit I is also phosphorylated at Ser, Thr and Tyr residues on complex IV isolated from rat hearts (Helling et al. 2008). In studies of myocardial ischemia-related injury, Prabu et al. (Prabu et al. 2006) found that that hypoxia reduced complex IV function coincidentally with hyperphosphorylation of several subunits on complex IV, including subunit 1. Complex IV activity could be rescued by PKA inhibitors. Subsequent phosphorylation site mapping, by mass spectrometry, implicated Ser 115/116 of subunit 1 as well as sites within matrix facing subunits IV and Vb (Fang et al. 2007).
Other prominent proteins of the mitochondrial intermembrane space that have emerged as phosphoproteins are cytochrome c, creatine kinase (CK) and the IMS-facing cavity of the adenine nucleotide transporter (ANT). Cytochrome c is tyrosine-phosphorylated in both heart and liver, albeit at different sites. In heart, cytochrome C is phosphorylated at Tyr 97, an exposed site within its membrane-tethering cardiolipin-binding domain (Lee et al. 2006). Creatine kinase from brain is phosphorylated at tyrosines, which when modeled onto the octameric crystal structure, lie at the radial periphery and the ends of the molecule respectively, prompting Lewandrowski et al. (Lewandrowski et al. 2008) to speculate that phosphorylation may mitigate the interaction of CK with cardiolipin in the mitochondrial membrane. ANT from brain is phosphorylated at tyrosines 190 and 194 within its ADP-binding pit that is accessible to the IMS. These tyrosines form the basis of a “tyrosine ladder” that guides ADP towards its binding site at the bottom of a deep pocket within the protein. Phosphorylation of ANT at Tyr194 has also been found in rat hearts preconditioned with isofluorane (Feng et al. 2008). Given their location, phosphorylation of these tyrosines might be predicted to influence nucleotide transport (Lewandrowski et al. 2008).
Phosphorylation in the mitochondrial inner membrane and matrix
Phosphorylation as a regulatory modality in the mitochondrial matrix has been appreciated for some time (Burnett and Kennedy 1954; Linn et al. 1969). One of primary targets, under tight regulation, is the pyruvate dehydrogenase complex (PDC) that catalyzes the conversion of pyruvate, a product of glycolysis, to Acetyl-CoA, a substrate for the Krebs cycle that provides electrons to the electron transport chain. Specifically, the E1 subunit of PDC is phosphorylated at Ser264, Ser271, and Ser204 (known as sites 1, 2 and 3) by the pyruvate dehydrogenase kinases (Patel and Korotchkina 2006). There are 4 isoforms of the PDKs that are part of the atypical family of Ser/Thr kinases whose primary sequences are reminiscent of prokaryotic histidine kinases (Hanks and Hunter 1995; Popov et al. 1993). Their expression varies by tissue, with the heart expressing PDK1, PDK2 and PDK4 (Bowker-Kinley et al. 1998). The action of the PDKs is counteracted by the pyruvate dehydrogenase phosphatases (PDP1 and PDP2), which consists of a catalytic and a regulatory subunit. The PDPs belong to the PP2C family of phosphatases that differ from the PPP family (PP1, PP2A & PP2B) in that they have a requirement for Mg2+ as a cofactor and stimulated by Ca2+ (Roche et al. 2001).
Catabolism of the branched-chain amino acids valine, leucine, and isoleucine, is also subject to regulation by phosphorylation (Harris et al. 1995). The amino acids are first converted to the alpha-keto acids by branched-chain aminotransferase, then decarboxylated by the enzyme branched chain dehydrogenase (BCDH). This enzyme is regulated by branched chain dehydrogenase kinase, another member of the atypical kinase family. Far less well characterized is the antagonizing phosphatase, also whose biochemical properties would classify as a member of the PP2C family.
Conventional wisdom has long held that regulation of PDC and BCDH activity constitutes the alpha and the omega of kinase/phosphatase action within the mitochondrial matrix. However, there is growing evidence from proteomic studies that there are more mitochondrial phosphoproteins than previously thought, particularly within the matrix and on the matrix-facing proteins of the inner mitochondrial membrane (see (Horbinski and Chu 2005; Pagliarini and Dixon 2006)). Hopper et al. identified 45 phosphoproteins stained with the phosphospecific stain, ProQ-Diamond (Hopper et al. 2006). Interestingly, a phosphoproteomic screen of yeast mitochondria identified 80 phosphorylation sites on 48 proteins (Reinders et al. 2007). Both studies identify inner membrane and matrix proteins falling into several functional categories including transport, lipid metabolism, chaperones, redox enzymes and energy metabolism/oxidative phosphorylation. In the latter category two prominent targets have been F1FoATPase (see Kane et al., this issue), and complex I.
Complex I was posited as a potential phosphoprotein following the observations that NADH-ubiquinone dehydrogenase activity could be stimulated at least two-fold in fibroblasts treated with the cell-permeant dibutyryl-cAMP (Scacco et al. 2000). It has since been shown that complex I, in cells and mitochondria, undergoes cAMP-responsive phosphorylation on the 18-kDa (NDUFS4) subunit and a 10-kDa subunit (Papa et al. 2008). Phosphorylation site determination by mass spectrometry revealed that purified Complex I also has phosphorylation sites on the 42-kDa subunit, as well as on its B16.6, B14.5a, and B14.5b subunits (Palmisano 2007). Independent efforts to map the cAMP-responsive phosphorylation sites indicate that the ESSS, rather than the NDUFS4 subunit, is the 18-kDa phosphoprotein, and the 10-kDa target is the MWFE subunit (Chen et al. 2004). The former is thought to face the mitochondrial matrix, while that latter likely faces the intermembrane space (Chen et al. 2004). Interestingly, despite the cAMP-induced activation of complex I activity in cultured cells, similar activation was not observed for the purified complex phosphorylated in vitro. The discrepancy led to the discovery that cytosolic phosphorylation of NDUFS4 modulates its import into mitochondria and/or subsequent assembly of complex I (De Rasmo et al. 2008). This observation is not unique to Complex I. Indeed PKA-mediated targeting of proteins for mitochondrial import has been observed for p4502B1, cyp2E1 and glutathione S-transferase (Anandatheerthavarada et al. 1999; Robin et al. 2002, 2003).
Our understanding of mitochondrial phosphorylation has also benefited from efforts to understand cardioprotective preconditioning. Two decades of research have elucidated how the paracrine action of adenosine, bradykinin and opioids stimulates pro-survival kinase cascades that culminate at the mitochondrion where, by processes that are still poorly understood, the mitochondrial permeability transition is inhibited (Downey et al. 2007). As well-characterized as the signaling is, the total impact on the myocyte proteome is still being addressed. Arrell et al. used a 2D-gel proteomics strategy to identify changes to the proteomes of myocytes pharmacologically with Adenosine and the potassium channel opener, diazoxide (Arrell et al. 2006). Mitochondrial proteins constituted the bulk of the proteins that had been affected by the pharmacological treatments, some of which exhibited changes in spot pattern consistent with phosphorylation. This ultimately led to the identification of 5 new phosphorylation sites on the β subunit of the F1FoATPase (see Kane et al., this issue). Like the work of Sun et al. mentioned previously in the context of S-nitrosylation (Sun et al. 2006, 2007), this kind of proteomic study reaffirms that preconditioning entails many modifications rather than a single endpoint. The trick will ultimately be to sort the causal modifications from the epiphenomena.
Putting it all together: redox/kinase interplay in the mitochondrion
To be sure, we are still in the early days of the study of redox signaling and protein phosphorylation in mitochondria. No doubt the catalogue of nitrosylated, glutathionylated and phosphorylated targets in mitochondria will continue to grow as high throughput proteomic approaches are applied (e.g. (Derakhshan et al. 2007; Kettenhofen et al. 2007; Reinders et al. 2007)). Notwithstanding the groundwork yet to be laid on each front, the early indications suggest that there is likely to be some degree of interplay between these two signaling modalities.
As to how they may augment or impinge on each other, extra-mitochondrial cell signaling research offers several clues. ROS generation, for example, is a hallmark of mitogenic signaling to the nucleus, and ROS scavengers blunt cell proliferation. Part of the reason is that ROS inactivates cascade-inhibiting tyrosine phosphatases by targeting conserved reactive cysteines (Janssen-Heininger et al. 2008; Rhee 2006). Direct interaction between cytosolic redox enzymes with kinases is another mode of crosstalk. Specifically, binding of Thioredoxin to Apoptosis Signal Kinase 1 (ASK1) confers redox sensitivity upon the kinase (Matsuzawa and Ichijo 2008). In response to sustained oxidant stress, thioredoxin dissociates, freeing ASK1 to activate its downstream targets, ultimately leading to apoptosis. Conversely, phosphorylation can influence the activity of redox enzymes. Witness the observation that phosphorylation of peroxiredoxin2 by cdc2 kinase mitigates its H2O2-scavenging function (Rhee et al. 2005). To what extent might this kind of cross-modulation be at play within heart mitochondria and to what end? Let us end up by briefly considering recent examples of redox-mediated phosphorylation, and, reciprocally, phosphorylation-mediated ROS regulation in vitro.
One documented example would be modulation of the mitochondrial fuel mix through regulation of the PDC. Briefly, the PDKs are acutely responsive to changes to the local NADH/NAD and Acetyl-CoA/CoA ratios. PDKs are stimulated by NADH/Acetyl-CoA and inhibited by NAD/CoA (Bao et al. 2004; Patel and Korotchkina 2006). This redox balance is important to the mechanism of substrate feedback inhibition of the PDC in the fatty acid-fed state. The recent observation that the thiols of PDK are oxidized by endogenous ROS, thereby inhibiting the enzyme, provides a second consistent potential mechanism for the regulation of PDC (Hurd et al. 2007). Thus, bursts of ROS/RNS might inhibit PDK either directly via thiol modification or indirectly by oxidation of the NADH pool which is sensed by the lipoyl (PDK-activating) moiety on the E2 subunit of PDC.
Secondly, it had been well established that PDC is activated by Ca2+ through Ca2+-dependent dephosphorylation. The results by Hopper et al. (Hopper et al. 2006) indicate that the number of targets for Ca2+-dependent dephosphorylation is larger than previously thought. Specifically, matrix-resident MnSOD is dephosphorylated and activated in response to Ca2+. Thus, though elevated Ca2+ increases ROS production, it also activates a scavenging mechanism which ensures tight control of ROS, using the phosphorylation state of MnSOD as the intermediary signal. Similarly, Ca2+ dependent dephosphorylation of cytochrome c oxidase is thought to activate complex IV, which could influence mitochondrial ROS production (Bender and Kadenbach 2000; Lee et al. 2002). That ROS may be influenced by phosphorylation is also supported by the observation that incubation of saponin-skinned myocytes with the catalytic subunit of PKA increased mitochondrial flavoprotein fluorescence while partially depolarizing the membrane potential (Nagasaka et al. 2007). Finally, intramitochondrial tyrosine phosphorylation may also be regulated by ROS, particularly if there are resident mitochondrial tyrosine phosphatases which harbor conserved redox-sensitive cysteines.
Conclusion
A fundamental step toward understanding the relative roles of redox signaling and phosphorylation within heart mitochondria will be to identify the relevant parties, the complement of kinases, phosphatases and ROS scavengers. In the case of phosphorylation, it will be crucial to know which kinases and phosphatases are resident in mitochondria, and which are translocated transiently (Foster et al. 2008). Recently-published compendia of mitochondrial proteins will prove useful for hypothesis testing (Pagliarini et al. 2008), as will future efforts to mine the margins of the heart mitochondrial proteome. Technical and technological advances promise the high-throughput assessment and quantification of site specific phospho/redox modifications from which one can look for functional correlates.
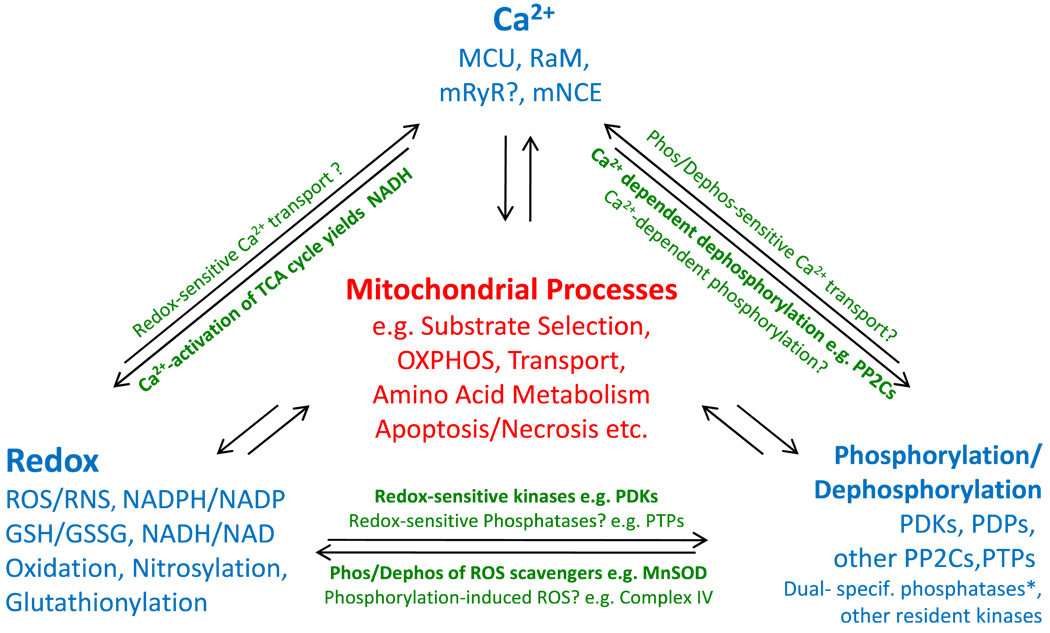
Fig. 1.
Interplay of Signaling Modalities in Mitochondria. Summary of examples that illustrate the degree of interdependency between mitochondrial Ca2+, redox balance, and phosphorylation (Inspired by the “Love-Hate Triangle” formalism coined by Brookes et al. 2004; * see Pagliarini et al. 2005, Rardin et al. 2008; abbreviations: MCU, mitochondrial Ca2+-uniporter; RaM, Rapid Mode Ca2+ transporter; mRYR, mitochondrial Ryanodine Receptor; mNCE, mitochondrial Na+/Ca2+ exchanger)
Acknowledgments
This work was supported by NIH grant P01 HL081427.
Contributor Information
D. Brian Foster, Email: dbrianfoster@jhmi.edu, Division of Cardiology, Department of Medicine, The Johns Hopkins University School of Medicine, Ross Research Building, Room 847, 720 Rutland Avenue, Baltimore, MD 21205, USA.
Jennifer E. Van Eyk, Division of Cardiology, Department of Medicine, The Johns Hopkins University School of Medicine, Ross Research Building, Room 847, 720 Rutland Avenue, Baltimore, MD 21205, USA
Eduardo Marbán, The Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Brian O’Rourke, Division of Cardiology, Department of Medicine, The Johns Hopkins University School of Medicine, Ross Research Building, Room 847, 720 Rutland Avenue, Baltimore, MD 21205, USA.
References
- Alto NM, Soderling J, Scott JD. J Cell Biol. 2002;158:659–668. doi: 10.1083/jcb.200204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Biswas G, Mullick J, Sepuri NB, Otvos L, Pain D, Avadhani NG. Embo J. 1999;18:5494–5504. doi: 10.1093/emboj/18.20.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arachiche A, Augereau O, Decossas M, Pertuiset C, Gontier E, Letellier T, Dachary-Prigent J. J Biol Chem. 2008:M709217200. doi: 10.1074/jbc.M709217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marban E, Van Eyk JE. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- Augereau O, Claverol S, Boudes N, Basurko MJ, Bonneu M, Rossignol R, Mazat JP, Letellier T, Dachary-Prigent J. Cell Mol Life Sci. 2005;62:1478–1488. doi: 10.1007/s00018-005-5005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Zhang J, Wang G-W, Zheng Y-T, Xiu JX, Cardwell EM, Bolli R, Ping P. Circ Res. 2002;90:390–397. doi: 10.1161/01.res.0000012702.90501.8d. [DOI] [PubMed] [Google Scholar]
- Baines CP, Song C-X, Zheng Y-T, Wang G-W, Zhang J, Wang O-L, Guo Y, Bolli R, Cardwell EM, Ping P. Circ Res. 2003;92:873–880. doi: 10.1161/01.RES.0000069215.36389.8D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H, Kasten SA, Yan X, Hiromasa Y, Roche TE. Biochemistry. 2004;43:13442–13451. doi: 10.1021/bi0494875. [DOI] [PubMed] [Google Scholar]
- Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. J Biol Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- Bender E, Kadenbach B. FEBS Lett. 2000;466:130–134. doi: 10.1016/s0014-5793(99)01773-1. [DOI] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Hess DT, Stamler JS. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borutaite V, Budriunaite A, Brown GC. Biochim Biophys Acta Bioenerg. 2000;1459:405–412. doi: 10.1016/s0005-2728(00)00178-x. [DOI] [PubMed] [Google Scholar]
- Bowker-Kinley MM, Davis WI, Wu P, Harris RA, Popov KM. Biochem J. 1998;329:191–196. doi: 10.1042/bj3290191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes PS, Levonen A-L, Shiva S, Sarti P, Darley-Usmar VM. Free Radic Biol Med. 2002;33:755–764. doi: 10.1016/s0891-5849(02)00901-2. [DOI] [PubMed] [Google Scholar]
- Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu S-S. Am J Physiol Cell Physiol. 2004;287:C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- Brown GC, Borutaite V. Cardiovasc Res. 2007;75:283–290. doi: 10.1016/j.cardiores.2007.03.022. [DOI] [PubMed] [Google Scholar]
- Burnett G, Kennedy EP. J Biol Chem. 1954;211:969–980. [PubMed] [Google Scholar]
- Burwell LS, Brookes PS. Antioxid Redox Signal. 2008;10:579–600. doi: 10.1089/ars.2007.1845. [DOI] [PubMed] [Google Scholar]
- Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Biochem J. 2006;394:627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-R, Blackstone C. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Chen L, Hahn H, Wu G, Chen C-H, Liron T, Schechtman D, Cavallaro G, Banci L, Guo Y, Bolli R, Dorn GW, II, Mochly-Rosen D. Proc Natl Acad Sci. 2001;98:11114–11119. doi: 10.1073/pnas.191369098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Fearnley IM, Peak-Chew SY, Walker JE. J Biol Chem. 2004;279:26036–26045. doi: 10.1074/jbc.M402710200. [DOI] [PubMed] [Google Scholar]
- Chen Y-R, Chen C-L, Pfeiffer DR, Zweier JL. J Biol Chem. 2007;282:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- Churchill EN, Mochly-rosen D. Biochem Soc Trans. 2007;035:1040–1042. doi: 10.1042/BST0351040. [DOI] [PubMed] [Google Scholar]
- Clementi E, Brown GC, Feelisch M, Moncada S. Proc Natl Acad Sci. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa NJ, Dahm CC, Hurrell F, Taylor ER, Murphy MP. Antioxid Redox Signal. 2003;5:291–305. doi: 10.1089/152308603322110878. [DOI] [PubMed] [Google Scholar]
- Costa ADT, Pierre SV, Cohen MV, Downey JM, Garlid KD. Cardiovasc Res. 2008;77:344–352. doi: 10.1093/cvr/cvm050. [DOI] [PubMed] [Google Scholar]
- Cribbs JT, Strack S. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm CC, Moore K, Murphy MP. J Biol Chem. 2006;281:10056–10065. doi: 10.1074/jbc.M512203200. [DOI] [PubMed] [Google Scholar]
- De Rasmo D, Panelli D, Sardanelli AM, Papa S. Cell Signal. 2008;20:989–997. doi: 10.1016/j.cellsig.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Derakhshan B, Wille PC, Gross SS. Nat Protocols. 2007;2:1685–1691. doi: 10.1038/nprot.2007.210. [DOI] [PubMed] [Google Scholar]
- Downey J, Davis A, Cohen M. Heart Fail Rev. 2007;12:181–188. doi: 10.1007/s10741-007-9025-2. [DOI] [PubMed] [Google Scholar]
- Dyson MT, Jones JK, Kowalewski MP, Manna PR, Alonso M, Gottesman ME, Stocco DM. Biol Reprod. 2008;78:267–277. doi: 10.1095/biolreprod.107.064238. [DOI] [PubMed] [Google Scholar]
- Erusalimsky JD, Moncada S. Arterioscler Thromb Vasc Biol. 2007;27:2524–2531. doi: 10.1161/ATVBAHA.107.151167. [DOI] [PubMed] [Google Scholar]
- Fang JK, Prabu SK, Sepuri NB, Raza H, Anandatheerthavarada HK, Galati D, Spear J, Avadhani NG. FEBS Lett. 2007;581:1302–1310. doi: 10.1016/j.febslet.2007.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhu M, Schaub MC, Gehrig P, Roschitzki B, Lucchinetti E, Zaugg M. Cardiovasc Res. 2008;80:20–29. doi: 10.1093/cvr/cvn161. [DOI] [PubMed] [Google Scholar]
- Finkel T. Curr Opin Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Forman HJ, Fukuto JM, Torres M. Am J Physiol Cell Physiol. 2004;287:C246–C256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- Foster DB, O’Rourke B, Van Eyk JE. Expert Rev Proteomics. 2008;5:633–636. doi: 10.1586/14789450.5.5.633. [DOI] [PubMed] [Google Scholar]
- Galmiche A, Fueller J. Biochim Biophys Acta Mol Cell Res. 2007;1773:1256–1262. doi: 10.1016/j.bbamcr.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Greco TM, Hodara R, Parastatidis I, Heijnen HFG, Dennehy MK, Liebler DC, Ischiropoulos H. Proc Natl Acad Sci. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW, Meister A. Proc Natl Acad Sci. 1985;82:4668–4672. doi: 10.1073/pnas.82.14.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Harada H, Becknell B, Wilm M, Mann M, Huang LJ-s, Taylor SS, Scott JD, Korsmeyer SJ. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- Harris RA, Popov KM, Zhao Y, Kedishvili NY, Shimomura Y, Crabb DW. Adv Enzyme Regul. 1995;35:147–158. doi: 10.1016/0065-2571(94)00020-4. [DOI] [PubMed] [Google Scholar]
- Harris RA, Hawes JW, Popov KM, Zhao Y, Shimomura Y, Sato J, Jaskiewicz J, Hurley TD. Adv Enzyme Regul. 1997;37:271–293. doi: 10.1016/s0065-2571(96)00009-x. [DOI] [PubMed] [Google Scholar]
- Helling S, Vogt S, Rhiel A, Ramzan R, Wen L, Marcus K, Kadenbach B. Mol Cell Proteomics. 2008;7:1714–1724. doi: 10.1074/mcp.M800137-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Nudelman R, Stamler JS. Nat Cell Biol. 2001;3:E46–E49. doi: 10.1038/35055152. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim S-O, Marshall HE, Stamler JS. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Hool LC, Corry B. Antioxid Redox Signal. 2007;9:409–435. doi: 10.1089/ars.2006.1446. [DOI] [PubMed] [Google Scholar]
- Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen R-F, Witzmann FA, Harris RA, Balaban RS. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbinski C, Chu CT. Free Radic Biol Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Antioxid Redox Signal. 2005a;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP. Biochem Soc Trans. 2005b;33:1390–1393. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Prime TA, Harbour ME, Lilley KS, Murphy MP. J Biol Chem. 2007;282:22040–22051. doi: 10.1074/jbc.M703591200. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Requejo R, Filipovska A, Brown S, Prime TA, Robinson AJ, Fearnley IM, Murphy MP. J Biol Chem. 2008;283:24801–24815. doi: 10.1074/jbc.M803432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C, Sinskey AJ, Lodish HF. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- Janssen-Heininger YMW, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zhuo Y, Guo W, Field J. J Biol Chem. 2005;280:24698–24705. doi: 10.1074/jbc.M413374200. [DOI] [PubMed] [Google Scholar]
- Jocelyn PC. Biochim Biophys Acta Bioenerg. 1975;396:427–436. doi: 10.1016/0005-2728(75)90148-6. [DOI] [PubMed] [Google Scholar]
- Jocelyn PC, Kamminga A. Biochim Biophys Acta Gen Subj. 1974;343:356–362. doi: 10.1016/0304-4165(74)90099-3. [DOI] [PubMed] [Google Scholar]
- Jones DP. Antioxid Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- Jones SP, Bolli R. J Mol Cell Cardiol. 2006;40:16–23. doi: 10.1016/j.yjmcc.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Jung C-H, Thomas JA. Arch Biochem Biophys. 1996;335:61–72. doi: 10.1006/abbi.1996.0482. [DOI] [PubMed] [Google Scholar]
- Kettenhofen NJ, Broniowska KA, Keszler A, Zhang Y, Hogg N. J Chromatogr B. 2007;851:152–159. doi: 10.1016/j.jchromb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil IS, Park J-W. J Biol Chem. 2005;280:10846–10854. doi: 10.1074/jbc.M411306200. [DOI] [PubMed] [Google Scholar]
- Lee I, Bender E, Kadenbach B. Mol Cell Biochem. 2002;234–235:63–70. [PubMed] [Google Scholar]
- Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Huttemann M. J Biol Chem. 2005;280:6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- Lee I, Salomon AR, Yu K, Doan JW, Grossman LI, Huttemann M. Biochemistry. 2006;45:9121–9128. doi: 10.1021/bi060585v. [DOI] [PubMed] [Google Scholar]
- Lewandrowski U, Sickmann A, Cesaro L, Brunati AM, Toninello A, Salvi M. FEBS Lett. 2008;582:1104–1110. doi: 10.1016/j.febslet.2008.02.077. [DOI] [PubMed] [Google Scholar]
- Li H, Degenhardt B, Tobin D, Yao Z-x, Tasken K, Papadopoulos V. Mol Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Linn TC, Pettit FH, Reed LJ. Proc Natl Acad Sci U S A. 1969;62:234–241. doi: 10.1073/pnas.62.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li H, Papadopoulos V. J Steroid Biochem Mol Biol. 2003;85:275–283. doi: 10.1016/s0960-0760(03)00213-9. [DOI] [PubMed] [Google Scholar]
- Mackay K, Mochly-Rosen D. J Mol Cell Cardiol. 2001;33:1301–1307. doi: 10.1006/jmcc.2001.1400. [DOI] [PubMed] [Google Scholar]
- Matsuzawa A, Ichijo H. Biochim Biophys Acta. 2008;1780:1325–1336. doi: 10.1016/j.bbagen.2007.12.011. [DOI] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D, Gordon AS. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- Murphy E, Steenbergen C. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaka S, Katoh H, Niu CF, Matsui S, Urushida T, Satoh H, Watanabe Y, Hayashi H. Circ J. 2007;71:429–436. doi: 10.1253/circj.71.429. [DOI] [PubMed] [Google Scholar]
- Ogbi M, Johnson JA. Biochem J. 2006;393:191–199. doi: 10.1042/BJ20050757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Dixon JE. Trends Biochem Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE. Mol Cell. 2005;19:197–207. doi: 10.1016/j.molcel.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano G, Sardanelli AM, Signorile A, Papa S, Larsen MR. Proteomics. 2007;7:1575–1583. doi: 10.1002/pmic.200600801. [DOI] [PubMed] [Google Scholar]
- Papa S, De Rasmo D, Scacco S, Signorile A, Technikova-Dobrova Z, Palmisano G, Sardanelli AM, Papa F, Panelli D, Scaringi R, Santeramo A. Biochim Biophys Acta. 2008;1777:719–728. doi: 10.1016/j.bbabio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Liu J, Culty M. Mol Cell Endocrinol. 2007;265–266:59–64. doi: 10.1016/j.mce.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Patel MS, Korotchkina LG. Biochem Soc Trans. 2006;34:217–222. doi: 10.1042/BST20060217. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Piedimonte G, Silvotti L, Chamaret S, Borghetti AF, Montagnier L. J Cell Biochem. 1986;32:113–123. doi: 10.1002/jcb.240320204. [DOI] [PubMed] [Google Scholar]
- Poderoso JJ. Arch Biochem Biophys. 2009;484(2):214–220. doi: 10.1016/j.abb.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Poderoso C, Converso DP, Maloberti P, Duarte A, Neuman I, Galli S, Maciel FC, Paz C, Carreras MaC, Poderoso JJ, Podestá EJ. PLoS ONE. 2008;3:e1443. doi: 10.1371/journal.pone.0001443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AW, Pula G, Hers I, Crosby D, Jones ML. Trends Pharmacol Sci. 2004;25:528–535. doi: 10.1016/j.tips.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Popov KM, Kedishvili NY, Zhao Y, Shimomura Y, Crabb DW, Harris RA. J Biol Chem. 1993;268:26602–26606. [PubMed] [Google Scholar]
- Prabu SK, Anandatheerthavarada HK, Raza H, Srinivasan S, Spear JF, Avadhani NG. J Biol Chem. 2006;281:2061–2070. doi: 10.1074/jbc.M507741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przedborski S, Ischiropoulos H. Antioxid Redox Signal. 2005;7:685–693. doi: 10.1089/ars.2005.7.685. [DOI] [PubMed] [Google Scholar]
- Rardin MJ, Wiley SE, Murphy AN, Pagliarini DJ, Dixon JE. J Biol Chem. 2008;283:15440–15450. doi: 10.1074/jbc.M709547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, van der Laan M, Rehling P, Sickmann A, Pfanner N, Meisinger C. Mol Cell Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- Rhee SG. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Chae HZ, Kim K. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Robin MA, Anandatheerthavarada HK, Biswas G, Sepuri NB, Gordon DM, Pain D, Avadhani NG. J Biol Chem. 2002;277:40583–40593. doi: 10.1074/jbc.M203292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin M-A, Prabu SK, Raza H, Anandatheerthavarada HK, Avadhani NG. J Biol Chem. 2003;278:18960–18970. doi: 10.1074/jbc.M301807200. [DOI] [PubMed] [Google Scholar]
- Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, Dong J, Turkan A, Kasten SA. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- Salvi M, Brunati AM, Bordin L, La Rocca N, Clari G, Toninello A. Biochim Biophys Acta Mol Cell Res. 2002;1589:181–195. doi: 10.1016/s0167-4889(02)00174-x. [DOI] [PubMed] [Google Scholar]
- Salvi M, Stringaro A, Brunati AM, Agostinelli E, Arancia G, Clari G, Toninello A. Cell Mol Life Sci. 2004;61:2393–2404. doi: 10.1007/s00018-004-4211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavati L, Lee I, Mathes I, Lottspeich F, Huttemann M. J Biol Chem. 2008 doi: 10.1074/jbc.M801954200. M801954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacco S, Vergari R, Scarpulla RC, Technikova-Dobrova Z, Sardanelli A, Lambo R, Lorusso V, Papa S. J Biol Chem. 2000;275:17578–17582. doi: 10.1074/jbc.M001174200. [DOI] [PubMed] [Google Scholar]
- Schlossmann J, Feil R, Hofmann F. Ann Med. 2003;35:21–27. doi: 10.1080/07853890310004093. [DOI] [PubMed] [Google Scholar]
- Sengupta R, Ryter SW, Zuckerbraun BS, Tzeng E, Billiar TR, Stoyanovsky DA. Biochemistry. 2007;46:8472–8483. doi: 10.1021/bi700449x. [DOI] [PubMed] [Google Scholar]
- Sinha SS, Shiva S, Gladwin MT. Trends Cardiovasc Med. 2008;18:163–172. doi: 10.1016/j.tcm.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Smith FD, Langeberg LK, Scott JD. Trends Biochem Sci. 2006;31:316–323. doi: 10.1016/j.tibs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- Sun J, Steenbergen C, Murphy E. Antioxid Redox Signal. 2006;8:1693–1705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Morgan M, Shen R-F, Steenbergen C, Murphy E. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- Thomas DD, Liu X, Kantrow SP, Lancaster JR. Proc Natl Acad Sci USA. 2001;98:355–360. doi: 10.1073/pnas.011379598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrens JF. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S, Masutani H, Nakamura H, Tanaka T, Ueno M, Yodoi J. Antioxid Redox Signal. 2002;4:405–414. doi: 10.1089/15230860260196209. [DOI] [PubMed] [Google Scholar]
- Wahllander A, Soboll S, Sies H, Linke I, Muller M. FEBS Lett. 1979;97:138–140. doi: 10.1016/0014-5793(79)80069-1. [DOI] [PubMed] [Google Scholar]
- Wang H-G, Rapp UR, Reed JC. Cell. 1996a;87:629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- Wang H-G, Takayama S, Rapp UR, Reed JC. Proc Natl Acad Sci. 1996b;93:7063–7068. doi: 10.1073/pnas.93.14.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-L, Yeh S-F, Chang Y-I, Hsiao S-F, Lian W-N, Lin C-H, Huang C-YF, Lin W-J. J Biol Chem. 2003;278:37705–37712. doi: 10.1074/jbc.M304619200. [DOI] [PubMed] [Google Scholar]
- West MB, Hill BG, Xuan Y-T, Bhatnagar A. FASEB J. 2006;20:1715–1717. doi: 10.1096/fj.06-5843fje. [DOI] [PubMed] [Google Scholar]
- Wink DA, Miranda KM, Katori T, Mancardi D, Thomas DD, Ridnour L, Espey MG, Feelisch M, Colton CA, Fukuto JM, Pagliaro P, Kass DA, Paolocci N. Am J Physiol Heart Circ Physiol. 2003;285:H2264–H2276. doi: 10.1152/ajpheart.00531.2003. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wong W, Scott JD. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hogg N. Free Radic Biol Med. 2005;38:831–838. doi: 10.1016/j.freeradbiomed.2004.12.016. [DOI] [PubMed] [Google Scholar]