miR-375 Inhibits Differentiation of Neurites by Lowering HuD Levels (original) (raw)
Abstract
Neuronal development and plasticity are maintained by tightly regulated gene expression programs. Here, we report that the developmentally regulated microRNA miR-375 affects dendrite formation and maintenance. miR-375 overexpression in mouse hippocampus potently reduced dendrite density. We identified the predominantly neuronal RNA-binding protein HuD as a key effector of miR-375 influence on dendrite maintenance. Heterologous reporter analysis verified that miR-375 repressed HuD expression through a specific, evolutionarily conserved site on the HuD 3′ untranslated region. miR-375 overexpression lowered both HuD mRNA stability and translation and recapitulated the effects of HuD silencing, which reduced the levels of target proteins with key functions in neuronal signaling and cytoskeleton organization (N-cadherin, PSD-95, RhoA, NCAM1, and integrin α1). Moreover, the increase in neurite outgrowth after brain-derived neurotrophic factor (BDNF) treatment was diminished by miR-375 overexpression; this effect was rescued by reexpression of miR-375-refractory HuD. Our findings indicate that miR-375 modulates neuronal HuD expression and function, in turn affecting dendrite abundance.
Posttranscriptional processes implicating mRNA transport, stability, and translation critically affect mammalian gene expression patterns and cell fate. These events are governed by two main types of mRNA-interacting factors, microRNAs (miRNAs) and RNA-binding proteins (RBPs). MicroRNAs are small, noncoding RNAs that associate with the RNA-induced silencing complex (RISC) and bind target mRNAs with partial complementarity, typically causing gene repression by lowering mRNA translation, stability, or both processes (12). MicroRNAs are involved in numerous physiological and pathological processes, including development, cell proliferation, apoptosis, energy metabolism, immune response, and tumorigenesis (10, 29, 45).
During embryonic development, the temporal expression of microRNAs critically influences differentiation of cell types in an organism. While ablation of specific microRNAs often does not lead to the total loss of proper development, it can cause measurable abnormalities (40). The abundance of microRNAs, their tissue distribution, and the developmental stages in which they are expressed impact dynamically upon the expression of target gene products. The evolutionarily conserved microRNA miR-375 was found to be expressed in many tissues, including the gastrointestinal system, and played an essential role in pancreatic islet development (28). miR-375 was shown to regulate the expression levels of 3′-phosphoinositide-dependent protein kinase 1 (PDK1), a key molecule involved in phosphatidylinositol 3-kinase (PI 3-kinase) signaling in pancreatic β cells (20). It was essential for normal glucose homeostasis, for maintaining α- and β-cell populations, and for the expansion of β-cells in response to increased insulin demand (42).
Here, we show that miR-375 expression was specifically repressed during the late stages of neuronal development. This finding led us to discover that miR-375 impaired dendrite formation and maintenance. We identified the RBP HuD, a member of the embryonic-lethal abnormal vision (elav)/Hu protein family, as a major effector of miR-375 on neurite outgrowth and dendritic maintenance. Like other elav/Hu members, including the ubiquitous HuR and the preferentially neuronal HuB and HuC (26), HuD contains three RNA recognition motifs (RRMs) through which it binds to mRNAs bearing U-rich and C-rich elements in their 3′ untranslated regions (UTRs) (16). Among the HuD target mRNAs are those that encode GAP-43, p21Waf1, acetylcholinesterase (AchE), and numerous other recently identified targets (16, 18, 19, 37). Through its association with target mRNAs, which generally enhances their half-lives, HuD was found to modulate neuronal differentiation, identity, and function (4, 7, 8, 39, 48). HuD is highly expressed in neuroblastomas and is associated with Parkinson's and Alzheimer's diseases (5, 9, 36), suggesting that alterations in HuD levels may affect genes implicated in these pathologies. However, the mechanisms that regulate HuD expression are largely unknown.
We report that miR-375 potently suppresses HuD expression, both by destabilizing HuD mRNA and by repressing HuD translation. These effects required the interaction of miR-375 with the HuD 3′ UTR and implicated the RISC. HuD downregulation, in turn, lowered the HuD target genes implicated in neuronal development and function and suppressed neurite outgrowth upon brain-derived neurotrophic factor (BDNF) treatment. We propose that miR-375 impairs neuronal function by potently repressing HuD levels, and hence HuD function, in gene regulation and neuronal differentiation, regeneration, and plasticity.
MATERIALS AND METHODS
Cell culture, treatment, and transfection.
BE(2)-M17 cells were cultured in Opti-MEM supplemented with 10% fetal bovine serum (FBS). SH-SY5Y, Neuro2a, and PC12 cells were cultured in Dulbecco's modified essential medium (DMEM) (Invitrogen) supplemented with 10% FBS and antibiotics. Lipofectamine 2000 (Invitrogen) was used to transfect cells with small RNAs (the control [Ctrl] small interfering RNAs [siRNAs] were AATTCTCCGAACGTGTCACGT [Qiagen], HuD siRNA [Santa Cruz Biotechnology], and miR-375 [Ambion]) and with plasmid DNA [pGFP, pGFP-HuD, and pGFP-HuD(mut)]. Treatment with BDNF (10 ng/ml) lasted 3 days. A plasmid expressing N-terminal myc-tagged HuD (pmyc-HuD), derived from pcDNA3 (pVector), was a generous gift from N. I. Perrone-Bizzozero.
Western blot analysis and rhodamine-phalloidin staining.
Whole-cell lysates were prepared using RIPA buffer supplemented with protease inhibitors (Roche), resolved, and transferred as described previously (1). Incubations with primary mouse monoclonal antibodies recognizing HuD, α-tubulin, PSD-95, RhoA, KCNQ2 (Santa Cruz Biotechnology), or β-actin (Abcam) were followed by incubations with the appropriate secondary antibodies (Amersham) and by detection using enhanced luminescence (Amersham). Staining of filamentous actin (F-actin) was performed using rhodamine-phalloidin (Invitrogen) according to the manufacturer's protocol.
Translation assays.
For polyribosome fractionation assays, cells were incubated with cycloheximide (Calbiochem; 100 μg/ml; 15 min), and cytoplasmic lysates (500 μl) were fractionated by centrifugation through 10 to 50% linear sucrose gradients and divided into 10 fractions for quantitative reverse transcription-PCR (RT-qPCR) analysis to determine the distribution of HuD and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs, as described previously (2, 30).
RNA and miRNA analyses.
Total cellular RNA was prepared directly from cells or tissues using TRIzol (Invitrogen). After RT using random hexamers and SSII reverse transcriptase (Invitrogen), real-time qRT-PCR analysis was performed using gene-specific primer pairs (Table 1) and SYBR green PCR master mix (Applied Biosystems), as described previously (3). Mature miR-375 and U1 snRNA were quantified using a QuantiMir detection assay (System Biosciences) with miR-375- and U1 snRNA-specific forward primers (Table 1).
TABLE 1.
Oligomers used for qPCR analysis
Immunohistochemistry.
Human tissue slides (Array II; BioChain Institute, Inc., Hayward, CA, and Pantomics, Inc.) were subjected to heat-induced epitope retrieval, incubation with primary antibody, and detection with the LSAB+ system (Dako, Carpinteria, CA). A monoclonal anti-HuD antibody (Santa Cruz) was used at 0.2 μg/ml. Stained tissue sections were imaged at ×200 total magnification using a ScanScope CS system (Aperio, Vista, CA). Images were analyzed as described previously (34).
Gene arrays, validation, and Ingenuity analysis.
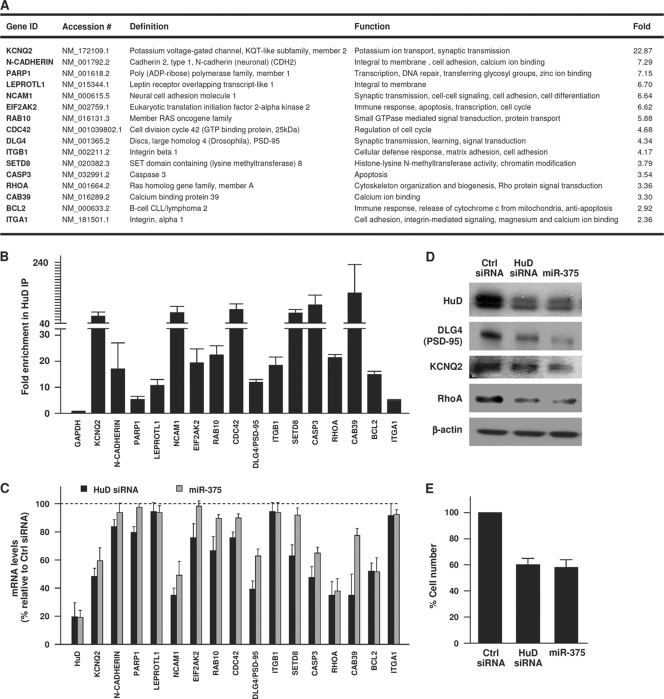
HuD-associated transcripts were isolated from BE(2)-M17 cells as described previously (50). The isolated RNA was subjected to Illumina array analysis, and the HuD target genes involved in neuronal development and functions were identified using Ingenuity software (see Fig. 6). The putative HuD-interacting RNAs identified were validated by RT-qPCR analysis with gene-specific primers, as indicated above.
FIG. 6.
miR-375 recapitulates the effect of HuD silencing upon target mRNAs. (A) HuD target mRNAs were identified by RNP IP analysis of BE(2)-M17 cells using anti-HuD antibody (relative to IgG control IP); after isolation of RNA in the IP material, the mRNAs preferentially associated with HuD were identified by Illumina microarray analysis and studied using Ingenuity software (enrichments are shown in the Fold column). The top target transcripts encoding proteins with roles in neuronal signaling, division, cytoskeletal organization, and survival are listed. (B) The interaction of HuD with putative target transcripts in panel A was validated by RNP IP analysis of HuD, followed by RT-qPCR amplification using gene-specific primer pairs. Transcript abundance was normalized to 18S rRNA signals and plotted as fold enrichment of the mRNAs relative to their levels in IgG IP. (C) Forty-eight hours after silencing of HuD or overexpression of miR-375, the levels of HuD-associated mRNAs were measured by RT-qPCR analysis, normalized to 18S rRNA, and plotted as a percentage of the mRNA levels measured in the Ctrl siRNA transfection group. (D) Forty-eight hours after transfection with the small RNAs shown, the levels of HuD, PSD-95, KCNQ2, RhoA, and loading control β-actin were assessed by Western blot analysis. (E) Numbers of BE(2)-M17 cells by 48 h after transfection with the small RNAs shown. The data in panels B, C, and E represent the means and SEM from three independent experiments.
RNA stability assays.
Transfected cells were treated with 2 μg/ml actinomycin D, and the levels of HuD mRNA in each transfection group were measured by RT-qPCR from total RNA using HuD-specific primer pairs (see above). GAPDH mRNA was measured as a control housekeeping transcript. The levels of HuD and GAPDH mRNAs were normalized to 18S rRNA levels and plotted as the percentage of mRNA remaining compared with the levels of the same mRNA at time zero.
Animal injections and brain tissue analysis.
C57BL/6 mice (Jackson Laboratories) were anesthetized (1.2% solution; 500 μl intraperitoneal [i.p.] avertin), and 1.5 μl of either Lenti-GFP or Lenti-GFP-miR-375 was injected into the dentate gyrus. Each animal was injected 4 times, with 2 injections per hippocampus (site 1: spatial coordinates from Bregma, anteroposterior, −2 mm; lateral, 1.5 mm; ventral, 1.5 mm; site 2: spatial coordinates from Bregma, anteroposterior, −3 mm; lateral, 2 mm; ventral, 2 mm). Four weeks after surgery, the mice were perfused with saline and paraformaldehyde. After cryoprotection in 30% sucrose, brain sections (40 m thick) were used for immunohistochemistry. All animal work was conducted according to relevant national and international guidelines.
RESULTS
Ectopic overexpression of miR-375 reduces dendrite density.
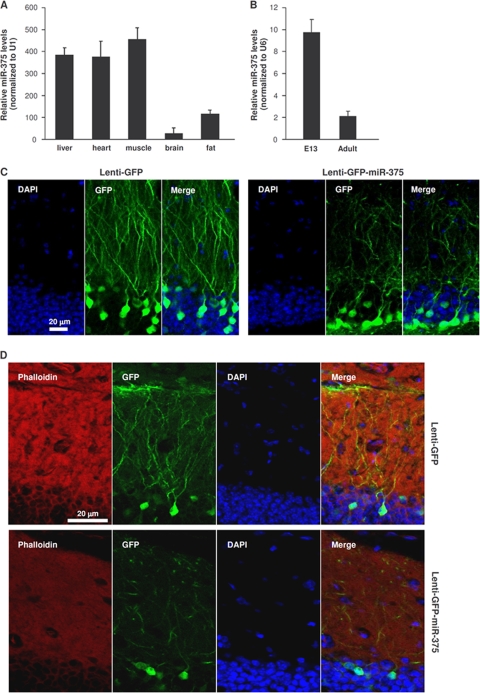
Analysis of miR-375 abundance in several adult mouse tissues by RT-qPCR revealed high expression of the microRNA in spleen, heart, and skeletal muscle; relatively lower expression in fat tissue; and the lowest levels of miR-375 in the brain (Fig. 1 A), in addition to the reported strong epithelial presence (31). Measurement of miR-375 levels in mouse embryos (embryonic day 13 [E13]) revealed high miR-375 abundance in the telencephalon and dramatically lower expression in adult cortex (Fig. 1B). To investigate the significance of the robust decline in miR-375 levels during development, we used a green fluorescent protein (GFP)-expressing lentiviral vector that also expressed miR-375 (Lenti-GFP-miR-375). Injection of the control vector Lenti-GFP into the mouse hippocampus revealed many green neurons (infected cells) with a high density of dendrites (Fig. 1C). In contrast, injection of Lenti-GFP-miR-375 led to infection of a comparable number of neurons, but the abundance of dendrites was greatly reduced (Fig. 1D), as was the extent of the cytoskeleton, as evidenced by staining with rhodamine-phalloidin, which interacts with F-actin.
FIG. 1.
Endogenous and ectopic expression of miR-375. (A) The levels of miR-375 in the mouse tissues shown were quantified by RT-qPCR analysis and normalized to the levels of U1 (also measured by RT-qPCR). (B) miR-375 abundance in embryonic telencephalon and in adult cortex were measured by RT-qPCR analysis and normalized to the levels of U6 (also measured by RT-qPCR). The error bars indicate standard deviations. (C) Lenti-GFP (control) or Lenti-GFP-miR-375 was injected into mouse hippocampus (dentate gyrus); 4 weeks later, tissue sections were prepared for analysis by confocal microscopy. DAPI (4′,6-diamidino-2-phenylindole), staining of nuclei; GFP, infected cells in a given plane of microscopy. (D) Rhodamine-phalloidin was used to stain the actin cytoskeleton in cells that were infected and processed as explained in the legend to panel C.
miR-375 is predicted to target HuD mRNA.
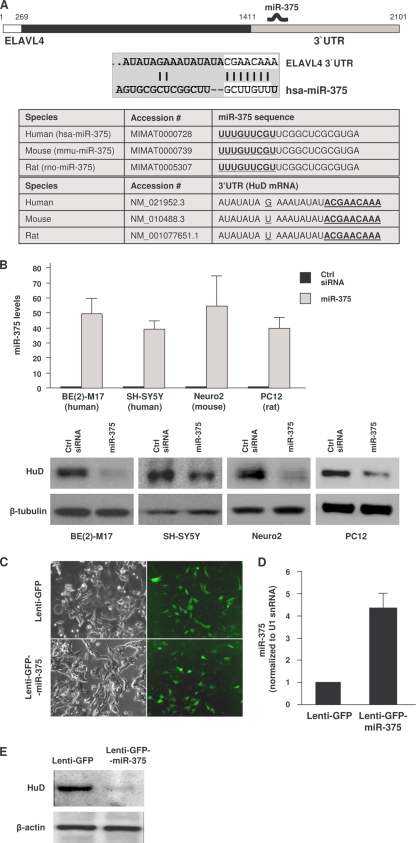
Using several microRNA analysis programs (e.g., TargetScan and miRBase), the highly conserved miR-375 was found to target numerous mammalian mRNAs, as determined through approaches previously reported for other miRNAs (2). One of these targets, encoding the primarily neuronal RNA-binding protein HuD, was chosen for further study (Fig. 2A). Despite the divergence between the human and rodent HuD 3′ UTRs (∼25% conservation between human and rat or mouse), the miR-375 site was extremely well conserved, particularly in the “seed” region (100% conserved), which is critical for microRNA function. By 48 h after transfection of a mimic of miR-375 RNA to overexpress miR-375 in human neuroblastoma BE(2)-M17 and SH-SY5Y cells, mouse Neuro2 cells, and rat PC12 cells (as monitored by RT-qPCR analysis), HuD abundance was markedly reduced (Fig. 2B), suggesting that HuD was a bona fide target of miR-375. Overexpression of microRNAs that were also predicted to target HuD (miR-21 or miR141) did not affect HuD levels (not shown), indicating that this effect was specific for miR-375. At the target site on the HuD mRNA, the single-nucleotide substitution in mouse and rat, located 8 bases away from the seed region (G in human, U in mouse and rat), did not affect the repressive influence of miR-375 on HuD expression in mouse Neuro2a or rat PC12 cells (Fig. 2B). Similarly, injection of BE(2)-M17 cells with Lenti-GFP-miR-375 reduced the abundance of HuD (Fig. 2C to E). Together, these data indicate that miR-375 suppresses HuD protein expression in different species.
FIG. 2.
miR-375 overexpression lowers HuD abundance. (A) (Top) Schematic of HuD mRNA depicting the 5′ UTR (white), coding region (black), and 3′ UTR (gray). The interaction between the human (hsa)-miR-375 and the 3′ UTR of HuD mRNA (ELAVL4) was predicted by TargetScan, with the seed region nucleotides marked by white rectangles. (Bottom) Conservation among several species of the miR-375 sequence and the interaction site on the HuD 3′ UTR. (B) Forty-eight hours after transfection of the indicated cell lines (human and rodent) with either control (Ctrl) siRNA or miR-375 mimic, the levels of mature miR-375 were measured using RT-qPCR and normalized to U1 snRNA. The levels of HuD and loading control β-tubulin were assessed by Western blot analysis. The error bars indicate standard deviations. (C to E) BE(2)-M17 cells were infected with Lenti-GFP or Lenti-GFP-miR-375; 5 days later, green fluorescence was examined (C), the levels of miR-375 were assessed by RT-qPCR analysis (D), and the abundance of HuD protein was assessed by Western blot analysis (E).
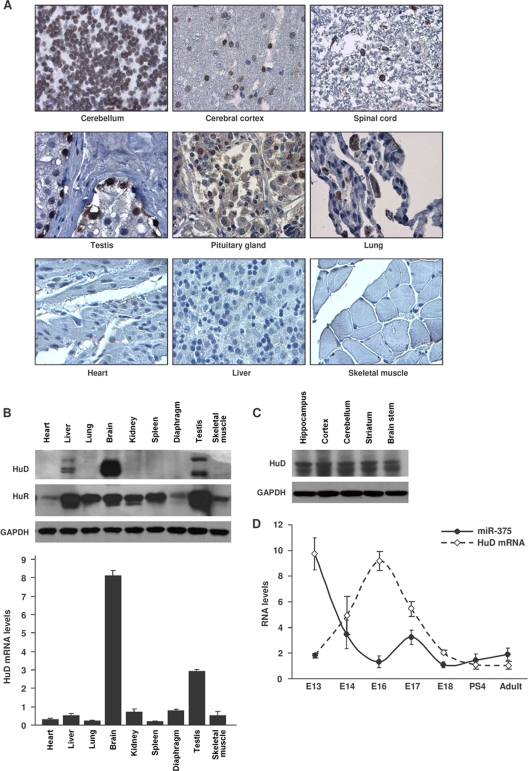
Tissue microarrays revealed that HuD was expressed in neuronal tissues, in agreement with previous reports examining its tissue distribution (23), but it was also detected in several other tissues, such as testis, pituitary gland, and lung; it was not detected in numerous other tissues, such as heart, liver, or skeletal muscle (Fig. 3A). These findings were confirmed by Western blot analysis of HuD and by RT-qPCR detection of HuD mRNA in mouse tissues, although moderate HuD signals were seen in mouse liver (Fig. 3B); in contrast, the ubiquitous RBP HuR was detected in all of the tissues (Fig. 3B) (34). HuD was detected throughout the mouse brain (Fig. 3C); it migrates with sizes ranging between 37 and 43 kDa, as a result of alternative splicing. Not unexpectedly, miR-375 was highly expressed in tissues such as liver, heart, and muscle (Fig. 1A) but was lower in the brain, while the opposite was true for HuD mRNA and protein (Fig. 3B). Moreover, HuD expression was low in early stages of the developing rat brain, increased markedly by E16, declined by birth, and remained low in the adult (25); miR-375 displayed an opposite expression pattern during embryogenesis, showing strong expression in early embryonic telencephalon and declining by E16 (Fig. 3D). These findings suggested that HuD levels correlated inversely with miR-375 abundance during embryonic neuronal development.
FIG. 3.
HuD expression in mouse and human tissues. (A) Immunohistochemical analysis of HuD expression in a human tissue array. (B) (Top) The levels of HuD, HuR, and loading control GAPDH in the mouse tissues indicated were assessed by Western blot analysis. (Bottom) The levels of HuD mRNA in the same tissues were assessed by RT-qPCR analysis and normalized to 18S rRNA. The error bars indicate standard deviations. (C) Levels of HuD in different mouse brain regions, as assessed by Western blot analysis. (D) miR-375 and HuD mRNA abundance in mouse telencephalon and cortex. RNA in the telencephalons of mice at the indicated developmental stages (cortex in adults) was used for quantification by RT-qPCR. miR-375 levels were normalized to U6, and HuD mRNA levels were normalized to 18S rRNA. The data are the means ± standard deviations from 3 mice.
HuD mRNA is a direct target of miR-375 via RISC.
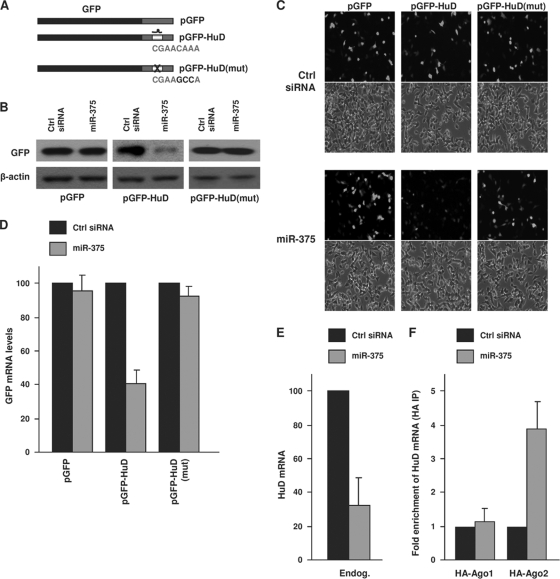
Next, we investigated if miR-375 directly regulates HuD expression by interacting with the predicted site within the HuD 3′ UTR. We generated constructs that expressed the heterologous reporter protein GFP alone (pGFP) or expressed GFP encoded by chimeric mRNAs bearing the intact miR-375 target site [pGFP-HuD] or the miR-375 site with 3 mutated seed region nucleotides, to disrupt the interaction with miR-375 [pGFP-HuD(mut)] (Fig. 4A). By 48 h after transfection of BE(2)-M17 cells with pGFP, GFP signals were strong and were unaffected by miR-375 overexpression (Fig. 4B). While cells transfected with pGFP-HuD also expressed high levels of GFP, miR-375 overexpression strongly reduced GFP abundance (Fig. 4B). Overexpression of miR-375 lowered the levels of the GFP reporter specifically through the HuD miR-375 site, since GFP expressed in the pGFP-HuD(mut) transfection group was refractory to miR-375 overexpression (Fig. 4B). The pattern of GFP abundance was reflected in the GFP fluorescence of the transfected cultures, as GFP-positive cells were detected in all plasmid groups under control conditions (Fig. 4C, Ctrl siRNA), were selectively reduced after miR-375 overexpression in the pGFP-HuD group, and remained elevated in the other two plasmid transfection groups. These results support the view that miR-375 selectively represses GFP expression through the HuD miR-375 site.
FIG. 4.
miR-375 targets the HuD 3′ UTR. (A) Schematic of plasmids pGFP, pGFP-HuD, and pGFP-HuD(mut), the last bearing 3 mutant nucleotides in the HuD site corresponding to the miR-375 seed region. (B and C) BE(2)-M17 cells were cotransfected with the plasmids shown in panel A and with either Ctrl siRNA or miR-375, as indicated; 48 h after transfection, GFP expression levels were assessed by Western blot analysis (B) and by green fluorescence (C). The data are representative of three independent experiments. (D) The levels of GFP, GFP-HuD, and GFP-HuD(mut) mRNAs in cells transfected as described in the legend to panel B were assessed by RT-qPCR analysis and normalized to 18S rRNA abundance. (E) By 48 h after transfection of Ctrl siRNA or miR-375, the levels of endogenous HuD mRNA were assessed by RT-qPCR analysis and normalized to 18S rRNA levels. (F) Forty-eight hours after transfection of Ctrl siRNA or miR-375, together with plasmids expressing tagged Argonaute proteins (pHA-Ago1 or pHA-Ago2), the levels of HuD mRNA associated with Argonaute proteins were assessed by IP using HA-coated beads, followed by HuD mRNA (and control UBC mRNA) detection using RT-qPCR. The data in panels D, E, and F are the means and standard errors of the mean (SEM) from three independent experiments.
We sought to discover if miR-375 acted primarily by lowering HuD mRNA levels or HuD translation. As shown in Fig. 4D, miR-375 overexpression in BE(2)-M17 cells potently reduced the levels of GFP-HuD mRNA, while GFP and GFP-HuD(mut) mRNAs were refractory to this intervention (Fig. 4D). HuD mRNA levels were also reduced by miR-375 overexpression (Fig. 4E). To study whether miR-375 elicited these actions by recruiting the HuD mRNA to the RISC, miR-375 was overexpressed in BE(2)-M17 cells that had been transfected with vectors to express Argonaute (Ago) proteins, critical components of the RISC, as hemagglutinin (HA)-tagged Ago1 or Ago2. Following immunoprecipitation (IP) of ribonucleoprotein (RNP) complexes using HA-coated beads, RNA was isolated from the IP materials and used to measure the enrichment of HuD mRNA in HA-Ago IP samples. As shown in Fig. 4F, HuD mRNA was highly enriched in the HA-Ago2 IP samples after miR-375 overexpression, in keeping with the reported role of Ago2 in mRNA degradation (35). To determine if miR-375 generally induces mRNA decay of predicted target mRNAs, we tested its effects upon other putative targets; miR-375 overexpression lowered HuD and RASD1 mRNAs, but not HNF1B or PTPN4 mRNAs (not shown), suggesting that miR-375 promotes the destabilization of only a subset of target mRNAs. Together, these findings indicate that miR-375 promotes the recruitment of HuD mRNA to Ago2/RISC and suggest that Ago2 mediates the repression of HuD by miR-375.
miR-375 suppresses HuD mRNA stability and translation.
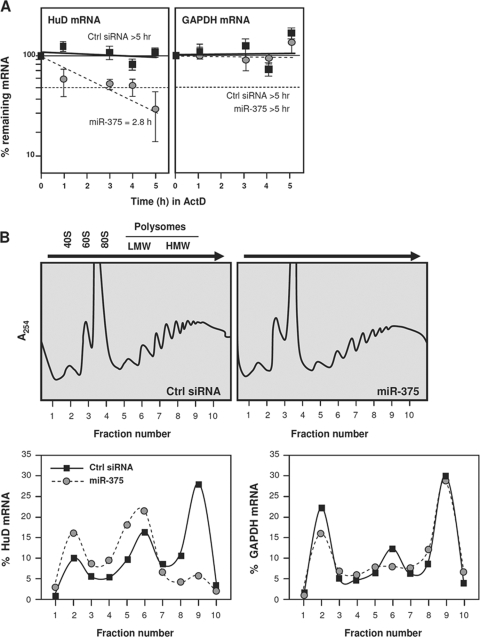
We further investigated if miR-375 reduces HuD mRNA stability, suppresses translation, or elicits both effects. Twenty-four hours after transfection of BE(2)-M17 cells with control (Ctrl) siRNA or the miR-375 mimic, the cells were treated with actinomycin D, and the half-life of HuD mRNA was measured. HuD mRNA was very stable in untreated cells (Ctrl siRNA group), but its half-life was potently reduced (∼4 h) after miR-375 overexpression; in contrast, the half-life of the housekeeping GAPDH mRNA was not influenced by miR-375 overexpression (Fig. 5A). As miRNAs can also suppress mRNA translation, we assayed HuD translation by using two different methodologies. First, we quantified the fraction of HuD mRNA associated with the translational machinery in each transfection group. Cytoplasmic extracts from the Ctrl siRNA and ectopic miR-375 overexpression groups were fractionated on sucrose gradients, and the relative abundance of HuD mRNA in each fraction was used to measure the association of HuD mRNA with the cellular polysomes. As shown, in Ctrl siRNA cells, HuD mRNA levels were very low in nontranslating and low-translating fractions of the gradient (fractions 1 to 5, where free RNA and 40S and 60S subunits, as well as 80S monosomes, are found), and HuD mRNA was found in the actively translating fractions of the gradient (fractions 6 to 10, spanning low- and high-molecular-weight polysomes) (Fig. 5B). In miR-375-transfected cells, the HuD mRNA showed a leftward shift on the gradient, indicating that HuD mRNA associated with smaller polysomes or with no polysomal components and further suggesting that miR-375 suppressed the initiation of HuD translation. The repression of HuD translation was further assessed by monitoring the rate of HuD biosynthesis, as measured by a brief incubation of BE(2)-M17 cells with l-[35S]methionine and l-[35S]cysteine. The incorporation of radiolabeled amino acids into immunoprecipitated HuD confirmed that de novo HuD translation was lower in miR-375-transfected cells (data not shown). Together, these results indicate that miR-375 represses HuD expression both by lowering HuD mRNA stability and by repressing HuD translation.
FIG. 5.
miR-375 represses HuD mRNA stability and translation. (A) Twenty-four hours after transfection with either Ctrl siRNA or miR-375, BE(2)-M17 cells were treated with actinomycin D for the times indicated. RNA was then isolated; subjected to RT-qPCR analysis to measure the levels of HuD mRNA, GAPDH mRNA (encoding a housekeeping protein), and 18S rRNA (for normalization); and plotted on a semilogarithmic scale. mRNA half-lives were calculated as the times needed for each mRNA to reach one-half of its abundance at time zero. The error bars indicate standard deviations. (B) Lysates prepared from cells that were transfected as described in the legend to panel A were fractionated through sucrose gradients (top), and the relative distributions (percent) of HuD mRNA (left) and housekeeping GAPDH mRNA (right) were studied by RT-qPCR analysis of RNA in each of 10 gradient fractions. The arrows indicate the direction of sedimentation; fraction 1 had no ribosomal components. 40S and 60S, small and large ribosome subunits, respectively; 80S, monosome; LMW and HMW, low- and high-molecular-weight polysomes, respectively. The data are representative of 3 independent experiments.
HuD is an effector of miR-375 function.
Given that HuD is potently repressed by miR-375, we hypothesized that inhibiting HuD might help to implement the phenotype of miR-375 overexpression (Fig. 1). To study this possibility, we first examined the targets of HuD in BE(2)-M17 cells. RNP IP followed by microarray analysis of associated mRNAs revealed numerous HuD target transcripts. Figure 6 A lists the major HuD-associated mRNAs encoding proteins with functions in neuronal physiology, including those that participate in neuronal excitability (the K+ channel subunit KCNQ2), neurite outgrowth (N-cadherin, NCAM1, and β1-integrin), signaling (Rab10), cytoskeletal organization (RhoA), and survival (Bcl-2), as determined by Ingenuity analysis. These interactions were individually validated by RNP IP analysis of HuD, followed by single-mRNA detection of the target transcripts identified on the arrays (Fig. 6B). HuD appeared to have a broad stabilizing influence upon these mRNAs, as silencing HuD using siRNA lowered the abundance of most target mRNAs (Fig. 6C and D). It was previously reported that HuD stabilized GAP-43 mRNA, a target transcript (46), although GAP-43 mRNA was not identified in our analysis. Importantly, the influence of HuD silencing on target mRNA levels closely mimicked the effect of miR-375-mediated reduction upon the same target mRNAs (e.g., KCNQ2, NCAM, PSD-95, Casp3, RHOA, and Bcl-2 mRNAs), as well as the levels of a small set of encoded proteins (Fig. 6C and D). Lowering HuD or increasing miR-375 did not affect the steady-state levels of other HuD target mRNAs (e.g., N-cadherin, PARP1, LEPROTL1, ITGB1, and ITGA1 mRNAs); the specific influence of HuD upon these targets awaits further analysis. The altered gene expression patterns elicited by silencing HuD or overexpressing miR-375 were accompanied by profound physiological changes. BE(2)-M17 cells proliferated more slowly, as assessed by direct cell counting 48 h after the transfections, when there was no appreciable cell toxicity (Fig. 6E). Taken together, these findings strongly support the notion that HuD is a major effector of changes in the expression of genes with pivotal roles in neuronal morphology and function.
Impacts of miR-375 and HuD on neuronal function and dendrite morphology.
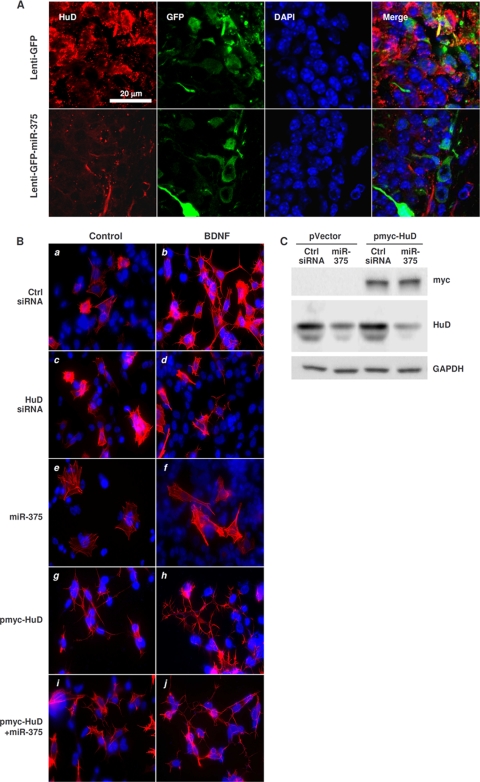
By 4 weeks after injection of Lenti-GFP-miR-375 into adult mouse hippocampus, there was a marked decrease in the density of dendrites in infected dentate granule neurons (Fig. 1B), accompanied by reduction in the levels of HuD (Fig. 7A) and RhoA (not shown) in the infected neurons. These findings are significant because the RhoA mRNA is a prominent HuD target mRNA (Fig. 6A and B), since its levels declined after HuD silencing or miR-375 overexpression in BE(2)-M17 cells (Fig. 6C), and because RhoA plays a pivotal role in cytoskeletal organization and signal transduction.
FIG. 7.
Consequences of overexpressing miR-375 and modulating HuD levels in vivo and in BDNF-treated cells. (A) Four weeks after infection of mouse hippocampus with Lenti-GFP or Lenti-GFP-miR-375, tissue sections (40 μm thick) were prepared. GFP expression was visualized by fluorescence and HuD abundance by immunofluorescence. (B) Forty-eight hours after transfection with the small RNAs and plasmids shown, BE(2)-M17 cells were treated with BDNF (10 ng/ml); 72 h later, the cytoskeleton was visualized using rhodamine-phalloidin (red) and the nuclei using DAPI (blue). (C) Western blot analysis of the expression of HuD, myc-tagged HuD (myc), and loading control GAPDH 48 h after transfection of BE(2)-M17 cells with pVector (pcDNA3) or pmyc-HuD, together with either Ctrl siRNA or miR-375.
Finally, to study whether miR-375-regulated HuD was implicated in de novo neurite outgrowth, BE(2)-M17 cells that had been transfected for 48 h with Ctrl siRNA or miR-375 were cultured for 3 additional days in the presence of brain-derived neurotrophic factor (BDNF). BDNF treatment promoted the sprouting of new neurites (Fig. 7B), as detected by staining using rhodamine-phalloidin, which binds F-actin specifically. Silencing HuD (Fig. 7B, c and d) or overexpressing miR-375 (Fig. 7B, e and f) each diminished the appearance of neurites after BDNF treatment. Overexpression of HuD using a plasmid vector that contains only the HuD coding region with an N-terminal c-myc tag (a gift from N. I. Perrone-Bizzozero) (Fig. 7C) induced neurite outgrowth, particularly after BDNF treatment (Fig. 7B, g and h). Importantly, as HuD was overexpressed from an ectopic mRNA that lacked 3′ UTR sequences, miR-375 coexpression in these cells did not abolish the outgrowth of neurites (Fig. 7B, i and j). These results showed that a miR-375-refractory HuD could rescue the repression of neurite outgrowth by miR-375 and that the inhibition of de novo neurite formation (Fig. 7B, e and f) required the HuD 3′ UTR. Taken together, our findings indicate that HuD promotes neurite outgrowth in the presence of BDNF and that miR-375 inhibits this effect.
DISCUSSION
Posttranscriptional regulation of gene expression is increasingly recognized as an important general mechanism by which the function and morphological plasticity of dendrites, axons, and synapses are controlled locally in response to various environmental signals (44, 49). Our findings reveal pivotal roles for miR-375 and HuD, two different molecules that bind mRNAs encoding neuron-specific proteins, in the regulation of neurite outgrowth and differentiation in cultured neuronal cells and hippocampal granule neurons in vivo. There is an inverse relationship between the levels of expression of miR-375 and HuD during mouse brain development: the levels of miR-375 are highest in early embryos and decrease by E16, whereas HuD levels are low in early embryos and increase markedly by E16, when neurogenesis peaks (Fig. 1B). We showed that by binding to the HuD 3′ UTR, miR-375 impedes the translation of HuD, thereby suppressing the expression of multiple HuD target mRNAs that encode neuron-specific proteins. By acting in an antagonistic manner to regulate the stability and translation of proteins involved in neurite growth and in the stability of dendrites and synapses, miR-375 and HuD may control the formation, maintenance, and plasticity of neuronal circuits in the mammalian brain. Consistent with such roles for miR-375 and HuD, we found that overexpression of miR-375 or silencing of HuD abolished the ability of BDNF to stimulate neurite growth. In mature neurons, BDNF is critical for maintaining neuronal plasticity and neuronal outgrowth, but BDNF also has many developmental roles, including neuronal differentiation (17, 24, 27, 32, 33). We found that miR-375 disrupts BDNF-mediated neurite outgrowth in neuroblastoma cells (Fig. 7B), indicating that miR-375 and HuD can interact at the posttranscriptional level with BDNF signaling pathways that control neuronal morphology and function. Therefore, we propose that miR-375 inhibits premature neuronal differentiation and might help to maintain a proliferative phenotype of embryonic neural stem cells and progenitors. Ongoing efforts are aimed at examining the role of miR-375 in the developmental regulation of neuronal differentiation.
Our findings suggest that in differentiated neurons, low miR-375 abundance is critical for maintaining elevated HuD levels. In turn, HuD helps to keep a constitutive density of neurites in mouse hippocampus and to elicit de novo neurite outgrowth after BDNF treatment of cultured cells. miR-375 interacts with a specific HuD 3′ UTR site that was strikingly well conserved among rodent and human genes, despite the generally poor conservation of the 3′ UTRs. miR-375 lowered both HuD mRNA stability and translation; the resulting diminution of HuD levels reduced the expression of critical proteins implicated in neuronal signaling, survival, and cytoskeletal organization. The inhibition of neurite outgrowth by miR-375 was rescued by HuD overexpression from an HuD mRNA that lacks the miR-375 site.
Recent en masse analysis of HuD-associated mRNAs in mouse brain revealed that many HuD targets encoded proteins with vital roles in neuronal differentiation, cytoskeletal transport, and RNA metabolism (16), including many of the targets we identified here using human BE(2)-M17 cells. These findings fully support HuD's ability to promote neuronal development, synaptic plasticity, and nerve regeneration (reviewed in references 37 and 41). Accordingly, ectopic interventions to overexpress or downregulate HuD in cultured neuronal models revealed a role for HuD in the expression of target mRNAs and in controlling neuronal morphology (19). HuD-null mice show deficient neurogenesis, nerve development, and motor and sensory functions (4), while HuD-overexpressing mice displayed alteration in hippocampus physiology associated with impaired learning and memory (14).
With neuronal abnormalities arising from both elevations and reductions in HuD abundance, it is surprising that the mechanisms responsible for regulating HuD expression and function were largely unknown until now. In this regard, the discovery that miR-375 represses HuD expression provides a means to express high HuD levels in tissues with low miR-375 (e.g., brain) and low HuD levels in tissues that express miR-375 in abundance (e.g., heart and muscle). It should be noted that while HuD was previously believed to be strictly neuronal, it was detectable in other tissues, like testis, pituitary gland, and lung (Fig. 3A and B), suggesting that HuD could have functions in cells other than neurons. As individual- and species-specific variations appear to exist, a deeper analysis of HuD tissue distribution is warranted. Regarding expression and function, HuD was shown to be methylated by the coactivator-associated arginine methyltransferase 1 (CARM1) on Arg 236, a modification that reduces its ability to interact with a target transcript, the p21Waf1 mRNA (21). HuD was also found to bind the HuD mRNA (16), suggesting a possible self-regulatory loop akin to those shown for other RBPs, like TTP, AUF1, and HuR (11, 43, 47, 51). In models of nerve regeneration and contextual learning, HuD abundance was found to increase, but the mechanisms responsible were not elucidated (6, 13). In the rat, HuD protein levels were highest at ∼E16, declined perinatally, and remained low in the adult rat brain (25), precisely the opposite of the expression pattern that was observed for miR-375 in the developing mouse telencephalon (Fig. 3D). Thus, the finding reported here that miR-375 regulates HuD production provides a mechanism to explain the magnitude and tissue-specific abundance of HuD.
Analyzed as a group, neuronal elav/Hu proteins were shown to be phosphorylated by protein kinase C (PKC); this modification changes their distribution and interaction with GAP-43 mRNA, but the specific changes in HuD were not studied separately from the other neuronal elav/Hu proteins (38). Interestingly, exposure of cultured SH-SY5Y to Aβ(1-42), the product of APP (amyloid precursor protein) cleavage, strongly lowered the interaction of neuronal elav/Hu proteins with ADAM10 (a _d_isintegrin _a_nd _m_etalloproteinase 10), an integral membrane protein that acts as an α-secretase and thereby promotes the nonamyloidogenic processing of APP (5). Whether HuD specifically mediated this process and which precise mechanisms (transcriptional and/or posttranscriptional) control ADAM10 expression also remain to be investigated.
In mammalian cells, posttranscriptional gene regulation by mRNA-interacting factors is recognized as a major mechanism for determining the amount of protein expressed. Besides governing the amount and timing of the proteins expressed, mRNA-binding factors can effectively govern the location where the protein is synthesized. This feature is particularly relevant to the neuronal system, given the special morphology of neurons, where mRNAs are synthesized in the nucleus but the gene products are often required at the distal ends of dendrites and axons, far from the nucleus. The elucidation of the mechanisms that govern the transport and local translation of mRNAs in neuronal cells has become an area of intense research. Several RBPs have been implicated in neuronal mRNA transport, stability, and localized translation, including elav/Hu, AUF1, cytoplasmic polyadenylation element binding (CPEB), Musashi, zipcode-binding protein (ZBP1 and -2), K homology-type splicing regulatory protein (KSRP), NOVA-1 and -2, fragile X mental retardation protein (FMRP), and Staufen (Stau1 and -2) (reviewed by Bolognani and Perrone-Bizzozero [15]). A growing number of microRNAs have also been implicated in neuronal differentiation, integrity, and plasticity (miR-124a and miR-133b); in the formation of synapses in postmitotic neurons (miR-134 and miR-9a); and in circadian rhythms (miR-132 and miR-219) (reviewed by Gao [22]). In this context, maintenance of low miR-375 levels allows the expression of high levels of HuD, which can then coordinate the production of proteins that establish, restore, and maintain neuronal function. Since restoring HuD expression did not totally rescue the impairment in neuritogenesis (Fig. 7B), miR-375 likely controls the expression of important neuronal proteins in addition to HuD. Further study of miR-375-regulated targets will be important in order to fully understand how miR-375 coordinates dendrite formation and maintenance.
In conclusion, the finding that a single microRNA regulates expression of an RBP with a pivotal function in neurons represents an effective mechanism of “amplification” of a gene expression program and highlights the richness and complexity of posttranscriptional gene regulation in the nervous system. RBPs and microRNAs are increasingly recognized as vital mediators of gene expression in neuronal cells, as they tightly regulate the timing and specific compartment in which particular proteins are expressed. Additional examples of interplay between microRNAs and RBPs regulating neuronal gene expression are bound to emerge as we gain deeper understanding of the magnitude, timing, and spatial regulation of neuronal proteins expressed in response to endogenous and environmental signals.
Acknowledgments
This research was supported entirely by the National Institute on Aging-Intramural Research Program, National Institutes of Health, Z01-AG000518-5.
We thank F. E. Indig, J. L. Martindale, X. Yang, K. G. Becker, and the NIA microarray facility for assistance with the experiments. N. I. Perrone-Bizzozero provided reagents for this work.
Footnotes
▿
Published ahead of print on 28 June 2010.
REFERENCES
- 1.Abdelmohsen, K., R. Pullmann, Jr., A. Lal, H. H. Kim, S. Galban, et al. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25**:**543-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelmohsen, K., S. Srikantan, Y. Kuwano, and M. Gorospe. 2008. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U. S. A. 105**:**20297-20302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelmohsen, K., S. Srikantan, X. Yang, A. Lal, H. H. Kim, et al. 2009. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28**:**1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akamatsu, W., H. Fujihara, T. Mitsuhashi, M. Yano, S. Shibata, et al. 2005. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl. Acad. Sci. U. S. A. 102**:**4625-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amadio, M., A. Pascale, J. Wang, L. Ho, and A. Quattrone. 2009. nELAV proteins alteration in Alzheimer's disease brain: a novel putative target for amyloid-beta reverberating on AbetaPP processing. J. Alzheimers Dis. 16**:**409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson, K. D., M. A. Merhege, M. Morin, F. Bolognani, and N. I. Perrone-Bizzozero. 2003. Increased expression and localization of the RNA-binding protein HuD and GAP-43 mRNA to cytoplasmic granules in DRG neurons during nerve regeneration. Exp. Neurol. 183**:**100-108. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, K. D., M. A. Morin, A. Beckel-Mitchener, C. D. Mobarak, R. L. Neve, et al. 2000. Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J. Neurochem. 75**:**1103-1114. [DOI] [PubMed] [Google Scholar]
- 8.Anderson, K. D., J. Sengupta, M. Morin, R. L. Neve, C. F. Valenzuela, et al. 2001. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp. Neurol. 168**:**250-258. [DOI] [PubMed] [Google Scholar]
- 9.Ball, N. S., and P. H. King. 1997. Neuron-specific hel-N1 and HuD as novel molecular markers of neuroblastoma: a correlation of HuD messenger RNA levels with favorable prognostic features. Clin. Cancer Res. 3**:**1859-1865. [PubMed] [Google Scholar]
- 10.Baltimore, D., M. P. Boldin, R. M. O'Connell, D. S. Rao, and K. D. Taganov. 2008. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9**:**839-845. [DOI] [PubMed] [Google Scholar]
- 11.Banihashemi, L., G. M. Wilson, N. Das, and G. Brewer. 2006. Upf1/Upf2 regulation of 3′-untranslated region splice variants of AUF1 links nonsense-mediated and A+U-rich element-mediated mRNA decay. Mol. Cell. Biol. 26**:**8743-8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116**:**281-297. [DOI] [PubMed] [Google Scholar]
- 13.Bolognani, F., M. A. Merhege, J. Twiss, and N. I. Perrone-Bizzozero. 2004. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci. Lett. 371**:**152-157. [DOI] [PubMed] [Google Scholar]
- 14.Bolognani, F., S. Qiu, D. C. Tanner, J. Paik, N. I. Perrone-Bizzozero, et al. 2007. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol. Learn. Mem. 87**:**635-643. [DOI] [PubMed] [Google Scholar]
- 15.Bolognani, F., and N. I. Perrone-Bizzozero. 2008. RNA-protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 86**:**481-489. [DOI] [PubMed] [Google Scholar]
- 16.Bolognani, F., T. Contente-Cuomo, and N. I. Perrone-Bizzozero. 2010. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 38**:**117-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng, A., S. Wang, J. Cai, M. S. Rao, and M. P. Mattson. 2003. Nitric oxide acts in a positive feedback loop with BDNF to regulate neural progenitor cell proliferation and differentiation in the mammalian brain. Dev. Biol. 258**:**319-333. [DOI] [PubMed] [Google Scholar]
- 18.Chung, S., M. Eckrich, N. I. Perrone-Bizzozero, D. T. Kohn, and H. Furneaux. 1997. The Elav-like proteins bind to a conserved regulatory element in the 3′-untranslated region of GAP-43 mRNA. J. Biol. Chem. 272**:**6593-6598. [DOI] [PubMed] [Google Scholar]
- 19.Deschênes-Furry, J., N. I. Perrone-Bizzozero, and B. J. Jasmin. 2006. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. Bioessays 28**:**822-833. [DOI] [PubMed] [Google Scholar]
- 20.El Ouaamari, A., N. Baroukh, G. A. Martens, P. Lebrun, D. Pipeleers, et al. 2008. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 57**:**2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujiwara, T., Y. Mori, D. L. Chu, Y. Koyama, S. Miyata, et al. 2006. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol. Cell. Biol. 26**:**2273-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, F. B. 2008. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 31**:**20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Good, P. J. 1995. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. U. S. A. 92**:**4557-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh, A., and M. E. Greenberg. 1995. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron 15**:**89-103. [DOI] [PubMed] [Google Scholar]
- 25.Hambardzumyan, D., S. Sergent-Tanguy, V. Thinard, R. Bonnamain, M. Masip, et al. 2009. AUF1 and Hu proteins in the developing rat brain: implication in the proliferation and differentiation of neural progenitors. J. Neurosci. Res. 87**:**1296-1309. [DOI] [PubMed] [Google Scholar]
- 26.Hinman, M. N., and H. Lou. 2008. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65**:**3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein, R. 1994. Role of neurotrophins in mouse neuronal development. FASEB J. 8**:**738-744. [DOI] [PubMed] [Google Scholar]
- 28.Kloosterman, W. P., A. K. Lagendijk, R. F. Ketting, J. D. Moulton, and R. H. Plasterk. 2007. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 5**:**e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kloosterman, W. P., and R. H. Plasterk. 2006. The diverse functions of microRNAs in animal development and disease. Dev. Cell 11**:**441-450. [DOI] [PubMed] [Google Scholar]
- 30.Kuwano, Y., H. H. Kim, K. Abdelmohsen, R. Pullmann, Jr., J. L. Martindale, et al. 2008. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 28**:**4562-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, Y., D. Ridzon, L. Wong, and C. Chen. 2007. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8**:**166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, Y., K. Christian, and B. Lu. 2008. BDNF: a key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol. Learn. Mem. 89**:**312-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, B., P. T. Pang, and N. H. Woo. 2005. The yin and yang of neurotrophin action. Nat. Rev. Neurosci. 6**:**603-614. [DOI] [PubMed] [Google Scholar]
- 34.Masuda, K., B. S. Marasa, J. L. Martindale, M. K. Halushka, and M. Gorospe. 2009. Tissue- and age-dependent expression of RNA-binding proteins that influence mRNA turnover and translation. Aging 1**:**681-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meister, G., M. Landthaler, A. Patkaniowska, Y. Dorsett, G. Teng, et al. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15**:**185-197. [DOI] [PubMed] [Google Scholar]
- 36.Noureddine, M. A., X. J. Qin, S. A. Oliveira, T. J. Skelly, J. van der Walt, et al. 2005. Association between the neuron-specific RNA-binding protein ELAVL4 and Parkinson disease. Hum. Genet. 117**:**27-33. [DOI] [PubMed] [Google Scholar]
- 37.Pascale, A., M. Amadio, and A. Quattrone. 2008. Defining a neuron: neuronal ELAV proteins. Cell. Mol. Life Sci. 65**:**128-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pascale, A., M. Amadio, G. Scapagnini, C. Lanni, M. Racchi, et al. 2005. Neuronal ELAV proteins enhance mRNA stability by a PKCalpha-dependent pathway. Proc. Natl. Acad. Sci. U. S. A. 102**:**12065-12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascale, A., P. A. Gusev, M. Amadio, T. Dottorini, S. Govoni, et al. 2004. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc. Natl. Acad. Sci. U. S. A. 101**:**1217-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasquinelli, A. E., and G. Ruvkun. 2002. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18**:**495-513. [DOI] [PubMed] [Google Scholar]
- 41.Perrone-Bizzozero, N., and F. Bolognani. 2002. Role of HuD and other RNA-binding proteins in neural development and plasticity. J. Neurosci. Res. 68**:**121-126. [DOI] [PubMed] [Google Scholar]
- 42.Poy, M. N., J. Hausser, M. Trajkovski, M. Braun, S. Collins, et al. 2009. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U. S. A. 106**:**5813-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pullmann, R. Jr., H. H. Kim, K. Abdelmohsen, A. Lal, J. L. Martindale, et al. 2007. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 27**:**6265-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schratt, G. 2009. Fine-tuning neural gene expression with microRNAs. Curr. Opin. Neurobiol. 19**:**213-219. [DOI] [PubMed] [Google Scholar]
- 45.Stefani, G., and F. Slack. 2008. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9**:**219-230. [DOI] [PubMed] [Google Scholar]
- 46.Tanner, D. C., S. Qiu, F. Bolognani, L. D. Partridge, E. J. Weeber, et al. 2008. Alterations in mossy fiber physiology and GAP-43 expression and function in transgenic mice overexpressing HuD. Hippocampus 18**:**814-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tchen, C. R., M. Brook, J. Saklatvala, and A. R. Clark. 2004. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 279**:**32393-32400. [DOI] [PubMed] [Google Scholar]
- 48.Tiruchinapalli, D. M., M. D. Ehlers, and J. D. Keene. 2008. Activity-dependent expression of RNA binding protein HuD and its association with mRNAs in neurons. RNA Biol. 5**:**157-168. [DOI] [PubMed] [Google Scholar]
- 49.Ule, J., and R. B. Darnell. 2006. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 16**:**102-110. [DOI] [PubMed] [Google Scholar]
- 50.van der Brug, M. P., J. Blackinton, J. Chandran, L. Y. Hao, A. Lal, et al. 2008. RNA binding activity of the recessive Parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 105**:**10244-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi, J., N. Chang, X. Liu, G. Guo, L. Xue, et al. 2010. Reduced nuclear export of HuR mRNA by HuR is linked to the loss of HuR in replicative senescence. Nucleic Acids. Res. 38**:**1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]