Branch formation during organ development (original) (raw)
. Author manuscript; available in PMC: 2011 Nov 1.
Published in final edited form as: Wiley Interdiscip Rev Syst Biol Med. 2010 Nov-Dec;2(6):734–741. doi: 10.1002/wsbm.96
Abstract
Invertebrates and vertebrates use branching morphogenesis to build epithelial trees to maximize the surface area of organs within a given volume. Several molecular regulators of branching have recently been discovered, a number of which are conserved across different organs and species. Signals that control branching at the cellular and tissue levels are also starting to emerge, and are rapidly unveiling the physical nature of branch development. Here we discuss the molecular, cellular and physical processes that govern branch formation and highlight the major outstanding questions in the field.
Keywords: Patterning, Morphogenesis, Tissue, Signaling pathways, Mechanics
Branch formation is a morphogenetic process used extensively across the animal kingdom to construct organs comprised of elaborate epithelial networks, including the Drosophila trachea and salivary gland, and the vertebrate lung, kidney, salivary gland (Fig. 1) and mammary gland. Branched systems maximize surface area, used for the exchange and transport of gases and fluids, within a constrained volume. The spatial patterns of branched organs are highly complex but surprisingly organized. How subpopulations of cells are instructed to form branches and how the process is reiterated numerous times to form epithelial trees has fascinated scientists across disciplines for many years. Advanced genetic tools, culture models and imaging techniques have recently unveiled many signals that regulate branch formation.
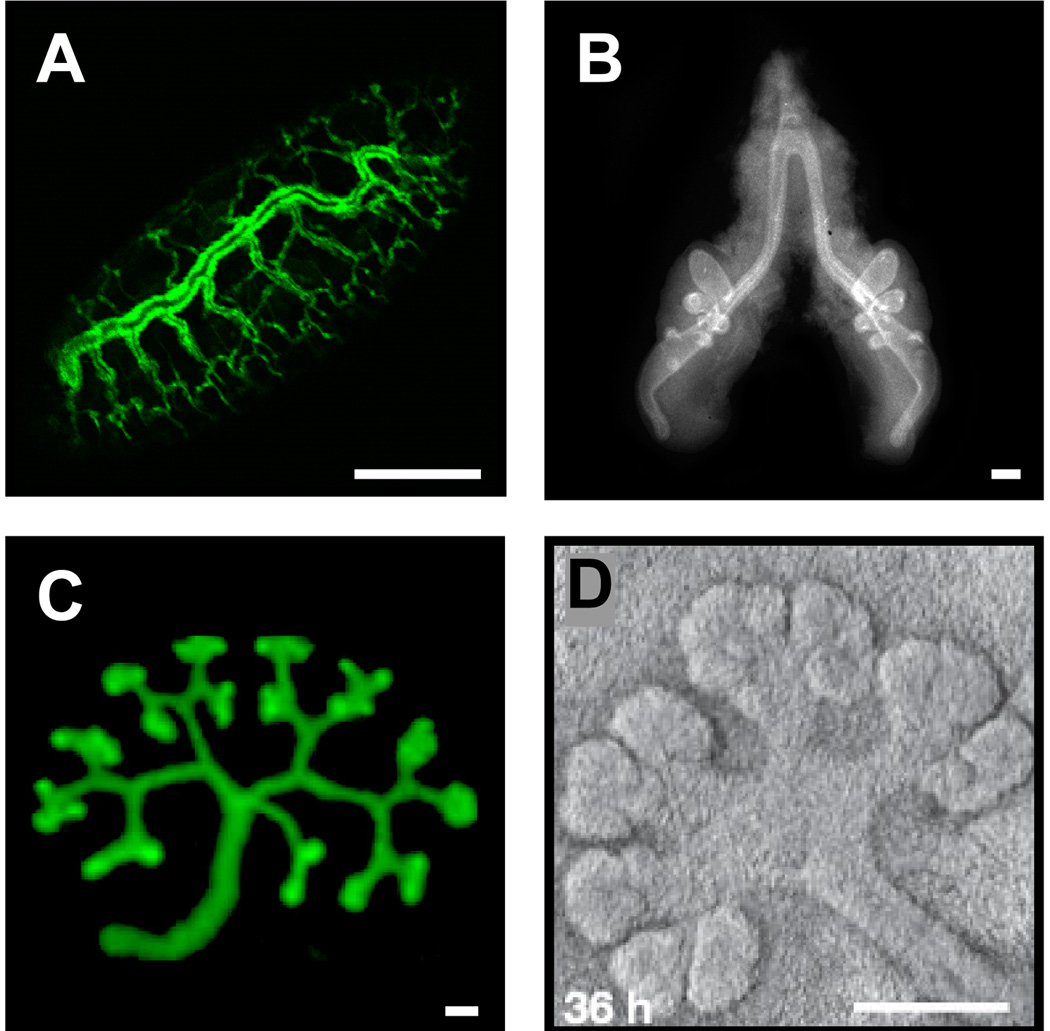
Figure 1.
Organs constructed by branch formation. (A) Drosophila trachea, (B) chicken lung, and mouse (C) kidney (reprinted from52 with permission from Elsevier, © 2009) and (D) salivary gland (reprinted with permission from Macmillan Publishers Ltd: Nature 423: 876–881, © 200629). Scale bars, 100 µm.
The morphology and organization of branches vary between organs. The branching patterns in the Drosophila trachea and mammalian lung are stereotyped and nearly identical between individuals1, 2, suggesting that branch formation in these organs is under “hardwired” genetic control. On the other hand, branching appears to be non-stereotyped in the mammary gland and prostate1, and in these organs cell fates and behaviors depend on the context. Despite the great variation of branching morphologies, a number of biological principles are conserved. Here, we cover some of the mechanisms that govern branch formation, list major unanswered questions and discuss potential systems biology strategies to address them effectively.
1. The knowns: conserved and unique aspects of branch formation
1.1 Molecular basis of branch formation
Perhaps the most conserved aspect of branch formation is that growth factors of mesenchymal origin act as instructive cues. Members of the fibroblast growth factor (FGF) family direct branching of the Drosophila trachea and mammalian lung, mammary and salivary glands (reviewed in1, 3). Multiple growth factors often regulate a single branching system. For instance, branching of the kidney is driven by glial cell line-derived neutrophic factor (GDNF), epidermal growth factor (EGF) and FGFs4, whereas EGF and hepatocyte growth factor (HGF) regulate branching of the mammary gland and lung (reviewed in1, 5). It is unclear what role these shared instructive cues play in different organs, or how they interact within a single organ to supply local information to subpopulations of cells.
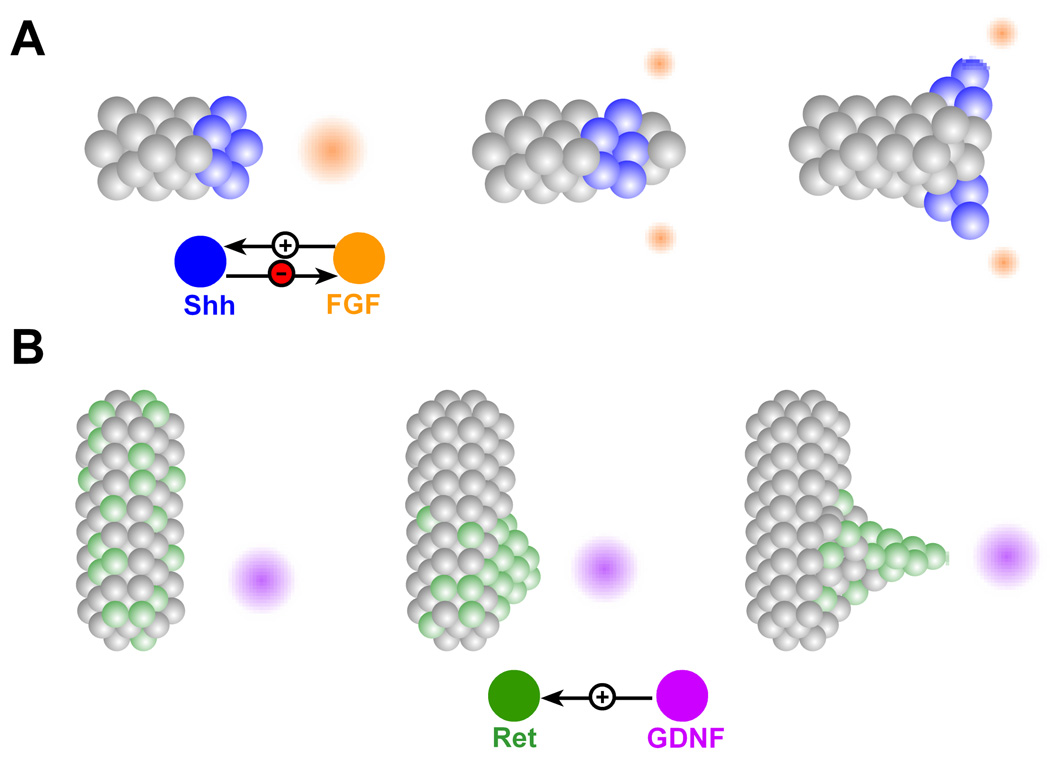
How can mesenchymally-derived growth factors induce branching? A relatively well documented mechanism is chemotaxis. During the development of the Drosophila trachea, clusters of mesenchymal cells produce the FGF molecule Branchless (Bnl) at stereotyped locations6. Epithelial cells extend filopodia toward the sources of Bnl, suggesting that Bnl induces branching by enhancing cell motility via cytoskeletal rearrangements7. A similar chemotactic mechanism is hypothesized to drive branch formation in the mammalian lung (Fig. 2A)3. Here, the levels and spatial profile of FGF10 are thought to be refined by Sonic hedgehog (Shh), which is expressed at the branch tips (reviewed in8). It has been postulated that FGF10 is locally down-modulated by Shh as the branch tip approaches. The FGF10 profile is thus split into two sources, leading to branch bifurcation. This interplay between Shh and FGF10 suggests a possible mechanism to space branches during morphogenesis of the lung airways. During kidney development, chemotaxis appears to be restricted to ureteric bud formation rather than branching. In this context, cells expressing the highest levels of the Ret receptor, initially randomly dispersed throughout the epithelium, migrate persistently towards the mesenchymal source of GDNF9, 10 (Fig. 2B). Once there, the cells form a cluster of high Ret activity, which swells outward, forming the ureteric bud. It is unclear, however, what role GDNF plays in branch initiation and guidance by chemoattraction in the subsequent stages of kidney development10.
Figure 2.
Branch formation by chemoattraction. (A) FGF source (orange) guides branch extension by enhancing motility of tip cells. FGF induces expression of Shh (blue), which in turn suppresses and splits the FGF source. The split source of FGF gives rise to branch bifurcation. (B) Cells expressing high levels of Ret (green), persistently migrate toward the source of GDNF (purple), forming a patch of high Ret activity, which ultimately forms the ureteric bud.
Conversely, there is no evidence for chemoattraction in specifying and guiding branches in the mammary gland. Nonetheless, as in all other branched organs, epithelial-stromal interactions are critical, and matrix metalloproteinases (MMPs) have emerged as key mediators. Elevated levels of both MMP2 and MMP14 have been observed at the tips of mammary and ureteric branches in vivo11, 12, and MMP activity is required for mammary branch formation13. MMPs could act in a path-clearing or signaling capacity: they could promote branching by degrading the extracellular matrix (ECM) adjacent to branch sites and facilitating cell invasion, or alter signaling by generating bioactive ECM fragments, cleaving cell-cell or cell-matrix adhesions, activating latent transforming growth factor (TGF)-β or releasing ECM-bound growth factors5, 11, 14. Indeed, a recent study demonstrated that MT2-MMP-mediated proteolysis of collagen IV releases NC1 domains, which regulate branching in the salivary gland by increasing epithelial proliferation15. Accordingly, in these organs MMPs might supply the patterning instructions, whereas FGF and other growth factors might serve a more global role.
In addition to positive instructive cues, branching organs are sculpted by pervasive inhibitory cues. TGFβ is a potent inhibitor of branching in the lung, kidney and mammary gland (reviewed in4). TGFβ is secreted by mammary epithelium and acts as an autocrine inhibitor in part by stimulating the synthesis and deposition of ECM along ducts16. Moreover, the concentration profile of TGFβ, dictated by the geometry of the existing epithelium, acts locally to supply patterning information by defining new sites of branching13. Bone morphogenetic proteins (BMPs) play a similar role in the lung and kidney: BMP4 directs proximal-distal differentiation of the lung tree17, 18, whereas BMP2 and BMP7 inhibit ureteric branching19.
Considering the downstream signaling cascades activated by the positive and negative cues discussed above reveals further overlap of the regulatory mechanisms employed by different organs. FGFs, EGF, HGF and GDNF all signal through the mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase cascades, whereas TGFβ and BMP mainly signal through Smads. However, even molecules that belong to the same family or signal through a common downstream pathway appear to contribute to branching in distinct ways. In the salivary and lacrimal glands, FGF7 induces formation of new branches whereas FGF10, which also signals through the FGFR2b receptor, leads to branch elongation20. In mouse mammary organoids, FGF7 induces growth and TGFα induces branching, even though both growth factors elicit their effects through MAPK21.
1.2 Cellular basis of branch formation
Even when the underlying signaling pathways are conserved, strikingly different cellular behaviors are used to drive branch formation. For instance, Drosophila tracheal branching is accomplished exclusively by cellular migration, as cell division is fully completed prior to branch outgrowth22. In contrast, cell division is critical for branch initiation and growth in vertebrates21, 23. The end buds of the pubertal mammary gland are highly proliferative24 and inhibiting proliferation prevents mammary branching21. Branching of different organs is accompanied and possibly driven by distinct cellular morphologies. Full epithelial polarization is maintained during ductal elongation in the zebrafish kidney and Drosophila salivary gland25, 26, whereas mammary end buds are multilayered and partially depolarized24. Reports of neo-expression of mesenchymal markers at the tips of mammary ducts in vivo and in culture13, 27 hint that mammary branching might occur through partial loss or alteration of epithelial character.
Whereas the organs discussed thus far use cell migration, division and shape change to drive branching via budding, the mammalian salivary gland initiates branches through clefting. Here, the epithelial cells undergo vigorous rearrangements, but the lack of choreographed motions has led to the conclusion that epithelial motility alone is insufficient for pattern formation28. Instead, it is the directional assembly of fibronectin that patterns branching, although cell motility may aid the process by allowing remodeling28, 29. Nonetheless, proliferation also plays a role in salivary gland branching. Although proliferation is not required for cleft initiation, it is increased in the distal regions of the epithelium and is critical for cleft progression and branch outgrowth30. Future studies will need to determine whether these distinct cellular behaviors (division, migration, clefting) are the final events in branching, or if they actively feed back to control the process.
1.3 Branching as a physical process
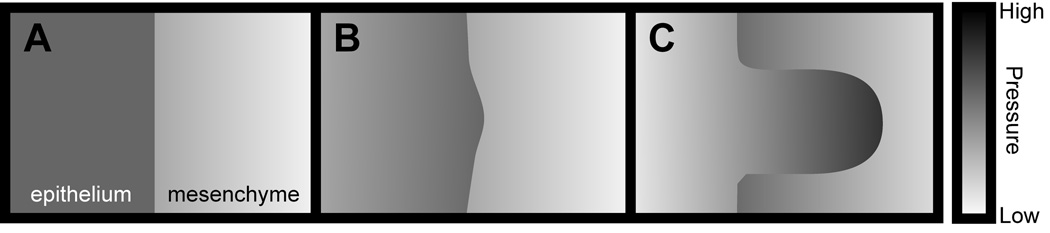
In addition to biochemical stimuli, developing organs receive and interpret biophysical and mechanical signals from the microenvironment. Many of the processes that drive branching – cell migration, shape change, rearrangement, budding, clefting and bifurcation – are fundamentally mechanical. In contrast to the different molecular regulators that are used by different organs, the putative physical mechanisms are universal. The process of branch formation can be thought of physically as a viscous fluid (epithelium) elongating and bifurcating against a second fluid (stroma or mesenchyme) of different rheological properties. This situation is modeled in physics with the free-boundary problem, a simple case of which is the phenomenon of viscous fingering31. Here, a given pressure gradient exists at the interface between the two fluids (Fig. 3). The formation of a small bulge in the inner fluid (the epithelium) redistributes the gradient, such that the bulge region experiences a sharper pressure drop, which propels it forward. As the bud grows, it displaces the mesenchyme laterally, thus reducing the pressure gradient and consequently the driving force for growth at what have now become the stalk regions. Instabilities at the tip can lead to bifurcation and generation of a complex self-similar structure reminiscent of the lung and kidney trees31.
Figure 3.
Branch formation by viscous fingering. (A) The pressure in the epithelium is initially uniform, whereas that of the mesenchyme decreases away from the epithelium. (B) A small bulge in the epithelium protrudes and encounters a sharper pressure drop, which drives further protrusion. (C) As the bud grows, it displaces the mesenchyme toward the stalk regions, reducing the pressure drop there and lowering the driving force for motion.
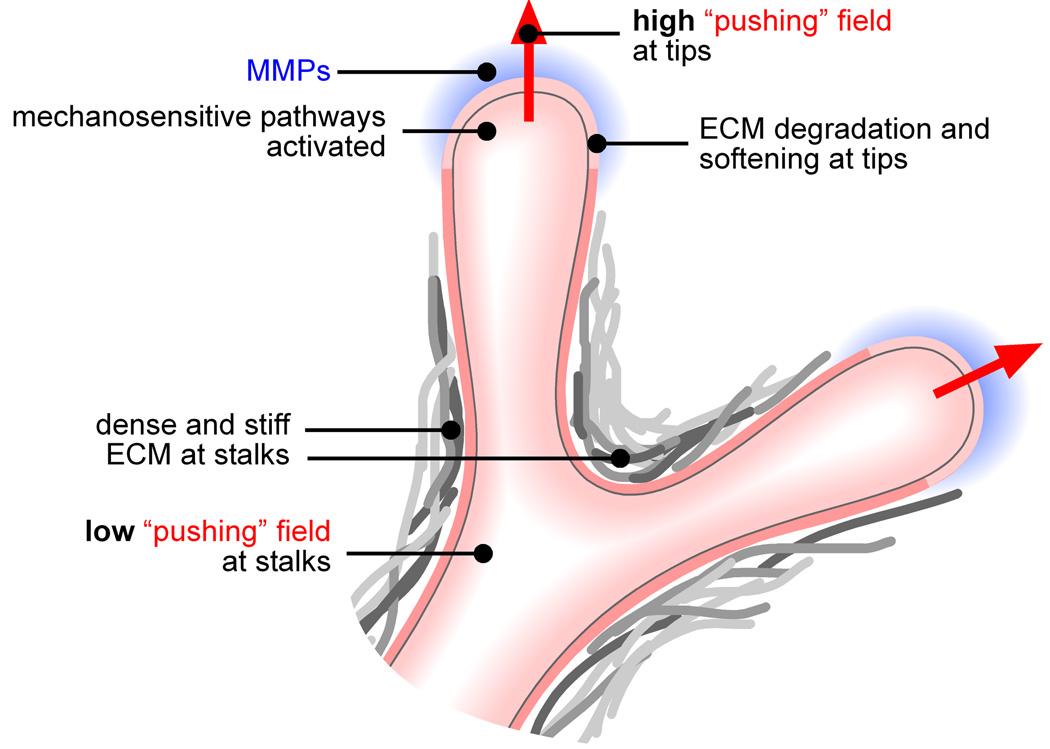
Cytoskeletal contraction has also been implicated in branch formation. Inhibiting actomyosin contractility prevents branching in the kidney, lung and salivary gland30, 32, 33. Conversely, activating contractility promotes branching in the lung34. Given reports that the basement membrane thins at sites of branch formation13, 33, Ingber has proposed a “run-in-a-stocking” mechanism, based on the postulate that contractility renders branching tissues in a state of isometric prestress. When the ECM is locally degraded, the adjacent prestressed epithelium is no longer physically hindered and invades in the form of a bud33. This prestress itself could lead to basement membrane thinning through local stimulation of MMP production35, 36, consistent with reports of elevated MMP2 and MMP14 at the leading edge of mammary and ureteric buds11, 12 and the tips of engineered mammary tubules37. High mechanical stress could also activate cell motility and invasion. Thus, physical fields could not only assist branch formation by mechanically propelling the epithelium forward, but also by controlling the biochemical regulators (summarized in Fig. 4).
Figure 4.
Mechanical control of branching. The high mechanical fields at the tips enhance branching by physically propelling the epithelium forward and by activating proteases and signaling pathways that enhance cellular motility and invasiveness.
2. Systems approaches to “connect the dots”
From the mechanisms discussed here, and those omitted due to space limitations, it is clear that branching requires various growth factors, morphogens, ECM molecules, proteases and mechanical signals from the microenvironment. It is unclear how these signals (identified separately by different investigators) interact to provide patterning information that makes a lung airway distinct from a kidney tubule. Perhaps the time is ripe to dissect the precise roles and interactions of the regulatory cues. Are the different signals coupled or independent? Is the regulation of branch formation hierarchical, such that removing a signal would induce a catastrophic collapse of the process, or is it structured redundantly? Are there a minimum number of cues that can orchestrate formation of branches with the correct histology and function? Many of these questions would likely be answered by taking a systems biology approach to the problem and organizing the critical signals into a “branching regulatory network”.
In order to describe branch formation using a systems perspective, it is imperative that we first rigorously delineate the sequence of events and morphologies during branching. The most comprehensive description of a branching program to date is the study by Metzger et al2. In this formidable effort, the authors mapped out the complete three-dimensional branching pattern of the mouse bronchial tree. In doing so, they uncovered a conserved set of three basic subroutines (domain branching, planar and orthogonal bifurcation). Each of the subroutines could in turn be described as a combination of four basic spatial operators (periodicity generator, domain specifier, bifurcator and rotator). To now fully define the branching regulatory network, we must investigate the molecular control mechanisms that execute each level of the branching program.
Large-scale approaches to analyze genetic changes that accompany branching have been reported for several organs. DNA microarrays have been used to describe the temporal gene expression profiles during kidney development in vivo and in culture38, 39. A genome-wide transcript analysis identified genes with altered expression levels between the terminal end bud, duct and stromal microenvironments of the mammary gland27. A separate study distinguished the gene expression levels in the cleft and bud regions of the salivary gland40. These efforts should be extended to define the full spatiotemporal gene expression profiles for each organ. The resulting information would instruct better-targeted perturbation of gene expression to examine both the effect of molecular signals on the branching phenotype and their interactions. As these large amounts of data accumulate, the challenge will lie in interpretation and synthesis into plausible multi-scale models and regulatory networks of branch formation. To that end, sophisticated in silico approaches will likely prove helpful, if not necessary.
Surprisingly, there have been few efforts to computationally model the process of branch formation. Hartmann and Miura modeled in vitro lung branching using the free-boundary problem described above41. A different model described the salivary epithelium and the surrounding mesenchyme as fluids of different viscosities, and demonstrated that a mesenchymally-localized force field, which could arise from fibroblast contraction, is sufficient to induce clefts in the epithelium42. The authors improved this two-dimensional model, in which the force was located at a single point and in a fixed direction, by devising a three-dimensional model which tracked the concentration of the contractile fibroblasts in the mesenchyme43. Both models suggest that, whereas salivary epithelium does branch when cultured in the absence of mesenchyme, the latter significantly increases the rate of branching. Nevertheless, these models do not describe the budding mechanism employed by other organs, and the presence of an advecting mesenchymal force field has yet to be confirmed in vivo.
Perhaps the most complex model of branching thus far is that developed by Hirashima et al44. Instead of modeling the epithelium as a continuous medium, the authors used the cellular Potts model to describe individual cells and cellular behaviors such as growth, division and chemotaxis, all under the control of a split GDNF source. The model suggests that the balance of cell proliferation and chemotaxis is the major determinant of branch shape. While this model is commendable for incorporating biochemical and cellular complexity, it does not provide a mechanism for the localization of GDNF and the transduction of the chemical signal into motile and invasive behaviors.
The shortcomings of past approaches motivate the need for models that reflect the a priori conviction of systems biology that no process can be understood predictively until a majority of its components are included in the analysis45. In that sense, a comprehensive understanding of the topology of the branching network is unlikely until the models incorporate the full molecular and physical complexity seen in vivo. In devising such models, connections must be drawn between spatiotemporal profiles of gene expression and the cellular processes that perform branching (proliferation, migration, invasion, shape change). Although the field is in its infancy and data acquisition should certainly continue, the present body of experimental knowledge is probably sufficient to begin connecting the dots. Tentative maps have already been composed for organs such as the salivary gland46. As the downstream-most processes of clefting, budding and shape change are ultimately physical, tentative molecular networks should be coupled with a physical component to fully simulate branching in silico.
First, such models could provide tractable platforms for testing the plausibility of proposed network topologies. For instance, the regulatory network for branching in the lung should yield structures that recapitulate the four branching modes described by Metzger et al2, in the correct sequences and in the correct spatial and temporal registration. Nigam and colleagues have recently postulated that branching regulatory networks resemble autocatalytic networks, in which complex emergent properties arise from the interaction of several members within the pathway47–51. Given that gene expression changes between the T-shaped ureteric bud and the multiple rounds of branching are modest39, the authors consider the possibility that the set of proteins interact in the context of an autocatalytic network thus giving rise to the iterative tip and stalk generation. Could the existence of such autocatalytic networks also explain how a set of conserved molecular regulators interact to yield emergent organ-specific patterns of branching? Comprehensive computational models can potentially shed light on this question and others. Further, sophisticated models could play a predictive role by identifying connections not readily apparent from previous experimental work and thereby direct future experiments. Finally, detailed simulations of branch formation could help elucidate where and how normal developmental mechanisms are disrupted in disease and even unveil strategies for reconstructing branched organs ex vivo.
Conclusion
The past decade has seen great progress in identifying the molecular signals that globally regulate branch formation. Surprisingly, many of these signals are used even by evolutionarily distant organs such as the fly tracheal system and the mammalian lung, kidney and mammary gland. Nevertheless, the conserved set of regulatory molecules effect branch formation via distinct cellular behaviors and there are enough unique molecular aspects across different organs to still keep us far from a unified theory of branching. Further, major questions about how cell behaviors are patterned remain unanswered. We still do not fully understand how branch sites are selected and what determines the correct spacing between individual branches. Reinforcing the classic genetic, pharmacologic and tissue recombination techniques used to study branch formation with detailed maps of the branching program in each system, large-scale gene expression screens and computational models that recapitulate the molecular complexity and the physical phenomena observed in vivo will likely prove helpful, if not necessary, for addressing the outstanding questions in this important process.
Acknowledgements
We thank Jiyong Kwak for technical assistance. The authors are supported by the NIH (GM083997 and CA128660), Susan G. Komen for the Cure, and the David & Lucile Packard Foundation. CMN holds a Career Award at the Scientific Interface from the Burroughs Welcome Fund.
Contributor Information
Nikolce Gjorevski, Email: gorevski@princeton.edu, Department of Chemical Engineering, Princeton University.
Celeste M. Nelson, Email: celesten@princeton.edu, Departments of Chemical Engineering and Molecular Biology, Princeton University.
References
- 1.Lu PF, Werb Z. Patterning Mechanisms of Branched Organs. Science. 2008;322:1506–1509. doi: 10.1126/science.1162783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affolter M, Bellusci S, Itoh N, Shilo B, Thiery JP, Werb Z. Tube or not tube: Remodeling epithelial tissues by branching morphogenesis. Dev Cell. 2003;4:11–18. doi: 10.1016/s1534-5807(02)00410-0. [DOI] [PubMed] [Google Scholar]
- 4.Davies JA. Do different branching epithelia use a conserved developmental mechanism? Bioessays. 2002;24:937–948. doi: 10.1002/bies.10161. [DOI] [PubMed] [Google Scholar]
- 5.Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development. 2005;132:3923–3933. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutherland D, Samakovlis C, Krasnow MA. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro C, Ebner A, Affolter M. In vivo Imaging reveals different cellular functions for FGF and Dpp signaling in tracheal branching morphogenesis. Dev Cell. 2002;2:677–683. doi: 10.1016/s1534-5807(02)00171-5. [DOI] [PubMed] [Google Scholar]
- 8.Cardoso WV. Molecular regulation of lung development. Am Rev Physiol. 2001;63:471–494. doi: 10.1146/annurev.physiol.63.1.471. [DOI] [PubMed] [Google Scholar]
- 9.Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, et al. Ret-Dependent Cell Rearrangements in the Wolffian Duct Epithelium Initiate Ureteric Bud Morphogenesis. Dev Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shakya R, Watanabe T, Costantini F. The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Wiseman BS, Sternlicht MD, Lund LR, Alexander CM, Mott J, Bissell MJ, Soloway P, Itohara S, Werb Z. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol. 2003;162:1123–1133. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanwar YS, Ota K, Yang Q, Wada J, Kashihara N, Tian Y, Wallner EI. Role of membrane-type matrix metalloproteinase 1 (MT-1-MMP), MMP-2, and its inhibitor in nephrogenesis. Am J Physiol. 1999;277:F934–F947. doi: 10.1152/ajprenal.1999.277.6.F934. [DOI] [PubMed] [Google Scholar]
- 13.Nelson CM, VanDuijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sternlicht MD. Key stages in mammary gland development - The cues that regulate ductal branching morphogenesis. Breast Canc Res. 2006;8 doi: 10.1186/bcr1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebustini IT, Myers C, Lassiter KS, Surmak A, Szabova L, Holmbeck K, Pedchenko V, Hudson BG, Hoffman MP. MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev Cell. 2009;17:482–493. doi: 10.1016/j.devcel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silberstein GB, Strickland P, Coleman S, Daniel CW. Epithelium-dependent extracellular matrix synthesis in transforming growth factor-beta 1-growth-inhibited mouse mammary gland. J Cell Biol. 1990;110:2209–2219. doi: 10.1083/jcb.110.6.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- 18.Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999;126:4005–4015. doi: 10.1242/dev.126.18.4005. [DOI] [PubMed] [Google Scholar]
- 19.Gupta IR, Piscione TD, Grisaru S, Phan T, Macias-Silva M, Zhou X, Whiteside C, Wrana JL, Rosenblum ND. Protein kinase A is a negative regulator of renal branching morphogenesis and modulates inhibitory and stimulatory bone morphogenetic proteins. J Biol Chem. 1999;274:26305–26314. doi: 10.1074/jbc.274.37.26305. [DOI] [PubMed] [Google Scholar]
- 20.Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, Patel VN, Lang RA, Mohammadi M. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fata JE, Mori H, Ewald AJ, Zhang H, Yao E, Werb Z, Bissell MJ. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGF alpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol. 2007;306:193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samakovlis C, Manning G, Steneberg P, Hacohen N, Cantera R, Krasnow MA. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development. 1996;122:3531–3536. doi: 10.1242/dev.122.11.3531. [DOI] [PubMed] [Google Scholar]
- 23.Fisher CE, Michael L, Barnett MW, Davies JA. Erk MAP kinase regulates branching morphogenesis in the developing mouse kidney. Development. 2001;128:4329–4338. doi: 10.1242/dev.128.21.4329. [DOI] [PubMed] [Google Scholar]
- 24.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerman BE, Cheshire AM, Andrew DJ. From fate to function: the Drosophila trachea and salivary gland as models for tubulogenesis. Differentiation. 2006;74:326–348. doi: 10.1111/j.1432-0436.2006.00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasilyev A, Liu Y, Mudumana S, Mangos S, Lam PY, Majumdar A, Zhao JH, Poon KL, Kondrychyn I, Korzh V, et al. Collective Cell Migration Drives Morphogenesis of the Kidney Nephron. Plos Biol. 2009;7:101–114. doi: 10.1371/journal.pbio.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouros-Mehr H, Werb Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev Dyn. 2006;235:3404–3412. doi: 10.1002/dvdy.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- 29.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 30.Daley WP, Gulfo KM, Sequeira SJ, Larsen M. Identification of a mechanochemical checkpoint and negative feedback loop regulating branching morphogenesis. Dev Biol. 2009;336:169–182. doi: 10.1016/j.ydbio.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleury V, Watanabe T, Nguyen T, Unbekandt M, Warburton D, Dejmek M, Nguyen MB, Lindner A, Schwartz L. Physical mechanisms of branching morphogenesis in animals. In: Davies JA, editor. Branching Morphogenesis. Georgetown: Landes Bioscience; 2005. pp. 202–234. [Google Scholar]
- 32.Michael L, Sweeney DE, Davies JA. A role for microfilament-based contraction in branching morphogenesis of the ureteric bud. Kidney International. 2005;68:2010–2018. doi: 10.1111/j.1523-1755.2005.00655.x. [DOI] [PubMed] [Google Scholar]
- 33.Moore KA, Polte T, Huang S, Shi B, Alsberg E, Sunday ME, Ingber DE. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev Dynam. 2005;232:268–281. doi: 10.1002/dvdy.20237. [DOI] [PubMed] [Google Scholar]
- 34.Moore KA, Huang S, Kong YP, Sunday ME, Ingber DE. Control of embryonic lung branching morphogenesis by the Rho activator, cytotoxic necrotizing factor 1. J Surg Res. 2002;104:95–100. doi: 10.1006/jsre.2002.6418. [DOI] [PubMed] [Google Scholar]
- 35.Prajapati RT, Chavally-Mis B, Herbage D, Eastwood M, Brown RA. Mechanical loading regulates protease production by fibroblasts in three-dimensional collagen substrates. Wound Rep Regen. 2000;8:226–237. doi: 10.1046/j.1524-475x.2000.00226.x. [DOI] [PubMed] [Google Scholar]
- 36.Ruddy JM, Jones JA, Stroud RE, Mukherjee R, Spinale FG, Ikonomidis JS. Differential Effects of Mechanical and Biological Stimuli on Matrix Metalloproteinase Promoter Activation in the Thoracic Aorta. Circ. 2009;120:S262–S268. doi: 10.1161/CIRCULATIONAHA.108.843581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM. Self-organization of engineered epithelial tubules by differential cellular motility. Proc Natl Acad Sci USA. 2009;106:14890–14895. doi: 10.1073/pnas.0901269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart RO, Bush KT, Nigam SK. Changes in global gene expression patterns during development and maturation of the rat kidney. Proc Natl Acad Sci U S A. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart RO, Bush KT, Nigam SK. Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development. Kidney Int. 2003;64:1997–2008. doi: 10.1046/j.1523-1755.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- 40.Sakai T, Larsen M, Yamada KM. Microanalysis of gene expression in tissues using T7-SAGE: serial analysis of gene expression after high-fidelity T7-based RNA amplification. Curr Protoc Cell Biol. 2002 doi: 10.1002/0471143030.cb1903s16. Chapter 19: Unit 19 13. [DOI] [PubMed] [Google Scholar]
- 41.Hartmann D, Miura T. Modelling in vitro lung branching morphogenesis during development. J Theor Biol. 2006;242:862–872. doi: 10.1016/j.jtbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Lubkin SR, Li Z. Force and deformation on branching rudiments: cleaving between hypotheses. Biomech Model Mechanobiol. 2002;1:5–16. doi: 10.1007/s10237-002-0001-4. [DOI] [PubMed] [Google Scholar]
- 43.Wan XH, Li ZL, Lubkin SR. Mechanics of mesenchymal contribution to clefting force in branching morphogenesis. Biomech Model Mechanobiol. 2008;7:417–426. doi: 10.1007/s10237-007-0105-y. [DOI] [PubMed] [Google Scholar]
- 44.Hirashima T, Iwasa Y, Morishita Y. Dynamic modeling of branching morphogenesis of ureteric bud in early kidney development. J Theor Biol. 2009;259:58–66. doi: 10.1016/j.jtbi.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Davidson EH. Developmental biology at the systems level. BBA. 2009;1789:248–249. doi: 10.1016/j.bbagrm.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Jaskoll T, Melnick M. Embryonic salivary gland branching morphogenesis. In: Davies JA, editor. Branching Morphogenesis. Georgetown: Landes Bioscience; 2005. pp. 160–175. [Google Scholar]
- 47.Monte JC, Sakurai H, Bush KT, Nigam SK. The developmental nephrome: systems biology in the developing kidney. Curr Opin Nephrol Hypertens. 2007;16:3–9. doi: 10.1097/MNH.0b013e3280118a5a. [DOI] [PubMed] [Google Scholar]
- 48.Mossel E, Steel M. Random biochemical networks: the probability of self-sustaining autocatalysis. J Theor Biol. 2005;233:327–336. doi: 10.1016/j.jtbi.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Wolkenhauer O, Ullah M, Wellstead P, Cho KH. The dynamic systems approach to control and regulation of intracellular networks. FEBS Lett. 2005;579:1846–1853. doi: 10.1016/j.febslet.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Nigam SK, Shah MM. How does the ureteric bud branch? J Am Soc Nephrol. 2009;20:1465–1469. doi: 10.1681/ASN.2008020132. [DOI] [PubMed] [Google Scholar]
- 51.Nigam SK, Wu W, Bush KT. Organogenesis forum lecture: In vitro kidney development, tissue engineering and systems biology. Organogenesis. 2008;4:137–143. doi: 10.4161/org.4.3.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michos O. Kidney development: from ureteric bud formation to branching morphogenesis. Curr Opin Genet Dev. 2009;19:484–490. doi: 10.1016/j.gde.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]