Obesity-linked phosphorylation of PPARγ by cdk5 is a direct target of the anti-diabetic PPARγ ligands (original) (raw)
. Author manuscript; available in PMC: 2011 Jan 1.
Published in final edited form as: Nature. 2010 Jul 22;466(7305):451–456. doi: 10.1038/nature09291
Abstract
Obesity induced in mice by high-fat feeding activates the protein kinase cdk5 in adipose tissues. This results in phosphorylation of the nuclear receptor PPARγ, a dominant regulator of adipogenesis and fat cell gene expression, at serine 273. This modification of PPARγ does not alter its adipogenic capacity, but leads to dysregulation of a large number of genes whose expression is altered in obesity, including a reduction in the expression of the insulin-sensitizing adipokine, adiponectin. The phosphorylation of PPARγ by cdk5 is blocked by anti-diabetic PPARγ ligands, such as rosiglitazone and MRL24. This inhibition works both in vivo and in vitro, and surprisingly, is completely independent of classical receptor transcriptional agonism. Similarly, inhibition of PPARγ phosphorylation in obese patients by rosiglitazone is very tightly associated with the anti-diabetic effects of this drug. These data strongly suggest that cdk5-mediated phosphorylation of PPARγ may be involved in the pathogenesis of insulin-resistance, and present an opportunity for development of an improved generation of anti-diabetic drugs through PPARγ.
Keywords: phosphorylation, PPARγ, cdk5, p35, adipogenesis, adipose tissue, rosiglitazone, MRL24
Adipose tissue is at the center of the Metabolic Syndrome. Excessive body fat defines obesity and leads to insulin resistance, dyslipidemia, type 2 diabetes, certain cancers and cardiovascular disease. Understanding the molecular pathways that link adipose tissue biology to this staggering array of pathologies is of paramount scientific and medical importance. Adipose cells and tissues secrete a variety of cytokines and cytokine-like molecules, called adipokines, which have positive or negative effects on insulin sensitivity, circulating lipid levels, and appetite1. In obesity, TNF-α, IL-1 and resistin are secreted from adipose tissues and suppress insulin action on peripheral tissues2–4. Conversely, adipose tissues from lean individuals secrete higher levels of adiponectin, a circulating protein which has insulin-sensitizing effects on liver and other tissues5–7.
The nuclear receptor PPARγ can be thought of as a “master” gene of fat cell biology and differentiation, being both necessary and sufficient to drive conversion of fibroblastic precursors into fat cells8–10. Other key transcription factors, including C/EBPs and EBF proteins, regulate the expression of PPARγ11,12. Thiazolidinedione drugs (such as rosiglitazone and pioglitazone) are synthetic ligands functioning as strong agonists on PPARγ, and are potent insulin-sensitizing agents13. While the structural basis for the insulin sensitizing effects of these drugs is unclear, their actions result in decreased expression of insulin resistance-inducing adipokines including TNF-α, IL-1 and resistin, and increased production of the insulin-sensitizing hormone, adiponectin14,15. Unfortunately, PPARγ agonists can have long-term adverse effects on the health of certain patients, such as increased body weight, fluid retention, and increased risk of heart failure16.
Several important aspects of PPARγ action remain confusing and unresolved. First, there is no general deficiency in PPARγ function in obesity or insulin-resistant states. Hence, it is not clear why synthetic activation of a receptor should give such dramatic anti-diabetic effects. Second, while anti- potency diabetic of the PPARγ ligand drugs correlates very well with their binding affinities17, some ligands with full agonist action, like rosiglitazone, have powerful insulin-sensitizing actions, while other compounds with poor agonist activities, such as the benzyl indole MRL24, retain very good anti-diabetic effects18.
We show here that phosphorylation of PPARγ by the protein kinase cdk5 neither activates nor suppresses general receptor transcriptional activity, but changes the expression of specific genes like adiponectin. Cdk5-mediated phosphorylation of PPARγ is linked to obesity induced in mice by high-fat feeding. Several anti-diabetic PPARγ ligands, with or without classical agonist properties, directly inhibit this action of cdk5 on PPARγ and restore a more normal, non-diabetic pattern of gene expression. In addition, inhibition of this PPARγ phosphorylation in humans by rosiglitazone is closely associated with its anti-diabetic effects. This unusual pharmacology suggests a surprising model for a cdk5-PPARγ link in the pathogenesis of obesity/diabetes and for the therapeutic action of PPARγ ligands in these disorders.
Results
Pro-inflammatory cytokines are secreted from both fat cells and immune cells residing in adipose tissue, particularly when animals or humans become obese2. We noted with interest that the primary amino acid sequence of PPARγ contained a consensus site for phosphorylation by the protein kinase cdk5 at serine 273 of PPARγ2 (Supplementary Fig. 1a). This protein kinase is not regulated by cyclins, but is instead activated by p35/25, which are targets of numerous cytokines and pro-inflammatory signals19. When murine PPARγ was incubated in vitro with cdk5 and its cofactor p35 (Fig. 1a), the nuclear receptor was phosphorylated as efficiently as histone H1, a known substrate of the cdk5/p35 complex. Mutation of serine-273 to alanine completely blocked phosphorylation by cdk5, indicating that there were no other cdk5 sites in PPARγ detected by an antibody that recognizes phosphorylated cdk5 consensus sites (Fig. 1a). Other members of the cdk protein family did not phosphorylate PPARγ (Supplementary Fig. 1b). Cdk5 also phosphorylated PPARγ at Ser-273 in cells, as shown by co-transfection of the kinase with wild-type (wt) and mutant PPARγ (Supplementary Fig. 1c). Finally, cdk5 did not modify murine PPARα or PPARδ (Supplementary Fig. 1d).
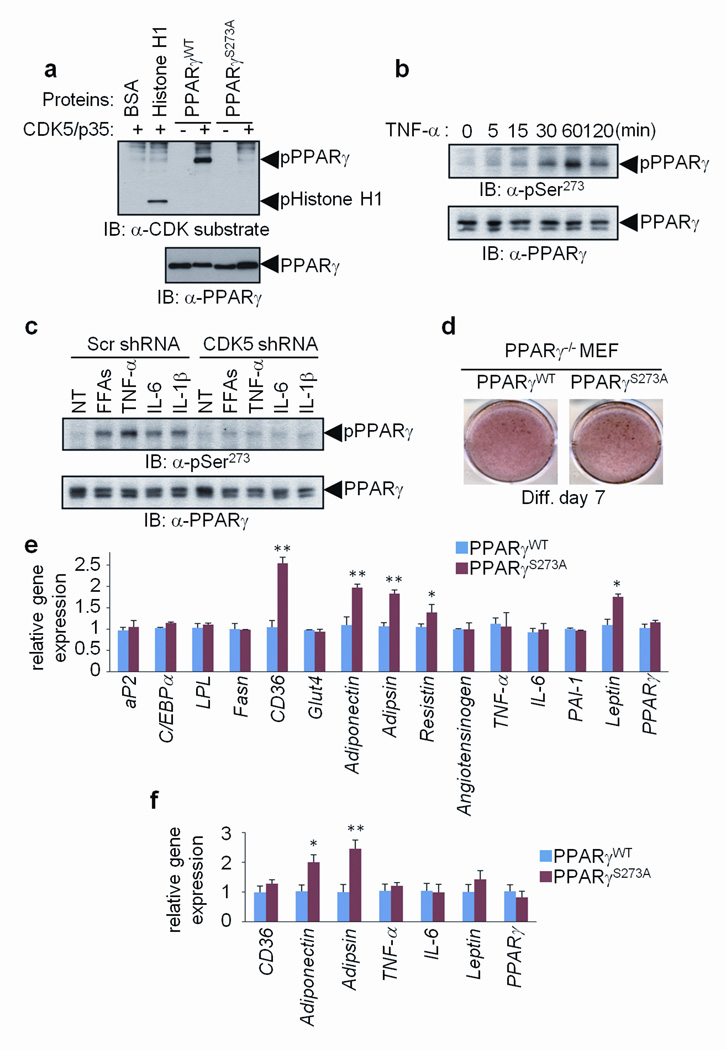
Figure 1. Specific fat cell gene dysregulation by the cdk5-mediated S273 phosphorylation of PPARγ.
a, In vitro CDK assays performed using cdk5/p35 with either wild type (WT) or S273A mutated PPARγ. b, Phosphorylation of PPARγ in differentiated 3T3-L1 adipocytes stimulated with TNF-α for the indicated times. c, Phosphorylation of PPARγ in cells expressing scrambled or CDK5 shRNA stimulated with indicated cytokines. NT, no treatment. d, Staining of PPARγ-null fibroblasts expressing WT or S273A mutant PPARγ with Oil-Red-O. e, Gene expression in these cells was analyzed by real-time quantitative PCR (qPCR) for expression of various genes (n=3). f, mRNA expression in transplanted fat pads was analyzed by qPCR (n=5) (Error bars are s.e.m.; *p<0.05, **p<0.01).
Obesity is characterized by elevated circulating and local levels of pro-inflammatory cytokines and free fatty acids, and cdk5 is activated by various cytokines19,20. Fig. 1b illustrates that treatment of 3T3-L1 adipocytes with TNF-α caused phosphorylation at Ser-273 of PPARγ. Furthermore, the phosphorylation of PPARγ at Ser-273 occurred upon treatment of fat cells with IL-6 or FFAs (Supplementary Fig. 2a and b). These modifications were greatly suppressed by an shRNA directed against murine cdk5 (Fig. 1c and Supplementary Fig. 2c) or roscovitine, a selective cdk5 inhibitor (Supplementary Fig. 2d). Although cytokines and FFA may affect other phosphorylation sites within PPARγ, these results strongly suggest that the phosphorylation at Ser-273 of PPARγ is occurring specifically through cdk5.
We next asked how the Ser-273 phosphorylation of PPARγ altered the ability of this receptor to affect adipogenesis and gene expression within differentiated adipocytes. Wild-type and S273A mutant alleles of PPARγ had the same transcriptional activity on a PPAR-response element (Supplementary Fig. 3) when expressed in fibroblasts previously engineered to completely lack this receptor (Fig. 1d, Supplementary Fig. 4a)21. These initial experiments took advantage of the considerable serum-induced basal level of cdk5 activity in cultured cells22. While treatment of cells with cytokines and FFAs induced a more robust activation of cdk5, these treatments also lead to de-differentiation of adipocytes23. The mutant and wt PPARγ alleles both drove PPARγ-mediated adipogenesis with equal efficiency (Fig. 1e). While most classical adipocyte-selective genes, like aP2 and _C/EBP_α, were expressed to equal levels, certain transcripts including the key fatty acid transporter cd36, and the adipokines adiponectin, adipsin and leptin were sensitive to mutation of the cdk5 site in PPARγ (Fig. 1e). Mutation of the cdk5 site also increased the secretion of adiponectin into the culture medium (Supplementary Fig. 4b). To understand how regulation of Ser-273 affects expression of adiponectin and other specific genes, we compared the chromatin association of phosphorylated and non-phosphorylated PPARγ (Supplementary Fig. 5). There were no differences in DNA binding by the phosphorylation of PPARγ, suggesting that other factors, such as co-regulator recruitment to PPARγ, may be differentially regulated in a phosphorylation-dependent manner. We next compared the ability of mutant and wild-type PPARγ alleles to alter fat cell gene expression in vivo. Adiponectin and adipsin were markedly dysregulated, being elevated in transplanted fat pads expressing mutant vs. wt PPARγ (Fig. 1f). While both cd36 and leptin were expressed at slightly higher levels in cells expressing mutant receptor, this did not reach statistical significance.
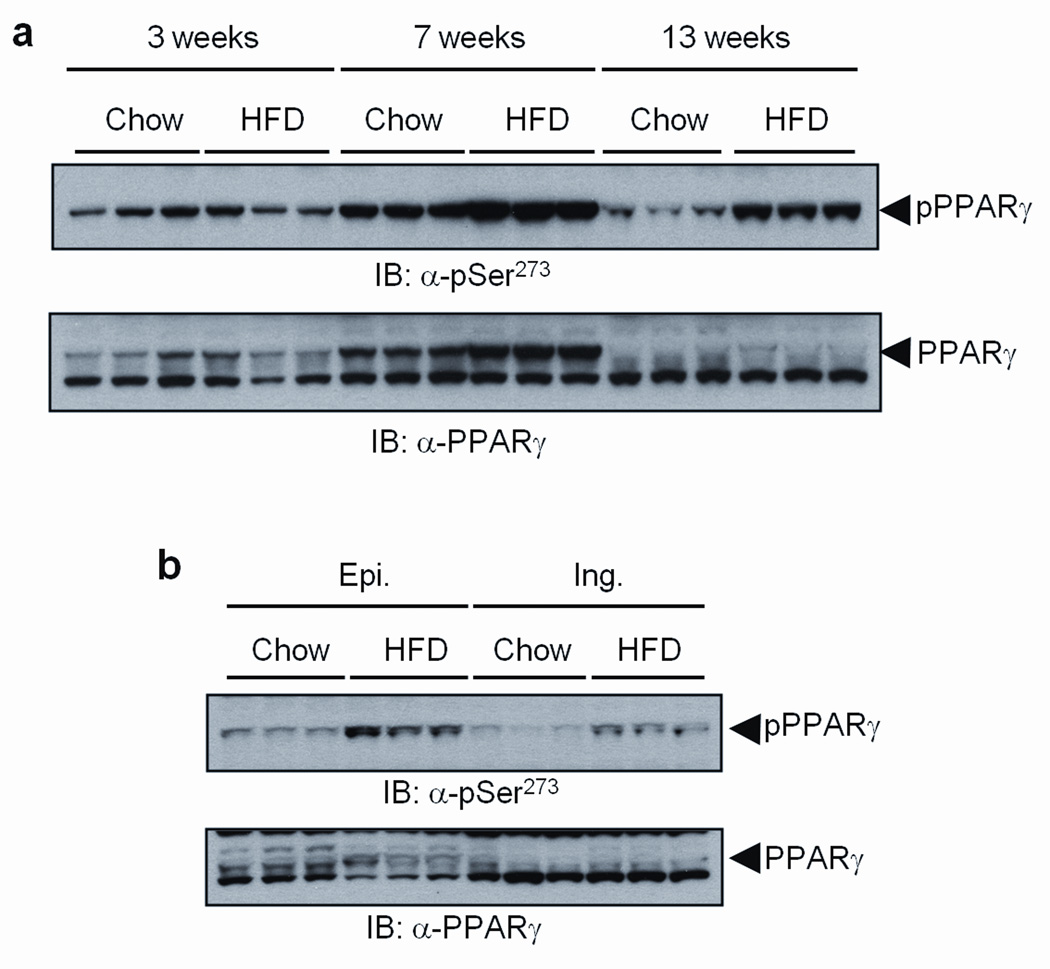
It is notable that adiponectin and adipsin are inappropriately regulated in obesity14,15. This led us to ask whether cdk5 modification of PPARγ is activated in adipose tissues of obese mice. Adipose tissues were harvested at several time points from mice placed on either a standard chow or a high fat/high sugar diet containing 60% kcal from fat. As shown in Supplementary Table 1, overt hyperinsulinemia was apparent at 13 weeks. There was detectable basal levels Ser-273 phosphorylation of PPARγ in chow fed animals, with no increase in this modification after 3 weeks on high fat diet (Fig. 2a, Supplementary Fig. 6a). High-fat feeding for 7 weeks led to increased phosphorylation of PPARγ compared to chow-fed controls, with the difference becoming more pronounced after 13 weeks. Similarly, there was no phosphorylation of cdk5 after 3 weeks of high fat feeding; however, activated cdk5 was easily observed after 7 and 13 weeks of the HFD (Supplementary Fig. 6c) along with increased levels of the cleaved p25 protein, the more stable form of the activating subunit for cdk519. We also compared the cdk5-mediated modification of PPARγ in two different white fat depots: inguinal fat, a type of subcutaneous fat, and epididymal fat, a visceral depot24. Fig. 2b and Supplementary Figs. 6b and d illustrate that the increase in phosphorylation by cdk5 occurred in both white fat depots, with greater intensity in the epididymal depot.
Figure 2. CDK5-mediated phosphorylation of PPARγ is increased in fat tissues of high fat diet fed mice (HFD).
a, White adipose tissue (epididymal) from mice on HFD for the indicated time was analyzed with phospho-S273 PPARγ and PPARγ antibodies. b, Epididymal (Epi.) or inguinal (Ing.) fat tissue from 13 weeks HFD mice was analyzed with phospho-S273 antibody.
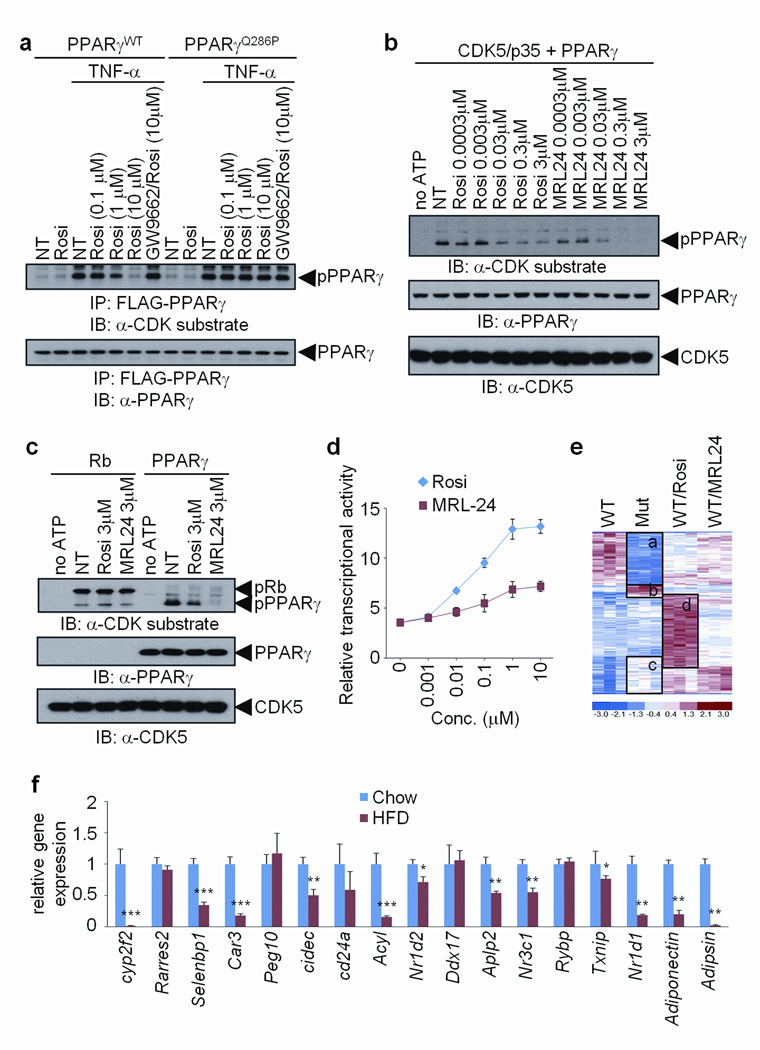
Anti-diabetic drugs of the thiazolidinedione (TZD) class, such as rosiglitazone, are agonist ligands for PPARγ, improving insulin sensitivity in both mice and humans10. To ask whether anti-diabetic PPARγ ligands alter cdk5-mediated phosphorylation of this receptor, we treated fat cells expressing wt PPARγ with TNF-α, rosiglitazone, or a combination of these two agents. Fig. 3a shows that rosiglitazone inhibited Ser-273 phosphorylation at approximately 1 µM, similar to the dose required for other PPARγ-mediated activities in adipose cells13. GW9662, a PPARγ antagonist25, completely blocked this effect of rosiglitazone. We also examined the effect of these compounds on the cdk5-mediated phosphorylation of a naturally occurring mutant form of PPARγ (Q286P) that no longer directly binds known ligands26. Rosiglitazone could not interfere with cdk5 phosphorylation in this case, implying that direct binding of the ligand was required for inhibition. To address whether ligands work directly to inhibit cdk5-mediated phosphorylation, we mixed purified PPARγ, cdk5/p35 and rosiglitazone under conditions to achieve modification in vitro. Rosiglitazone blocked Ser273-phosphorylation in vitro, with a half-maximally effective dose of about 30 nM (Fig. 3b), near the Kd of this compound for PPARγ binding13. Importantly, this inhibition is not caused by a general inhibition of cdk5 activity, since incubation with rosiglitazone did not inhibit the ability of cdk5 to phosphorylate Rb (Fig. 3c).
Figure 3. Anti-diabetic PPARγ ligands block CDK5-mediated phosphorylation of PPARγ.
a, TNF-α-induced phosphorylation of PPARγ in 3T3-L1 adipocytes expressing either WT or Q286P mutant of PPARγ treated with rosiglitazone and/or GW9662. b and c, In vitro CDK5 assay with either rosiglitazone or MRL24. d, Transcriptional activity of a PPAR-derived reporter gene in response to rosiglitazone or MRL24 (n=3). e, Microarray analysis of differentiated PPARγ-null fibroblasts expressing WT (NT, rosiglitazone or MRL24 treated) or S273A mutant PPARγ. f, mRNA expression of genes regulated by the phosphorylation of PPARγ in epididymal fat tissue of mice on either chow or HFD for 13 weeks (n=5). Error bars are s.e.m.; *p<0.05, **p<0.01, ***p<0.001.
MRL24 is a non-TZD compound with high affinity for PPARγ, with excellent anti-diabetic activity in mice18, but poor agonist properties toward PPARγ in transcription and adipogenesis assays. As reported previously18 and shown in Fig. 3d, MRL24 showed weak transcriptional activity compared to rosiglitazone. On the other hand, MRL24 was very effective at blocking cdk5-mediated phosphorylation of PPARγ (Fig. 3b); 30 nM of MRL24 inhibited the modification of PPARγ as effectively as 300 nM of rosiglitazone, both in vitro and in cells (Fig. 3b, Supplementary Figs. 7a and b). We also examined the ability of several other published PPARγ ligands that have been shown to have poor agonist properties, but strong anti-diabetic activities in vivo27–29. As shown in Supplementary Fig. 8, Mbx-102, BVT.13 and nTZDpa were all effective at inhibiting the cdk5-mediated phosphorylation of PPARγ.
To systematically compare changes in gene expression caused by both the Ser-273 mutation in PPARγ and specific PPARγ ligands, we performed Affymetrix analyses of gene expression with RNA from cells expressing either wt or mutant PPARγ, plus wt cells treated with rosiglitazone or MRL24. As shown in Fig. 3e, unsupervised clustering of these data resulted in several notable clusters of expression. The overlap in global regulation of gene expression between the mutant form of PPARγ and the PPARγ ligands is highly significant (p<0.005 for each drug vs. mutation). Genes exhibiting differential expression between wt and mutant PPARγ segregated into four major groups. One group, labeled “a”, were genes whose expression was decreased by Ser-273 mutation. This same group of genes was also suppressed by both rosiglitazone and MRL24, though neither ligand functioned as dramatically as did the non-phosphorylatable mutant of PPARγ. This is understandable as it is unlikely that ligand treatment caused complete dephosphorylation of PPARγ comparable to the mutant protein. The very small cluster labeled “b” contained Ahnak nucleoprotein and proteolipid protein (plp) 1, which are critical for myelination30,31, and sorting nexin 5 (Snx5), which is important for vesicle trafficking32. A larger cluster, labeled “c”, represented genes were decreased by cdk5-mediated phosphorylation of PPARγ. Several of the genes known to be dysregulated in obesity, including adiponectin and adipsin, were present in this cluster. Rosiglitazone increased essentially all of these genes, but Fig. 3e indicates that this occurred as part of a very large gene set increased by rosiglitzone action. Indeed, genes induced most dramatically by rosiglitazone (labeled “d”) did not correspond to the cdk5 mutation-induced gene set and were largely the classic genes of adipogenesis, like aP2 and lipoprotein lipase (Lpl). In sharp contrast, the gene cluster strongly increased by MRL24 was much smaller than that induced by rosiglitazone, and corresponded remarkably well to the gene set induced by the mutation in the cdk5 site of PPARγ.
Taken together, the data presented in Figs. 1–3 suggest that cdk5 modification of PPARγ could be a major source of gene dysregulation and pathology of adipose tissues in obesity. To directly address this, we created refined gene sets regulated by the cdk5 modification of PPARγ, using the concept of principal component analysis of gene expression data (Supplementary Fig. 9). Fig. 3f shows that the great majority of these genes had dysregulated expression in an obesity-dependent manner. In total, of the 17 genes most significantly reduced by cdk5 phosphorylation of PPARγ, at least 12 of these genes were also decreased in obese mice. Interestingly, most of these genes were more significantly impacted in epididymal fat tissue compared to the inguinal depot (Supplementary Fig. 10), consistent with differential phosphorylation of PPARγ in the two depots (Fig. 2b). In addition, shRNA-mediated knock-down of cdk5 in cells resulted in marked dysregulation of the gene sets, particularly adiponectin and adipsin (Supplementary Fig. 11), further confirming that cdk5-mediated phosphorylation of PPARγ plays a significant role in the regulation of specific fat cell genes.
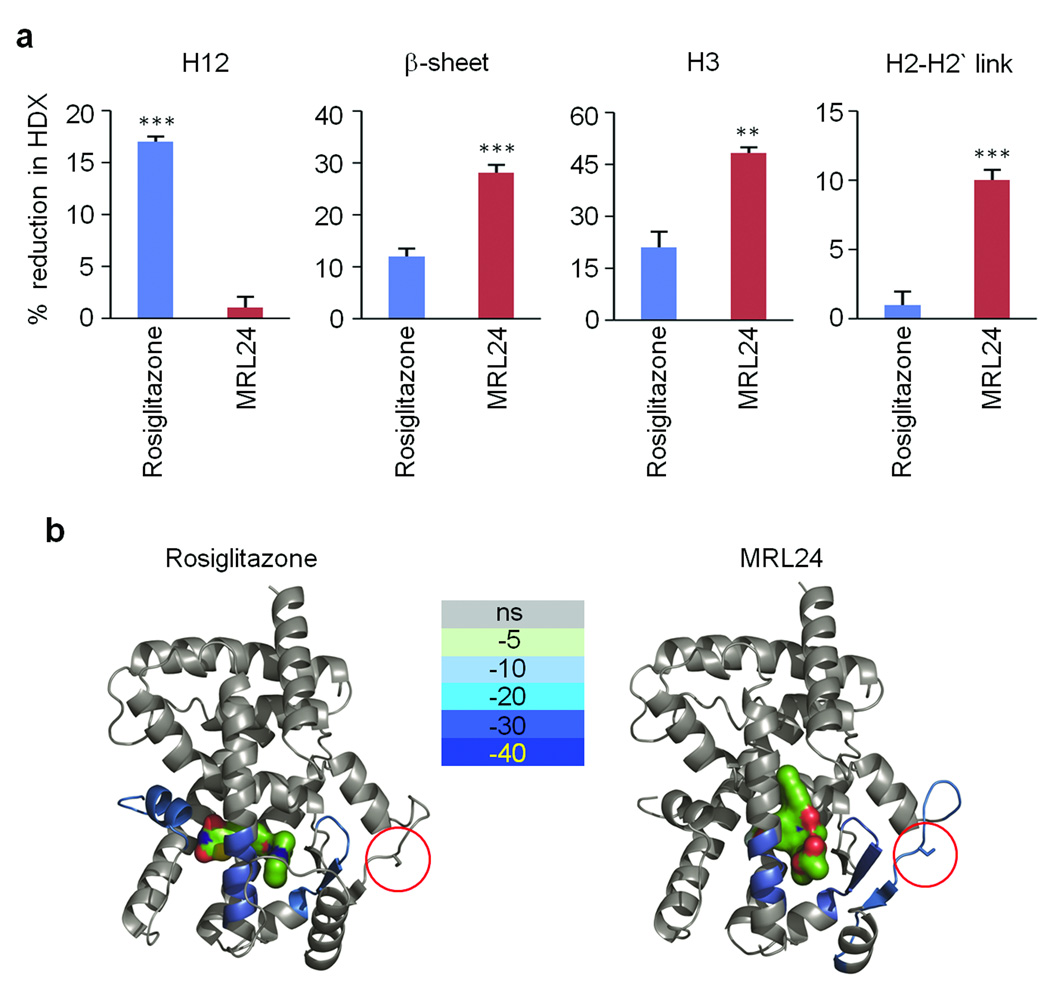
To develop a structural understanding of how PPARγ ligands affect cdk5-mediated phosphorylation of PPARγ, we turned to hydrogen/deuterium exchange (HDX) linked to mass-spectrometry33. As shown in Fig. 4a, rosiglitazone, but not MRL24 significantly reduced H/D exchange in helix 12 (H12), which contains the ligand-binding domain comprising part of the AF-2 surface of the receptor responsible for classical agonism34,35. On the other hand, MRL24 had a more marked impact on H/D exchange kinetics across H3 (aa 309–315), the β-sheet at 369–379, and the cdk5 site itself at Ser-273 in PPARγ. Rosiglitazone also affected the exchange across H3 and the beta sheet region, but did not significantly alter the H/D exchange across Ser-273. When these HDX data are mapped onto the known co-crystal structures of PPARγ with each of these ligands bound, it is clear that MRL24 reduced the dynamic nature of these regions to a greater extent than rosiglitazone (Fig. 4b). It is likely that the ligand-induced reduction in the dynamic nature of H3, the β-sheet, and the cdk5 site, “freezes” this region in a configuration less favorable to cdk5 phosphorylation. Since cdk5 phosphorylation of PPARγ does not affect chromatin occupancy of this receptor (Supplementary Fig. 2), we examined the possibility that the region near the cdk5 site might be an important location for ligand-gated coregulator interactions. As shown in Supplementary Fig. 12, HDX data shows that a model coactivator, SRC3, interacts with a PPARγ region far from helix 12, near Ser-273, when MRL24 is bound. Thus, specific coregulator modulation of PPARγ by modifications at serine 273 is highly plausible.
Figure 4. Differential HDX MS data for PPARγ-LBD ± rosiglitazone and MRL24.
a, Histograms showing the percent reduction in HDX for Helix 3 (IRIFQGCQF), the β-sheet region (ISEGQGFMTRE), Helix 12 (QEIYKDLY) and the Helix 2-2’ link region containing the site of CDK5 phosphorylation (KTTDKSPFVIYDM). Values are calculated relative to the measured %D value for apo PPARγ-LBD (n=4; error bars are s.e.m.; **p<0.01, ***p<0.001). b, HDX data for the four peptides of interest are plotted over the structures of PPARγ-LBD bound with rosiglitazone (left, PDB:2PRG) and MRL24 (right, PDB:2Q5P). Percent reduction in HDX relative to unliganded receptor is colored according to the key. Red circle indicates Ser273 residue of PPARγ.
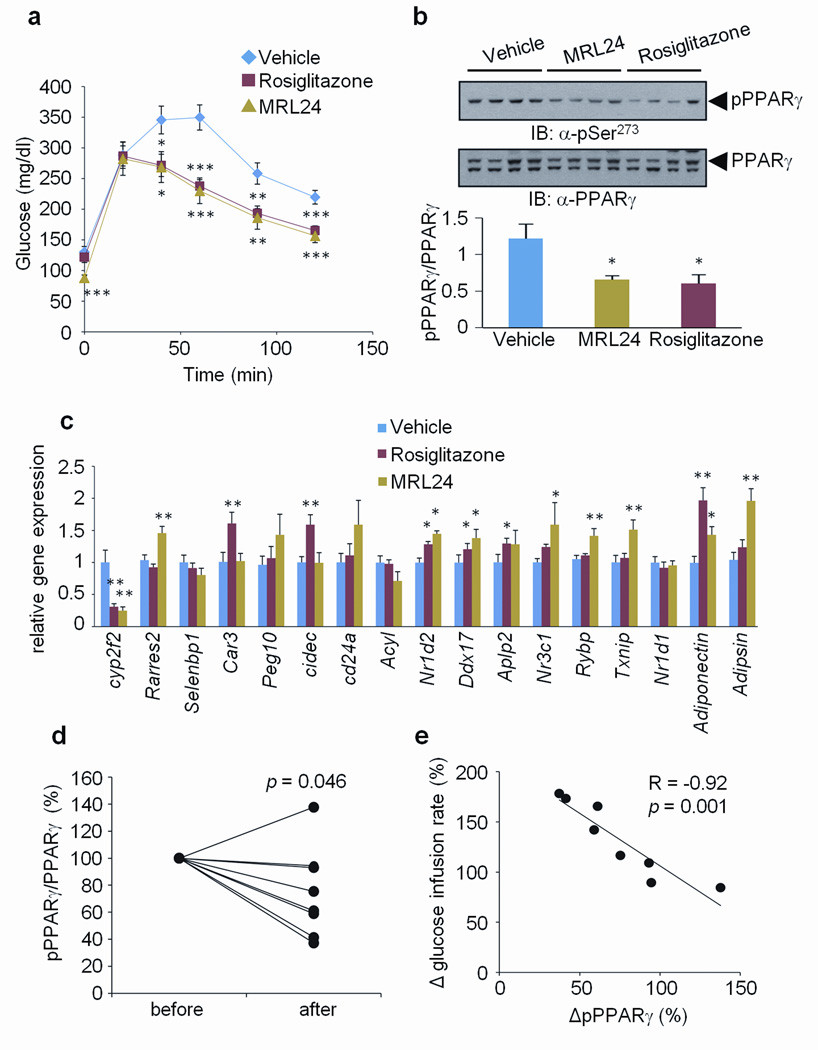
Next, we asked whether the PPARγ ligands alter cdk5-mediated phosphorylation of PPARγ in vivo. As shown in Fig. 5a and Supplementary Fig. 13, treatment with either rosiglitazone or MRL24 at 10 mg/kg for 7 days dramatically improved glucose tolerance of high-fat fed mice and reduced fasting insulin levels without changing body weight. Importantly, both of these compounds reduced cdk5-mediated phosphorylation of PPARγ in the adipose tissue of every mouse treated (Fig. 5b). These drugs did not affect the status of another known phosphorylation site, Ser112 (Supplementary Fig. 14). Furthermore, 12 of the 17 genes most significantly controlled by cdk5 action on PPARγ (as described above) were altered by the action of one or both drugs (Fig. 5c). These data demonstrate that anti-diabetic PPARγ ligands inhibit cdk5 phosphorylation of PPARγ in vivo and reverse changes in gene expression linked to this modification. Consistent with these results, treatment of roscovitine significantly suppressed cdk5-mediated phosphorylation and most of the gene set regulated by the phosphorylation of PPARγ (Supplementary Fig. 15). However, we cannot exclude the possibility that weight loss in the treatment group may be influencing changes in phosphorylation and gene expression.
Figure 5. Correlation between the inhibition of phosphorylation and improvement of insulin sensitivity by anti-diabetic PPARγ ligands.
a, Glucose-tolerance tests in 16-week HFD mice treated with vehicle, rosiglitazone or MRL24 (n=10). b, phosphorylation of PPARγ in WAT. c, The expression of gene sets regulated by PPARγ phosphorylation in WAT (Error bars are s.e.m.; *p<0.05, **p<0.01, ***p<0.001). d, Changes of phosphorylation of PPARγ in human fat biopsies which were treated with rosiglitazone for 6 months. before, non-treated; after, 6 months treatment. e, Correlation between the changes of PPARγ phosphorylation normalized to total PPARγ protein and the changes of glucose infusion rate measured by clamp. The data is presented as percent change after 6 months of rosiglitazone treatment.
Finally, we investigated whether humans undergoing therapy with a PPARγ ligand showed decreased phosphorylation of PPARγ at the cdk5 site in adipose tissue, and how this correlated with improvements in systemic insulin sensitivity. As shown in Fig 5d, Ser-273 phosphorylation of PPARγ was generally decreased in subcutaneous fat biopsies from patients following rosiglitazone treatment. However, one patient displayed increased phosphorylation and two patients showed only a modest decrease. Notably, patients who had little or no reduction in PPARγ phosphorylation also displayed little improvement in clinical parameters such as fasting insulin and fasting glucose with rosiglitazone treatment (Supplementary Tables 2 and 3). To investigate the relationship of these effects to changes in insulin sensitivity, we compared the final, euglycemic, hyperinsulinemic clamped glucose infusion rate with the relative change in PPARγ phosphorylation for each patient. As shown in Fig. 5e, there was a powerful, negative correlation (r=−0.92 and _p_=0.001) between changes in glucose infusion rate and changes in PPARγ phosphorylation. Thus, in this patient cohort, improvement of insulin sensitivity using rosiglitazone was tightly coupled to decreased phosphorylation of PPARγ at the cdk5 site.
Discussion
The data presented here suggest a unified framework for understanding the relationship between fat cell dysfunction in obesity and anti-diabetic therapies through PPARγ (Supplementary Fig. 16). High fat diets in vivo, activate the protein kinase cdk5. Cdk5-mediated phosphorylation of PPARγ at Ser-273 dysregulated expression of a subset of genes including a number of key metabolic regulators including adipsin, the first fat cell-selective gene whose expression is altered in obesity36 and adiponectin, a central regulator of insulin sensitivity in vivo. Ser-273 phosphorylation did not alter the chromatin occupancy of PPARγ, suggesting that other mechanisms, such as differential recruitment of co-regulators, may cause these differences in target gene expression. Anti-diabetic PPARγ ligands inhibited Ser-273 phosphorylation and reversed associated changes in gene expression. Crucially, inhibition of cdk5-mediated phosphorylation of PPARγ by ligands altered the conformation/dynamic nature of the protein, such that Ser-273 was no longer modified by the protein kinase. In fact, a PPARγ ligand with minimal agonist activity, such as MRL24, did this as well or better than a full agonist, such as rosiglitazone. Given that both have potent anti-diabetic activities; it is likely that a substantial portion of the therapeutic benefits of PPARγ ligands in metabolic disease is through inhibition of cdk5-mediated phosphorylation. The studies presented in Fig. 5d and e also indicate that this specific phosphorylation was closely coupled to the therapeutic effects of rosiglitazone in man. It is worth noting that the therapeutic doses used in animals did not lead to a complete inhibition of the cdk5 phosphorylation of PPARγ, so more robust therapy might be obtained with other, more narrowly targeted ligands.
Based on these results, the role of agonism in the actions of PPARγ ligand drugs is quite unclear. It is possible that a small degree of agonism further enhances the beneficial effects of blocking cdk5-mediated phosphorylation of PPARγ. On the other hand, it seems likely that at least some of the problematic side-effects of these drugs, including weight gain and fluid retention16, may occur through classical agonist actions. This can be better assessed as ligands with various degrees of agonism are evaluated in both preclinical models and human subjects. More broadly, the ability of “partial agonists” to alter protein phosphorylation may be true for other members of the nuclear receptor family, possibly allowing for identification of new drugs.
How is cdk5 activated in adipose tissues of obese mice? Both cytokines and fatty acids are elevated in adipose tissues in most obese states, and both may contribute to cdk5 activation in the experiments preformed here. Cytokines and other stimulators of cdk5 typically work via cleavage of the cdk5 partner p35 into a more active and more stable p25 subunit19. While the extracellular stimulator that functions on fat cells as a consequence of high fat feeding is not established, it is clear from Supplementary Fig. 6 that the activation of cdk5 in obesity is accompanied by enhanced formation of p25. Whether high fat feeding and obesity lead to activation of cdk5 in other non-adipose tissues is a very important question, since the negative effects of obesity extend far beyond the metabolic syndrome to cancer and neurodegeneration37,38. Neurodegeneration has been associated with the abnormal activation of cdk5 in the CNS39, so further studies are required to determine if cdk5 is activated in the brain and other tissues in obesity.
Methods Summary
Cell Culture
PPARγ-null mouse embryonic fibroblasts (MEFs)21 were cultured in Dulbecco's modified Eagle's medium and 10% fetal bovine serum. FLAG-PPARγ and FLAG-PPARγ S273A were subcloned into pMSCV-puro retrovial vector (Stratagene). Following infection of the cells with the retrovirus, cells expressing the ectopic protein were selected by incubation with 2 µg/ml puromycin. Adipocyte differentiation on 3T3-L1 or MEFs was induced by treating cells with 1 µM dexamethasone, 0.5 mM isobutylmethylxanthine, and 850 nM insulin for 48 h and cells were switched to the maintenance medium containing 850 nM insulin.
In vitro kinase assay
Immuno-purified WT or S273A mutant of PPARγ were incubated with active cdk kinases in kinase assay buffer containing ATP for 15 min at 30°C. Positive controls, either purified histone H1 (Millipore) or Rb (Cell Signaling Technology) were used. Several PPARγ ligands were pre-incubated with substrates for 30 min before the assay was performed.
Gene expression analysis
Total RNA was isolated from cells or tissues using Trizol reagents (Invitrogen). The RNA was reverse-transcribed using ABI reverse transcription kit. Quantitative PCR reactions were performed with SYBR green fluorescent dye using an ABI9300 PCR machine. Relative mRNA expression was determined by the ΔΔ-Ct method using tata-binding protein (TBP) levels.
Animals
All animal experiments were performed according to procedures approved by Beth Israel Deaconess Medical Center’s Institutional Animal Care and Use Committee. 4 to 5 week-old male C57BL/6J mice were obtained from the Jackson Laboratory. Mice were fed a regular diet (10% kcal fat, D12450B, Research Diets Inc.) or a high fat diet (60% kcal fat, D12492, Research Diets Inc.) as indicated. For glucose tolerance tests, mice were intraperitoneally (i.p.) injected with 10 mg/kg rosiglitazone or MRL24 for 6 days, and fasted overnight before i.p. injection of 2 g/kg D-glucose.
Methods
Cell Culture
3T3-L1 and HEK-293 cells were obtained from ATCC. PPARγ-null mouse embryonic fibroblasts (MEFs)21 were cultured in Dulbecco's modified Eagle's medium and 10% fetal bovine serum. FLAG-PPARγ and FLAG-PPARγ S273A were subcloned into pMSCV-puro retroviral vector (Stratagene). For retrovirus production, Phoenix packaging cells were transfected with 10µg retroviral vectors40. After 48h, the viral supernatant was collected and filtered. Following infection of the cells with the retrovirus, cells expressing the ectopic protein were selected by incubation with 2 µg/ml puromycin. Adipocyte differentiation on 3T3-L1 or MEFs was induced by treating cells with 1 µM dexamethasone, 0.5 mM isobutylmethylxanthine, and 850 nM insulin for 48h and cells were switched to the maintenance medium containing 850 nM insulin. Lipid accumulation in the cells was detected by Oil Red O staining. The amount of secreted adiponectin into cell medium was analyzed by ELISA (Millipore). All chemicals for cell culture were obtained from Sigma unless otherwise indicated.
DNA constructs and shRNA of CDK5
HA-WT CDK5, HA-KD CDK5 and Myc-p35 were obtained from Addgene. Murine PPARα or PPARδ were subcloned into FLAG-pcDNA3.1 (Invitrogen). The sequence used for lentivial shRNA expression vector (pLKO.1; Open Biosystems) targeting CDK5 was 5’-CGGGAGATCTGTCTACTCAAA-3’. For lentivirus production, HEK-293T cells (ATCC) were transfected with 10 µg lentiviral vectors40. Following infection of the cells with the lentivirus, cells were selected by incubation with 2 µg/ml puromycin.
In vitro kinase assay
Active cdk5/p35, cdk1/cdc2, cdk2/cyclin A, cdk2/cyclin E or cdk4/cyclin D1 kinases were purchased from Millipore or Cell Signaling Technology. In vitro CDK kinase assay was performed according to the manufacturer's instructions (Cell Signaling Technology). Briefly, 1 µg of immuno-purified WT or S273A mutant of PPARγ were incubated with active CDK kinase in kinase assay buffer (25 mM Tris-HCl pH 7.5, 5 mM beta-glycerophosphate, 2 mM dithiothreitol (DTT), 0.1 mM Na3VO4, 10 mM MgCl2) containing 20 µM ATP for 15 min at 30°C. Positive control for assay, purified histone H1 (Millipore) or Rb (cell signaling technology) were used. Several PPARγ ligands were pre-incubated with substrates for 30 min, and the assay was performed. Phosphorylation of substrates after SDS–PAGE was analyzed with anti-CDK substrate antibody to detect phospho-Ser in a K/R-S-P-K/R motif which is the concensus motif for cdk substrate proteins (Cell Signaling Technology).
Preparation of cell or tissue lysates, immunoprecipitation and immunoblotting
HEK-293 cells expressing CDK5 or PPARγ were collected after transfection. Total cell lysates were incubated with FLAG M2 agarose (Sigma) at 4°C. Immunoprecipitates or total cell lysates were analyzed with anti-CDK substrate, FLAG or HA (Roche) antibodies. Differentiated 3T3-L1 adipocytes were treated with TNF-α (50 ng/ml), IL-6 (50 ng/ml), IL-1β (50 ng/ml) or FFAs (400 µM palmitic acid and oleic acid mixtures) for the indicated times, and cell lysates were analyzed with phospho-specific or PPARγ antibodies. 3T3-L1 adipocytes were pre-treated with various PPARγ ligands, and incubated with TNF-α. For tissue lysates, WAT from mice was homogenized in RIPA buffer (50 mM Tris pH7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS with protease and phosphatase inhibitors). For western blotting, a rabbit polyclonal phospho-specific antibody against PPARγ Ser273 was produced by New England Peptides with a synthetic phosphopeptide corresponding to residues surrounding Ser273 of PPARγ (Ac-KTTDKpSPFVIYDC-amide). Total tissue lysates were analyzed with anti-PPARγ, phospho-CDK5 (Y15), CDK5 and p35 antibodies (Santa Cruz).
Reporter gene assay
HEK-293 cells were transfected with pDR-1 luciferase reporter plasmid, PPARγ, RXRa and pRL-renillin using Lipofectamine 2000 (Invitrogen). Following an overnight transfection, the cells were treated with rosiglitazone or MRL24 for 24h. The cells were harvested and reporter gene assays were carried out using the Dual-Luciferase kit (Promega). Luciferase activity was normalized to renillia activity.
Gene expression analysis
Total RNA was isolated from cells or tissues using Trizol reagents (Invitrogen). The RNA was reverse-transcribed using ABI reverse transcipton kit. Quantitative PCR reactions were performed with SYBR green fluorescent dye using an ABI9300 PCR machine. Relative mRNA expression was determined by the ΔΔ-Ct method normalized to tata-binding protein (TBP) levels. The sequences of primers used in this study are found in Supplementary Table 4.
Generation of fat pads in nude mice
PPARγ-null fibroblasts (1 × 107) stably expressing WT or S273A mutant of PPARγ were implanted subcutaneously into 7–8 week-old male NCR nude mice (Taconic) according to the previous methods (n=5 mice per group)41. 6 weeks after injection, the fat pads were isolated for the analysis of gene expression.
Microarray analysis
Total RNA was isolated from PPARγ-null fibroblasts expressing WT or S273A mutant of PPARγ or WT cells treated with 1µM rosiglitazone or MRL24 for 24h. Array hybridization and scanning were performed by the Dana-Farber Cancer Institute Microarray Core Facility using Affymetrix GeneChip Mouse Genome 430 2.0 arrays according to established methods42. The array data were analyzed using the DNA-Chip Analyzer (dChip) sofware43. The statistical significance of differences in gene expression was assessed by an unpaired _t_-test (p<0.05). Global overlap of genes was statistically assessed using Chi-Square statistics. To create refined gene sets regulated by cdk5 phopshorylation of PPARγ, we first calculated _p_-value as well as fold-change of gene expression in WT versus S273A mutant cells, and we plotted -log _p_-value versus log2 fold-change. From this list of genes, we selected 53 genes which were changed in magnitude (≥1.4 fold difference) and statistical significance (p<0.05). The selected genes were validated in cells or transplanted fat pads using by qPCR, the resulting gene set, consisting of 17 genes, was analyzed in WAT of mice using qPCR.
Animals
All animal experiments were performed according to procedures approved by Beth Israel Deaconess Medical Center’s Institutional Animal Care and Use Committee. 4 to 5-week-old male C57BL/6J mice were obtained from the Jackson Laboratory. Mice were fed a regular diet (10% kcal fat, D12450B, Research Diets Inc.) or a high fat diet (60% kcal fat, D12492, Research Diets Inc.) as indicated time periods. All mice initiated the HFD at the same age (4–5 week old), and harvested them at the indicated HFD time. For glucose tolerant tests, mice were intraperitoneally (i.p.) injected daily 10 mg/kg rosiglitazone or MRL24 for 6 days, and fasted overnight prior to i.p. injection of 2 g/kg D-glucose. Glucose was measured in tail vain blood at intervals after glucose injection using a Truetrack glucometer. Serum insulin concentrations were determined by ELISA (Crystal Chem).
Human study
We utilized fat biopsies from subjects enrolled in a parallel-group randomized and controlled trial to evaluate the effects of rosiglitazone and an exercise program on insulin sensitivity and endothelial function in patients with impaired glucose tolerance over a 6-month period. Consecutive patients referred to the University of Leipzig Heart Center, Germany, were invited to participate if they displayed impaired glucose tolerance (2h OGTT plasma glucose >7.8 and <11.1 mmol/l)44. Exclusion criteria included diabetes type 1 or 2, concomitant medication, unstable angina, coronary artery disease, myocardial infarction within preceding 3 months, ejection fraction <40%, significant heart valve disease, severe metabolic disorders, severe disorders in lipoprotein metabolism, thyroid disorders, alcohol or drug abuse, pregnancy, and participation in another trial. Included patients received 4 mg rosiglitazone daily (Avandia; GlaxoSmithKline, London, U.K.) for 6 months. At day 1 and after 6 months, patients were clinically examined, euglycemic-hyperinsulinemic clamps were performed, and blood samples were obtained. All baseline blood samples were collected between 8 and 10 am after an 12 hours overnight fast. Plasma insulin was measured with an enzyme immunometric assay for the IMMULITE automated analyzer (Diagnostic Products Corporation, Los Angeles, CA, USA). Serum total- HDL-, LDL-cholesterol, triglycerides, free fatty acids, adiponectin, were measured as previously described45. Insulin sensitivity was assessed with the euglycaemic-hyperinsulinaemic clamp method using a previously described protocol46. Fat biopsies were analyzed with anti-phospho-Ser273 and PPARγ antibody. Statistical analyses were performed using SPSS statistics 17.0 and Pearson’s two-sided correlation was used for statistical tests. All correlations with p<0.05 yielded similar results using Spearman´s rank correlation.
Supplementary Material
1
Acknowledgments
We thank to V. Mootha for help with the analysis of microarray data and critical comments. We thank M. Kirschner for a critical reading and comments of the manuscript. We are grateful to R. Gupta and P. Cohen for their critical comments on the manuscript. J.H.C., A.S.B., J.L.E., P.B., D.L., and B.M.S are supported by NIH DK31405. S.K. is supported by NIH grant DK087853. M.B. is supported by a grant from Deutsche Forschungsgemeinschaft (DFG) the Clinical Research group “Atherobesity” KFO 152 (project BL 833/1-1 to MB). P.R.G., M.J.C., and T.M.K. are supported in part by the by the Intramural Research Program of the National Institutes of Health National Institute of Mental Health [P.R.G., M.J.C., T.M.K. Grant U54-MH084512; H. Rosen PI], by the National Institutes of Health National Institute of General Medical Sciences [P.R.G., M.J.C. Grant R01-GM084041].
Footnotes
Conflicting interests statement. The authors declare that they have no competing financial interests.
Author Contributions
J.H.C. and B.M.S. conceived and designed the experiments. J.H.C., A.S.B., J.L.E., S.K., P.B., D.L., M.J.C., T.M.K, M.B., P.R.G performed the experiments. All authors analyzed the data. J.H.C., A.S.B., J.L.E., and B.M.S. wrote the manuscript.
Supplementary Information contains Supplementary Figures 1–16 and Tables 1–4. Microarray data has been deposited in Gene Expression Omnibus (GEO): GSE22033.
References
- 1.Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004;89:2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- 2.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 3.Lagathu C, Yvan-Charvet L, Bastard JP, et al. Long-term treatment with interleukin-1beta induces insulin resistance in murine and human adipocytes. Diabetologia. 2006;49:2162–2173. doi: 10.1007/s00125-006-0335-z. [DOI] [PubMed] [Google Scholar]
- 4.Steppan CM, Bailey ST, Bhat S, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 5.Berg AH, Combs TP, Du X, et al. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 6.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 1996;271:10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi T, Kamon J, Waki H, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 8.Morrison RF, Farmer SR. Hormonal signaling and transcriptional control of adipocyte differentiation. J. Nutr. 2000;130:3116S–3121S. doi: 10.1093/jn/130.12.3116S. [DOI] [PubMed] [Google Scholar]
- 9.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 10.Willson TM, Lambert MH, Kliewer SA. Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu. Rev. Biochem. 2001;70:341–367. doi: 10.1146/annurev.biochem.70.1.341. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez MA, Akerblad P, Sigvardsson M, et al. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol. Cell. Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, Bucher NL, Farmer SR. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol. Cell. Biol. 1996;16:4128–4136. doi: 10.1128/mcb.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann JM, Moore LB, Smith-Oliver TA, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 14.Trujillo ME, Scherer PE. Adipose tissue-derived factors: impact on health and disease. Endocr. Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 15.Sharma AM, Staels B. Review: Peroxisome proliferator-activated receptor gamma and adipose tissue--understanding obesity-related changes in regulation of lipid and glucose metabolism. J. Clin. Endocrinol. Metab. 2007;92:386–395. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- 16.Lipscombe LL, Gomes T, Levesque LE, et al. Thiazolidinediones and Cardiovascular Outcomes in Older Patients With Diabetes. JAMA. 2007;298:2634–2643. doi: 10.1001/jama.298.22.2634. [DOI] [PubMed] [Google Scholar]
- 17.Willson TM, Cobb JE, Cowan DJ, et al. The Structure−Activity Relationship between Peroxisome Proliferator-Activated Receptor Î3 Agonism and the Antihyperglycemic Activity of Thiazolidinediones. J. Med. Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 18.Acton JJ, 3rd, Black RM, Jones AB, et al. Benzoyl 2-methyl indoles as selective PPARgamma modulators. Bioorg. Med. Chem. Lett. 2005;15:357–362. doi: 10.1016/j.bmcl.2004.10.068. [DOI] [PubMed] [Google Scholar]
- 19.Dhavan R, Tsai LH. A decade of CDK5. Nat. Rev. Mol. Cell. Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 20.Utreras E, Futatsugi A, Rudrabhatla P, et al. Tumor necrosis factor-alpha regulates cyclin-dependent kinase 5 activity during pain signaling through transcriptional activation of p35. J. Biol. Chem. 2009;284:2275–2284. doi: 10.1074/jbc.M805052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen ED, Hsuq CH, Wang X, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musa FR, Takenaka I, Konishi R, et al. Effects of luteinizing hormone, follicle-stimulating hormone, and epidermal growth factor on expression and kinase activity of cyclin-dependent kinase 5 in Leydig TM3 and Sertoli TM4 cell lines. J. Androl. 2000;21:392–402. [PubMed] [Google Scholar]
- 23.Torti FM, Dieckmann B, Beutler B, et al. A macrophage factor inhibits adipocyte gene expression: an in vitro model of cachexia. Science. 1985;229:867–869. doi: 10.1126/science.3839597. [DOI] [PubMed] [Google Scholar]
- 24.Cinti S. The adipose organ. Prostaglandins Leukot Essent Fatty Acids. 2005;73:9–15. doi: 10.1016/j.plefa.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Leesnitzer LM, Parks DJ, Bledsoe RK, et al. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry. 2002;41:6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- 26.Sarraf P, Mueller E, Smith WM, et al. Loss-of-Function Mutations in PPAR[gamma] Associated with Human Colon Cancer. Mol. Cell. 1999;3:799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- 27.Berger JP, Petro AE, Macnaul KL, et al. Distinct properties and advantages of a novel peroxisome proliferator-activated protein [gamma] selective modulator. Mol. Endocrinol. 2003;17:662–676. doi: 10.1210/me.2002-0217. [DOI] [PubMed] [Google Scholar]
- 28.Gregoire FM, Zhang F, Clarke HJ, et al. MBX-102/JNJ39659100, a novel peroxisome proliferator-activated receptor-ligand with weak transactivation activity retains antidiabetic properties in the absence of weight gain and edema. Mol. Endocrinol. 2009;23:975–988. doi: 10.1210/me.2008-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostberg T, Svensson S, Selen G, et al. A new class of peroxisome proliferator-activated receptor agonists with a novel binding epitope shows antidiabetic effects. J. Biol. Chem. 2004;279:41124–41130. doi: 10.1074/jbc.M401552200. [DOI] [PubMed] [Google Scholar]
- 30.Hakak Y, Walker JR, Li C, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc. Natl. Acad. Sci. U S A. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salim C, Boxberg YV, Alterio J, et al. The giant protein AHNAK involved in morphogenesis and laminin substrate adhesion of myelinating Schwann cells. Glia. 2009;57:535–549. doi: 10.1002/glia.20782. [DOI] [PubMed] [Google Scholar]
- 32.Merino-Trigo A, Kerr MC, Houghton F, et al. Sorting nexin 5 is localized to a subdomain of the early endosomes and is recruited to the plasma membrane following EGF stimulation. J. Cell Sci. 2004;117:6413–6424. doi: 10.1242/jcs.01561. [DOI] [PubMed] [Google Scholar]
- 33.Maier CS, Deinzer ML. Protein conformations, interactions, and H/D exchange. Methods Enzymol. 2005;402:312–360. doi: 10.1016/S0076-6879(05)02010-0. [DOI] [PubMed] [Google Scholar]
- 34.Nolte RT, Wisely GB, Westin S, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 35.Bruning JB, Chalmers MJ, Prasad S, et al. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 36.Flier JS, Cook KS, Usher P, et al. Severely impaired adipsin expression in genetic and acquired obesity. Science. 1987;237:405–408. doi: 10.1126/science.3299706. [DOI] [PubMed] [Google Scholar]
- 37.Calle EE, Rodriguez C, Walker-Thurmond K, et al. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 38.Whitmer RA, Gunderson EP, Barrett-Connor E, et al. Obesity in middle age and future risk of dementia: a 27 year longitudinal population based study. BMJ. 2005;330:1360. doi: 10.1136/bmj.38446.466238.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patrick GN, Zukerberg L, Nikolic M, et al. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- 40.Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 41.Walkey CJ, Spiegelman BM. A functional peroxisome proliferator-activated receptor-gamma ligand-binding domain is not required for adipogenesis. J. Biol. Chem. 2008;283:24290–24294. doi: 10.1074/jbc.C800139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 43.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno-Navarrete JM, Martinez-Barricarte R, Catalan V, et al. Complement factor H is expressed in adipose tissue in association with insulin resistance. Diabetes. 59:200–209. doi: 10.2337/db09-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kloting N, Graham TE, Berndt J, et al. Serum retinol-binding protein is more highly expressed in visceral than in subcutaneous adipose tissue and is a marker of intra-abdominal fat mass. Cell Metab. 2007;6:79–87. doi: 10.1016/j.cmet.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Bluher M, Unger R, Rassoul F, et al. Relation between glycaemic control, hyperinsulinaemia and plasma concentrations of soluble adhesion molecules in patients with impaired glucose tolerance or Type II diabetes. Diabetologia. 2002;45:210–216. doi: 10.1007/s00125-001-0723-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1