TDP-43 regulates its mRNA levels through a negative feedback loop (original) (raw)
TDP-43 regulates its mRNA levels through a negative feedback loop
TAR DNA-binding protein-43 is a heterogeneous nuclear ribonucleoprotein involved in RNA metabolism, its aberrant localization and modification is also associated with a number of neurodegenerative diseases. Here, the TAR DNA-binding protein-43 protein negatively regulates its own mRNA levels, which may have consequences for pathogenesis.
Keywords: 3′ UTR, hnRNP, mRNA stability, neurodegeneration, RNA–protein interaction
Abstract
TAR DNA-binding protein (TDP-43) is an evolutionarily conserved heterogeneous nuclear ribonucleoprotein (hnRNP) involved in RNA processing, whose abnormal cellular distribution and post-translational modification are key markers of certain neurodegenerative diseases, such as amyotrophic lateral sclerosis and frontotemporal lobar degeneration. We generated human cell lines expressing tagged forms of wild-type and mutant TDP-43 and observed that TDP-43 controls its own expression through a negative feedback loop. The RNA-binding properties of TDP-43 are essential for the autoregulatory activity through binding to 3′ UTR sequences in its own mRNA. Our analysis indicated that the C-terminal region of TDP-43, which mediates TDP-43–hnRNP interactions, is also required for self-regulation. TDP-43 binding to its 3′ UTR does not significantly change the pre-mRNA splicing pattern but promotes RNA instability. Moreover, blocking exosome-mediated degradation partially recovers TDP-43 levels. Our findings demonstrate that cellular TDP-43 levels are under tight control and it is likely that disease-associated TDP-43 aggregates disrupt TDP-43 self-regulation, thus contributing to pathogenesis.
Introduction
The TAR DNA-binding protein (TDP-43) is a highly conserved heterogeneous nuclear ribonucleoprotein (hnRNP). Like other members of this family, it has been linked to different aspects of RNA processing (reviewed in Buratti and Baralle, 2008). Its function as a regulator of splicing is the one characterized in best detail, wherein TDP-43 recruitment to 3′ splice sites, rich in GU repeats inhibits exon recognition (Buratti et al, 2001). The abnormal cellular distribution and post-translational modification of TDP-43 are key markers for a group of neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U) (Arai et al, 2006; Neumann et al, 2006), now also referred to as TDP-43 proteinopathies. Accumulation of TDP-43 in ubiquitin-positive insoluble inclusions was subsequently found in a range of neurodegenerative pathologies, such as in some Alzheimer's disease (AD) cases (Amador-Ortiz et al, 2007; Higashi et al, 2007; Uryu et al, 2008) and in additional types of dementia (Higashi et al, 2007; Nakashima-Yasuda et al, 2007; Neumann et al, 2007; Geser et al, 2008). The connection between TDP-43 and disease was further strengthened by the isolation of TDP-43 mutations in ∼5% of patients with inherited and sporadic forms of ALS (Pesiridis et al, 2009). TDP-43 aggregation occurs in the cytoplasm and nuclear compartments of neurons and glial cells, and is often accompanied by nuclear clearance of the protein. TDP-43 isolated from TDP-43 proteinopathies is often modified through extensive ubiquitination, phosphorylation, and aberrant proteolysis (Neumann et al, 2006). The role of TDP-43 in pathogenesis is still unknown, but the lethal and paralytic phenotypes resulting from knockout transgenic mouse and fly models, respectively, highlight the importance of TDP-43 function (Feiguin et al, 2009; Wu et al, 2010). It remains to be seen whether the formation of TDP-43 inclusion bodies is toxic to cells per se, whether the TDP-43 insoluble aggregates sequester the protein resulting in cellular TDP-43 loss of function, or whether both phenomena contribute to pathogenicity.
Two RNA-binding domains, RRM1 and RRM2, are present in TDP-43 of which RRM1 is necessary and sufficient for nucleic acid-binding activity (Buratti and Baralle, 2001). The protein binds single-stranded RNA with high specificity for GU-rich sequences, measured in the low nanomolar range (Ayala et al, 2005). Phe 147 and 149 are located in the RNP-1 sequence of RRM1 forming a canonical binding platform for nucleic acid association (NMR structure:pdb2cqg). Substitution of these two key amino acids is sufficient to abolish RNA-binding and TDP-43 splicing regulatory activity (Buratti and Baralle, 2001; D'Ambrogio et al, 2009). The function of RRM2 is still unclear and does not appear to have a significant role in RNA interaction. In fact, its RNA-binding affinity is two orders of magnitude lower than that of RRM1 (Kuo et al, 2009). The splicing activity of TDP-43 also depends on the integrity of the glycine-rich C-terminus. More specifically, residues 321–366 within this domain recruit additional hnRNPs of the hnRNP-A and hnRNP-C families (D'Ambrogio et al, 2009).
We now show that TDP-43 controls its own homoeostasis in human cells by downregulating TDP-43 transcript levels. This control requires TDP-43 RNA-binding activity through the association to specific sequences in the 3′ UTR of the TDP-43 transcript as well as the presence of the 321–366 amino acid region. The autoregulatory mechanism seems to at least partly involve an exosome-mediated pathway of mRNA degradation.
Results
Induced expression of TDP-43 downregulates endogenous levels of the protein
We generated a HEK293 human kidney cell line to stably express a single copy of the TDP-43 cDNA on induction of the tetracycline promoter using the Flp-In technology (Invitrogen) as described (Glatter et al, 2009). The protein was tagged at the N-terminus either with a double tag (SH-TDP43) or with a FLAG tag (F-TDP43). Induced expression of the tagged transgenic forms of TDP-43 (TG-TDP43) resulted in a dramatic decrease in the levels of endogenous protein expression as seen by western blot (see Figure 1 WT). The same was not true on expression of TG-GFP in a similarly generated HEK293 cell line used as control (data not shown). After 24 h of tetracycline treatment, the levels of induced protein were similar to endogenous levels in untreated cells, consistent with the single-site integration provided by the Flp-In system. Longer times of induction (72 h) resulted in higher levels of TG-TDP43 and the consequent total depletion of endogenous TDP-43.
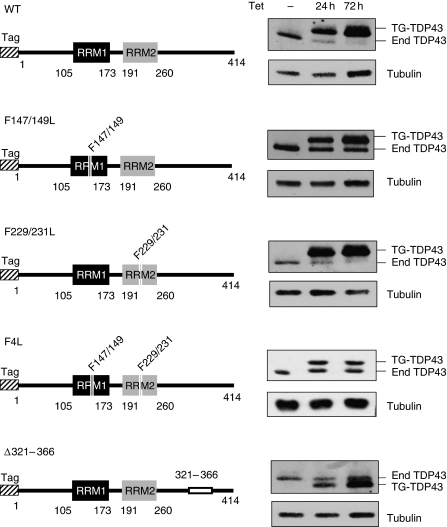
Figure 1.
Induced expression of wild-type and mutant TDP-43 in HEK293 cells. Tagged forms of TDP-43 (F-TDP43) were expressed for 24 or 72 h on tetracycline (Tet) induction (1 μg/ml). The diagrams on the left are a schematic representation of the different TDP-43 protein domains, numbering denotes amino acid residues. The N-terminal domain tag is represented by the hashed box and the RNA recognition motifs 1 and 2 are shown as black and grey boxes, respectively. Deletion of the C-terminal region in mutant Δ321–366 is depicted as a white box. The right panels are immunoblot analyses of the protein extracts with an antibody against TDP-43 that detects both endogenous (end) and tagged (TG-TDP43) protein. Tubulin was used as loading control.
The RRM1 and C-terminal domain of TDP-43 are necessary for autoregulation
To begin addressing the mechanism of self-regulation by TDP-43, we examined the importance of key TDP-43 regions (Figure 1). The variants were expressed as FLAG-tagged TDP-43 in the HEK293 Flp-In system. RRM1 and RRM2 were disrupted by double-site mutations F147/149L and F229/231L, respectively, and both RNA-binding domains were simultaneously targeted with the mutations F147/149/229/231L (F4L). In addition, the 321–366 region was deleted from the C-terminal domain (Δ321–366). Western blot analysis revealed that the RRM1 mutant failed to inhibit endogenous TDP-43 expression even after 72 h of induction (Figure 1 F147/149L), as did the mutant wherein both RRM1 and RRM2 were disrupted (F4L). The TDP-43 carrying mutations in RRM2 only (F229/231L), however, had no effect on the inhibition of endogenous TDP-43. Finally, in the case of the C-terminal deletion, the expression of variant Δ321–366 did not change endogenous TDP-43 levels after 72 h and failed to do so even after 120 h of transgene induction (Figure 1 and data not shown). These results indicate that autoregulation is mediated by an RNA–TDP-43 interaction and requires the recruitment of additional factors through the 321–366 C-terminal region.
TDP-43 self-regulation occurs at the transcript level
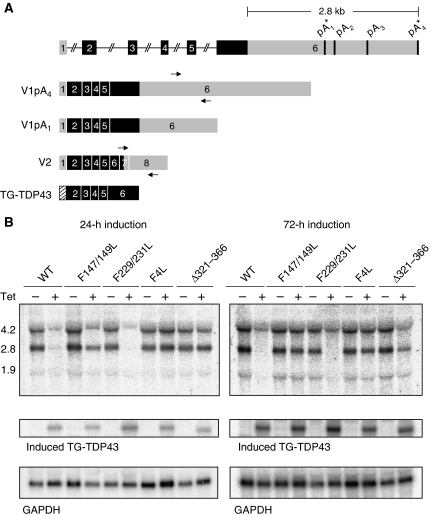
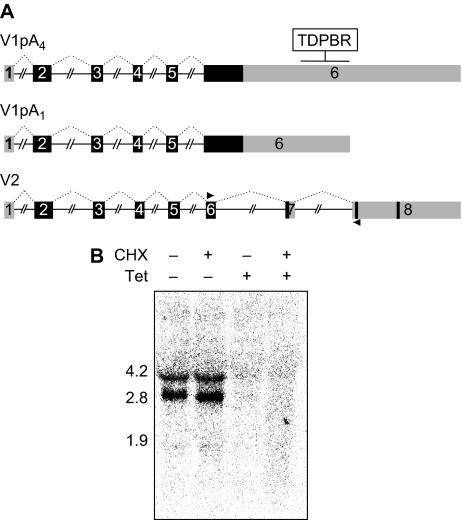
We next investigated whether TDP-43 overexpression similarly repressed endogenous transcript levels through northern blot analysis. Our experiments, using a probe specific for endogenous TDP-43 (indicated by arrows in Figure 2A), identified two main isoforms that correspond to the differential use of polyadenylation sites. Bioinformatic analysis of the 3′ UTR of TDP-43 predicted four polyadenylation signals located in the second half of the 3′ UTR (Figure 2A). Analysis of the TDP-43 ESTs reported in the database (UC Santa Cruz Genome Browser), however, indicated the near exclusive usage of polyadenylation sites labelled as pA1 and pA4 with predicted molecular weights of 2.8 and 4.2 Kb, respectively. Also found in the EST database is a minor form, we refer to as V2, that has a smaller molecular weight because of intron activation at the 3′ end of the gene. The two main isoforms detected in our northern blot assays (V1) are compatible with the use of pA1 and pA4 (Figure 2B, first lane from left, WT, Tet −).
Figure 2.
TDP-43 negative autogenous regulation occurs at the transcript level. (A) Schematic representation of the TDP-43 gene with coding exons and untranslated regions shown in black and grey, respectively. The various predicted polyadenylation signals are indicated, where (*) denotes the experimentally validated sites. The two principally expressed endogenous isoforms (V1pA1 and V1pA4) are shown including the transgenic TG-TDP43 transcript containing the coding sequences only. The V2 form corresponds to a minor isoform discussed in Supplementary Figure 2. (B) Northern blot analysis of control and after 24 and 72 h of induction. V1pA1 and V1pA4 correspond to the 2.8 and 4.2 Kb bands, respectively. Endogenous TDP-43 transcripts were detected by probing the region indicated by the arrows in (A), while TG-TDP43 was identified with a probe spanning exons 2 and 3. GAPDH detection was used as loading control.
A comparison of the endogenous TDP-43 transcript levels as a function TG-TDP43 expression by northern blot analysis showed a striking reduction of TDP-43 mRNA isoforms on tetracycline induction (Figure 2B second lane, WT, Tet +). Mutations in RRM1 (F147/149L and F4L) and the Δ321–366 deletion prevented significant regulation of endogenous TDP-43 mRNA after 24 and 72 h of TG-TDP43 expression (Figure 2B). Again, the RRM2 mutant was not different from wild-type. These findings are in agreement with the immunoblot results showing the requirement of RRM1 and the 321–366 region for the negative self-regulation of TDP-43.
TDP-43 autoregulation involves 3′ UTR-binding elements
The accumulation of TG-TDP43 over time observed in our western blot analyses (Figure 1) suggested that the inducible TDP-43, unlike endogenous TDP-43, was not subject to self-regulation through a negative feedback loop. A significant structural difference between endogenous and transgenic TDP-43 transcripts is the lack of the corresponding 3′ UTR in TG-TDP43 (Figure 2A), implying that this region may contain important regulatory elements. Of course, additional important differences exist between the wild-type transgene and the endogenous TDP-43 gene (i.e., presence of introns and different promoters), however, we first focused on the 3′ UTR region for several reasons. First of all, regarding the absence of introns, it must be taken into account that the 3′ UTR region of TDP-43 is highly conserved among different species while none of the intronic regions share an even similar degree of conservation (see Supplementary Figure 1). We also know that evolutionary conservation is probably very important for autoregulation as three heterozygous transgenic knockout mouse models have shown that +/− mice were observed to produce the same amount of TDP-43 protein and mRNA as the wild-type +/+ mice (Kraemer et al, 2010; Sephton et al, 2010; Wu et al, 2010). Second, regarding the use of different promoters, it should be considered that autoregulation is still observed in a heterologous reporter construct that uses a different promoter, which will be discussed later. Finally, and as a side note, the use of two different systems to produce stable cell lines also rules out an artifactual effect mediated by the protein tag sequences attached to the transgenic TDP-43 as these differed from each other in either system: FLAG and streptavidin-binding peptide fused to a hemagglutinin epitope tag.
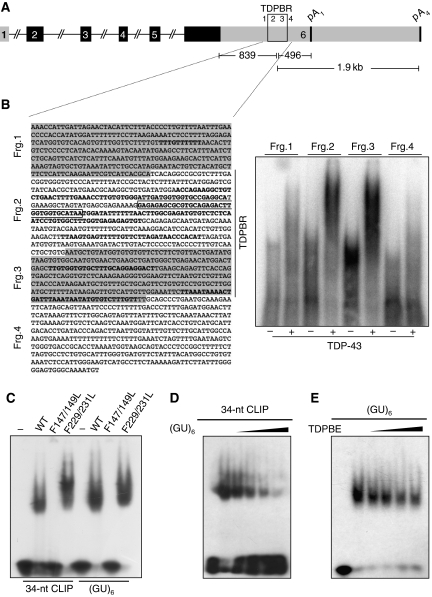
Regarding the 3′ UTR, recent cross-linking immunoprecipitation (CLIP) analysis performed in HEK293 cells determined that TDP-43 associated with several RNA sequences within the 3′ UTR of its own transcript in vivo (Figure 3) in a region that we called TDP-43-binding region (TDPBR, Figure 3A). To confirm TDP-43 binding to these sequences, we first of all performed electromobility shift assays (EMSAs) using fragments (fragment (frg.) 1–4) across this entire region. As shown in Figure 3B, right panel, fragments 2 and 3 that contain most CLIP sequences/clusters can efficiently bind TDP-43 while no binding could be observed for fragments 1 and 4 that contained only one or none (respectively) extended CLIP sequences/clusters.
Figure 3.
TDP-43 specifically binds its 3′ UTR region. (A) Schematic representation of the TDP-43 gene depicting the location and sequence of the TDPBR (B, fragments 2 and 3) within the 3′ UTR. Coding exons and untranslated regions are shown in black and grey, respectively. (B) In the left panel, the 3′ UTR region of TDP-43 with the different regions tested for TDP-43 binding (frgs. 1–4) is reported. In this panel, all the CLIP sequences/clusters reported from different experiments (J Ule and J Tollervey unpublished results) are highlighted in bold. The major 34-nt CLIP sequence seen in HEK 293 cells and used in comparative-binding analysis (C–E) is boxed. On the right, band-shift analysis using recombinant TDP-43 using these different fragments. (C) GST-TDP43 mutants were analyzed for binding to the major 34-nt CLIP sequence (boxed) and (GU)6 RNA as control. (D, E) Competition assays using constant levels of wild-type protein to bind radiolabelled (GU)6 as a function of increasing concentrations of unlabelled 34-nt CLIP sequence (D), or binding to radiolabelled 34-nt CLIP sequence as a function of increasing concentrations unlabelled (GU)6. Protein was absent from the first lane in each panel. ∼0.5 μg of the different recombinant proteins were used with 1 ng of the various 5′-labeled single-stranded RNA oligonucleotides. Competition analyses were carried out with 2.5, 3.75, 5, and 10 ng of unlabelled 34-nt CLIP sequence and (GU)6.
The 34-nucleotide boxed sequence (34 nt) in Figure 3B (frg. 2) was the one with the higher density of hits in the CLIP assay for TDP-43 in HEK 293 cells. However, as it was not a canonical (GU)n repetitive sequence that is known to be the preferred target of this protein, we considered necessary to study the 34 nt-binding efficiency compared with (GU)6. Figure 3C shows that wild-type TDP-43 and the RRM2 mutant (F229/231L) specifically bind the 34 nt sequence while the RRM1 mutant (F147/149L) did not produce a shift, as expected. These results coincide with our in vivo assays showing the lack of regulatory control on disruption of RRM1, and no apparent role of RRM2 (Figures 1 and 2). To compare the binding efficiency of TDP-43 with the 34 nt and (UG)6 RNAs, we then performed EMSA in the presence of competing amounts of unlabelled RNA. TDP-43 binding to the 34 nt was efficiently competed with increasing amounts of (UG)6 RNA, while higher concentrations of TDPBR RNA were necessary to see a reduction in TDP-43 association with (UG)6 (Figure 3D and E, respectively). Collectively, our data suggest that TDP-43 association with these sequences is specific, although it seems to occur with lower efficiency compared with UG repeat binding. A lower affinity may be desirable for regulatory sequences spread thorough a large region of the 3′ UTR that require multiple molecules of TDP 43 to induce a response.
Assessing the importance of the TDPBR region in a heterologous context
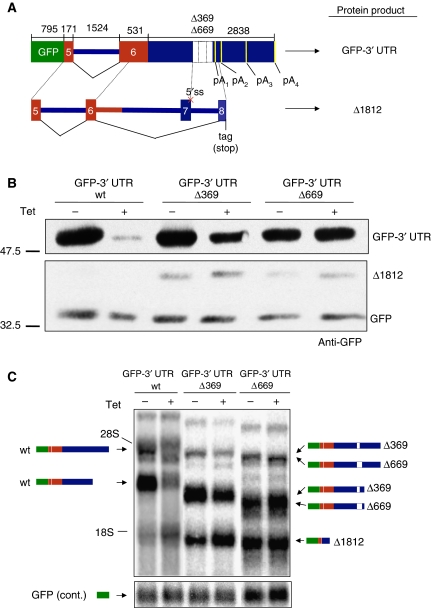
To test the self-regulatory function of the TDP-43-binding sites found in the TDP-43 mRNA 3′ UTR, we used a recombinant construct (GFP-3′ UTR wt) that contained exons 5 and 6, together with the 3′ UTR sequence of TDP-43, fused to the C-terminal end of a GFP reporter sequence (schematically summarized in Figure 4A). This construct provided the advantage to test the autoregulatory mechanism in a heterologous context and was also easier to manipulate than the endogenous gene. When transfected into our inducible HEK293 cell line, the GFP expression levels from the GFP-3′ UTR wt construct decreased following Tet induction (Figure 4B, upper panel), a result that was consistent with the conclusion that the downregulatory effect of TDP-43 did not involve additional TDP-43 exons or introns beside the sequence cloned in this system. Furthermore, no downregulation was observed on a transfected GFP wild-type protein used to normalize transfection efficiencies (Figure 4B, lower panel). We then engineered heterologous constructs wherein we progressively deleted the 3′ UTR across the region of the TDP-43-binding sites. Two constructs (GFP-3′ UTR Δ369 and Δ669, Figure 4A) clearly showed partial and complete loss of self-regulation when transfected into the inducible HEK293 cell line. In fact, it can be seen in Figure 4B that the downregulation of GFP protein expression levels following Tet induction was much reduced in the Δ369 construct compared with GFP-3′ UTR wt and eventually abolished in the Δ669 construct. Interestingly, our western blot analysis also highlighted the production of a smaller GFP-containing protein (Δ1812) of approximately 40 kDa in the lanes belonging to the GFP-3′ UTR Δ369 and Δ669 constructs (see below).
Figure 4.
A region in the 3′ UTR of TDP-43 mediates autoregulation (A) a schematic diagram of the GFP-3′ UTR wt and Δ369 and Δ669 constructs. Wild-type TDP-43 coding sequences are shown in grey. The lower panel shows the splicing events that lead to the Δ1812 mRNA and protein isoforms. (B) GFP protein expression when the GFP-3′ UTR wt and Δ369 and Δ669 constructs were transfected into the HEK293 stable cell lines following induction (+) of the TG-TDP43 transgene. Transfection efficiencies were normalized by co-transfecting a GFP expression vector (lower panel). In the GFP-3′ UTR Δ369 and Δ669 (−) and (+) lanes, the western blot also contains an additional 40 kDa protein band that was called Δ1812. The different proteins were detected with anti-GFP (C) shows a northern blot analysis of the transcripts derived from the GFP-3′ UTR wt, Δ369, and Δ669 constructs in the HEK293 stable cell lines before (−) and after Tet induction (+). All constructs produce two major mRNA species because of the usage of different polyadenylation sites. In addition to these species, the GFP-3′ UTR Δ369 and Δ669 constructs also produce an aberrantly spliced mRNA isoform (Δ1812) that accounts for the production of the aberrant protein present in the western blots (B).
In parallel, we performed northern blot analysis to confirm that autoregulation did not occur in the case of the Δ369 and Δ669 constructs at the mRNA level as expected. First of all, it should be noted that for all three constructs (wt, Δ369, and Δ669) we observed the presence of two major bands, which correctly mimic the endogenous mRNA species shown in Figures 2 and 5. In keeping with the results obtained with the endogenous TDP-43 transcript, Figure 4C shows that the two signals corresponding to the GFP-3′ UTR wt construct completely disappeared following Tet induction (this did not occur in the case of the control GFP used to normalize, see bottom panels). On the other hand, all the corresponding signals for the GFP-3′ UTR Δ369 and Δ669 constructs were progressively unaffected by Tet induction (note that these signals migrate lower than the GFP-3′ UTR wt transcript because of the substantial deletions in the TDPBR). In the deleted constructs, however, a new mRNA species is also visible (Δ1812). RT–PCR and sequencing analyses of this band found that it belongs to mRNA species resulting from alternative splicing of the last exon, similar to what occurs in the endogenous TDP-43 V2 form described in Figure 2A and Supplementary Figure 2. In the form Δ1812, exon and intron 7 were spliced out because of a deletion of the 5′ splice site of exon 7 in both Δ369 and Δ669 constructs (see lower scheme in Figure 4A). It is this band, labelled Δ1812 in Figure 4B, which accounted for the production of the short aberrant protein (the lower scheme in Figure 4A also shows the new coding frame).
Figure 5.
TDP-43 autoregulation is independent of NMD. (A) The existence of alternative splicing that activates the removal of two downstream introns (V2) was seen in the EST database. The shorter form V2 was seen to undergo NMD (Supplementary Figure 2), while northern blot analysis (B) of control and Tet-induced samples in the presence and absence of CHX treatment (50 μg/ml, for 3 h) did not detect the presence of V2. V2 should have a MW of 1.2 or 2.5 Kb depending on the poly A site usage. The probe was generated with the primers shown in (A) followed by purification of the product corresponding to V2.
Collectively, these results show that the sequence important to downregulate the TDP-43 mRNA is made up by a collection of closely localized-binding sites in the TDPBR region. Next, we decided to also investigate the molecular mechanisms involved in this self-regulatory process.
Nonsense-mediated decay
On the basis of previous autoregulatory mechanisms described for other hnRNPs (Wollerton et al, 2004; Rossbach et al, 2009), we first considered alternative splicing modifications coupled to nonsense-mediated decay (NMD) as a possible mechanism to explain TDP-43 self-regulation. PCR did not show changes in the pattern of TDP-43 mRNAs after Tet induction (Supplementary Figure 2). These analyses, however, did indicate the existence of a lower molecular weight form referred to as V2 in both control and induced cells (Figure 5A and Supplementary Figure 2). Cycloheximide (CHX) treatment showed that only V2 would be subject to NMD. However, from a functional point of view, it should be noted that the V2 mRNA is present at very low levels, and is clearly visible only with PCR (see Supplementary Figure 2B) while not on northern blot analysis (Figure 5B). At the same time, the corresponding protein derived from this isoform is not found in significant amounts after inhibition of the NMD process in the presence of TG-TDP43 induction (data not shown). Thus, although we cannot rule out a role of V2 and NMD in the regulation of TDP-43 through alternative pathways or in different systems wherein this isoform may be expressed at higher levels, we conclude that it is unlikely to mediate TDP-43 self-regulation in our cell lines.
Overexpression of TDP-43 promotes instability of its own transcript
As a second possibility, we examined the stability of the endogenous TDP-43 transcript as a function of TG-TDP43 expression in the HEK293 cell line. We first established the minimum time of Tet induction that would allow us to adequately measure endogenous transcript levels (data not shown). A 6-h period of induction led to a 20–30% decrease in the levels of endogenous TDP-43 by quantitative PCR (qPCR). Cells were then treated with actinomycin D (ActD) at 6 h after Tet induction to block transcription and were collected at different time points, along with the control-treated cells. As shown in Figure 6A, TG-TDP43 expression led to a decrease in the half-life of the endogenous transcript as measured by qPCR. We confirmed that TG-TDP43 transcript levels also decreased on Act D treatment (data not shown) by measuring TG-TDP43 levels with specific primers at the different time points, confirming the block of transcription under our experimental conditions. More importantly, this control indicated that the decrease in endogenous TDP-43 levels over time was not due to an increase in TG-TDP43 expression.
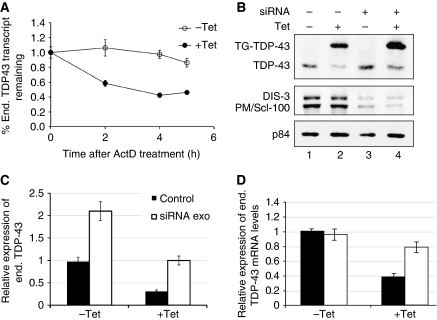
Figure 6.
TDP-43 overexpression promotes RNA instability in an exosome-dependent fashion. (A) Transcript half-life was estimated after ActD (5 μg/ml) treatment of HEK293 control and induced cells. Transcript levels at the indicated times were analyzed by qPCR. The relative levels of expression at the different time points were normalized according to the levels at time 0, (−) and (+) Tet induction (empty and filled circles, respectively). (B) Exosome components, PM/Scl-100 and DIS3, were simultaneously silenced in control and induced cells and their effect on TDP-43 expression levels was observed in western blot. (C) Quantification of endogenous TDP-43 protein levels under the different conditions was normalized against p84 levels. Error bars, s.e.m.; _n_=3 experiments for both panels. (D) mRNA expression levels of endogenous TDP-43 mRNA levels in −Tet and +Tet conditions. Both the transcript levels were normalized to the GAPDH mRNA levels and determined by real-time qPCR. The mRNA levels were quantified separately in triplicates and s.d. values obtained are shown on the bars.
Finally, we assessed the involvement of the exosome machinery in TDP-43 self-regulation by studying the role of two key exosome components PM/Scl-100 (Rrp6) and DIS3 (Rrp44), which are involved in nuclear and cytoplasmic RNA degradation in mammalian cells (Mitchell et al, 1997; van Hoof and Parker, 1999; Butler, 2002; Lejeune et al, 2003). Simultaneous knockdown of PM/Scl-100 and DIS3 reduced the inhibition of endogenous TDP-43 expression following TG-TDP43 induction. This was observed by comparing the difference in endogenous TDP-43 levels between –Tet and +Tet conditions in control-treated cells versus PM/Scl-100 and DIS3 knockdown cells (Figure 6B and C). Transfection of control siRNA in non-induced cells and after 24 h of Tet induction led to a 70% decrease in the levels of endogenous TDP-43 on Tet treatment, while exosome disruption by RNAi reduced TDP-43 by 50% following induction (compare ratios between lanes 2/1 and 4/3). These results may be underestimated because silencing of the exosome components was sufficient to cause a two-fold increase in endogenous TDP-43 levels in the absence of Tet induction after 48 h of RNAi treatment (Figure 6B, compare lanes 1 and 3). Using qPCR, we also measured the levels of endogenous TDP-43 mRNA following knockdown of the exosome components in −Tet and +Tet conditions (Figure 6D). These results show that increase in TDP-43 endogenous levels following knockout of the exosome components is clear only in the +Tet samples. This observation suggests that the endogenous TDP-43 mRNA substrates for exosome degradation are increased only when there are high amounts of total TDP-43 protein expressed in the cell.
Discussion
Regulation of protein levels through a negative feedback loop has been reported for a number of RNA processing factors including SR proteins SC35 and SF2, and hnRNP members PTB and hnRNP-L (Sureau et al, 2001; Wollerton et al, 2004; Lareau et al, 2007; Ni et al, 2007; Saltzman et al, 2008; Rossbach et al, 2009; Sun et al, 2010). In all these cases, the homoeostatic control of endogenous protein is efficiently and continuously kept in check through the downregulation of transcript levels by the protein molecules present in excess. Self-regulation of expression becomes particularly important for proteins whose abundance results in cellular toxicity. In this study, we report that TDP-43 acts as a regulator of its own expression at the transcript level in vivo through a mechanism that involves binding to the highly conserved 3′ UTR, specifically to an extended region that we have referred as TDPBR (Figure 7). In keeping with these results, TDP-43 mutants that do not bind RNA, lose the ability to control endogenous transcript levels. Interestingly, our observations indicate that the integrity of the TDP-43 C-domain in the region 321–366 is also necessary for this autoregulation. An understanding of the role of this domain during self-regulation will require further investigation and is likely to involve hnRNP recruitment as these proteins, primarily hnRNP-A2, have been shown to associate to this sequence. Moreover, the integrity of this interaction is required for TDP-43 functions such as the control of splicing (D'Ambrogio et al, 2009). Another interesting feature of this element is represented by the observation that the TDP-43-binding sequences in the TDPBR region are not made by continuous (UG)n repeats, which represent the highest affinity-binding sequence for this protein. However, in this particular case, a lower affinity of TDP-43 for these individual sequences spread thorough a large region of the 3′ UTR may be more desirable from a regulatory process point of view because multiple TDP-43 molecules would presumably be required to induce a response.
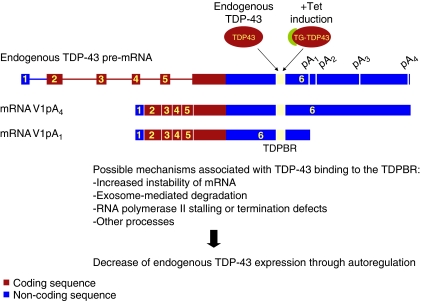
Figure 7.
Regulation of TDP-43 expression model. Endogenous or transgenic TDP-43 (TG-TDP43) protein binds to the TDPBR found in the 3′ UTR of TDP-43 transcripts. This TDP-43–RNA interaction promotes, along with other possible events, RNA instability and degradation resulting in lower levels of TDP-43 gene expression.
In the present work, we have also investigated the possible mechanisms associated with TDP-43 autoregulation. Our experiments show that in HEK293 cells, TDP-43 preferentially uses two polyadenylation sites that are ∼1.5 Kb apart (Figure 2). The usage of the proximal polyadenylation site (pA1, 2.8 Kb mRNA) appears to be slightly higher than the more distal pA4 site (4.2 Kb mRNA) (Figure 2) and the expression of TG-TDP43 does not seem to change the use of either polyadenylation site to a significant extent, as seen by northern blot and PCR analysis.
Regarding the mechanism of this autoregulation, it should be noted that several TDP-43-related proteins primarily depend on alternative splicing and NMD mechanisms to self-regulate (Wollerton et al, 2004; Rossbach et al, 2009). Although we have confirmed that this mechanism may account for the regulation of selected TDP-43 mRNA isoforms (i.e., V2) our findings suggest that TDP-43 self-regulation through the 4.2 and 2.8 kb transcripts does not depend on these systems, as shown by the lack of any additional significant mRNA species visible in northern blots after TG-TDP43 induction (Figure 2) and/or CHX treatment (see Figure 5 and Supplementary data). To establish the mechanism of regulation of these major transcripts, therefore, we examined the role of RNA stability by estimating the decay kinetics of TDP-43 mRNA (Figure 6). Expression of induced TDP-43 reduces the half-life of the endogenous transcript when compared with control conditions. Consistently, silencing of two principal exosome components, PM/Scl-100 and DIS3, also partially inhibits the TDP-43 negative feedback loop particularly in Tet + conditions wherein there is overexpression of TDP-43. At the moment, we cannot explain the non-correlation between stationary mRNA levels and increased endogenous TDP-43 protein levels in −Tet conditions (Figure 6C and D). In this case, it is possible that other factors play a role in modulating TDP-43 levels such as variations in translational or storage processes which will need further investigation.
The involvement of the exosome complex in TDP-43 autoregulation is also supported by the direct connection between TDP-43 and some components of this complex, as reported in the screening study on human mRNA degradation performed by Lehner and Sanderson (2004). Using a yeast two hybrid screening approach, these authors reported that TDP-43 interacts with the HRDC domain of PM/Scl-100 and Xrn2/Rat1.
It should be considered, moreover, that the binding of TDP-43 to its 3′ UTR is almost certainly not the only checkpoint that acts within cells to keep the levels of this protein constant. First of all, considering the very high degree of conservation of the >3000 long 3′ UTR of TDP-43, it is very likely that several other interactions will participate in the fine-tuning of this process. Second, considerable changes in the protein half-life have been recently demonstrated to occur in different cell lines, suggesting that regulation could also occur at a post-translational level (Ling et al, 2010, PNAS). Third, some kind of regulation at the transcription, or translational levels cannot be ruled out. Therefore, we expect that additional studies will be required to better unravel this issue. The recent reports of a great number of animal models based on exogenous TDP-43 overexpression will presumably allow the investigation of this process in greater detail.
The evidence that TDP-43 autogenous control that we describe in cell lines is also valid in animal models comes indirectly from a recent report on transgenic mice expressing the A315T TDP-43 mutant (Wegorzewska et al, 2009). The authors show an immunoblot of brain lysates wherein endogenous levels of TDP-43 are reduced in the presence of the FLAG-tagged protein. Although the authors do not comment on the reduction, this observation suggests that TDP-43 self-regulation is also active in the whole organism. More recently, a mouse model expressing wild-type human TDP-43 has confirmed that expression of the human protein can downregulate mouse TDP-43 protein and mRNA levels in a dose-dependent fashion see Figure 1C and Supplementary Figure 1C, D in Xu et al, 2010.
Our findings also generate a number of questions regarding the particular mechanism associated with TDP-43 transcript downregulation (Figure 7). For instance, does the change in RNA stability occur in the nucleus, cytoplasm or a particular subcellular compartment? Is it tied to transcription, translation, activation of 3′ UTR-linked degradation pathways or changes in the polyadenylated state of the transcript? These and other issues will require further studies to understand TDP-43 autoregulation and will probably shed light on the regulation of additional TDP-43-controlled transcripts. In fact, the abundance of TDP-43 target sequences in other genes located in regions similar to that of the TDPBR in the TDP-43 gene (J Ule, J Tollervey and A D'Ambrogio, unpublished observations), suggests that this regulatory mechanism by TDP-43 may not be restricted to its own transcript. Thus, it would be of great interest to find additional mRNA examples, particularly of motoneuron-specific transcripts that may explain the role of TDP-43 in neuromuscular junction development (Feiguin et al, 2009).
Taken together, our findings on TDP-43 autoregulation may also represent an important contribution to the understanding of the mechanism at the basis of TDP-43 proteinopathies. In fact, we hypothesize that disruptions of TDP-43 autoregulation would result in the aberrant accumulation of the protein. Accordingly, recent studies of Caenorhabditis elegans, mouse, and Drosophila models of TDP-43 overexpression show that it is sufficient to increase the levels of wild-type protein to obtain phenotypes resembling pathologies (Ash et al, 2010; Li et al, 2010; Tsai et al, 2010; Wils et al, 2010; Xu et al, 2010). It should also be considered that eventual compensatory mechanisms employed by the cell to overcome defects at the level of autoregulation may also end up in establishing some degree of cellular suffering. Thus, even if normal protein levels are successfully maintained under such conditions, this additional effort by the cell may prove to be detrimental in the long run. This consideration may contribute to explain why neurons, with their long life span and complex architecture, are especially targeted in these pathologies.
Our data also indicate that the C-terminal region 321–366 of TDP-43 is involved in the autoregulation. This suggested that TDP-43 mutations found in patients might interfere with its homoeostatic control by altering recruitment of the protein complexes required for self-regulation. However, no dramatic change of interactions with hnRNPs has been observed in vitro with mutations in this region tested up to now (D'Ambrogio et al, 2009) and, therefore, the effect of the 321–366 region in inhibiting autoregulation still needs to be clarified. In a wider perspective, however, of all the mutations that have been described in a fraction of patients affected by familial forms of ALS (recently reviewed by Pesiridis et al, 2009), only the D169G mutation may have potentially interfered with its autoregulatory process (Kabashi et al, 2008). This mutation is found in a region that may possibly affect the RNA-binding properties of TDP-43. However, to address this possibility we have now expressed this mutant as a GST–TDP43 fusion protein. Our experiments show that its RNA and hnRNP-binding abilities (both essential for autoregulation) do not seem to differ appreciably from those of the wild-type protein at least within the limitations of our experimental conditions (Supplementary Figure 3). Nonetheless, the possibility does exist that TDP-43 mutations may be interfering with the autoregulatory process, as higher levels of transcript expression have been recently associated with a mutation localized in the 3′ UTR region from patients suffering of ALS and FTD (Gitcho et al, 2009).
Finally, our observations may also explain the absence of reported mutations in the RNA-binding region of TDP-43. Any significant alteration in the RNA-binding ability of TDP-43 may be immediately deleterious, first of all, because of the disruption of the autoregulatory mechanism. Moreover, independently of the presence of mutations, we speculate that nuclear or cytoplasmic aggregation of TDP-43, typical of TDP-43 proteinopathies, may break the self-regulatory cycle by sequestering functional protein and, hence, increasing TDP-43 production. This work also indicates that studies aimed to investigate the role of TDP-43 variants that involve exogenous protein expression should maintain concentrations that are not much greater than endogenous levels.
Materials and methods
Plasmid construction
The TDP-43 cDNA vectors were generated to express wild-type TDP-43 tagged with either a streptavidin-binding peptide plus a hemagglutinin peptide (pSH-TDP43), or FLAG (pF-wt) at the amino-terminus. pSH-TDP43 was generated using the Gateway Technology from Invitrogen through amplification using GWTDPwtFW and RV primers and cloning into pDONR221. This vector was used to introduce SH-TDP43 into the pcDNA5/FRT/TO/SH/GW vector generously provided by M Gstaiger. The FLAG-tagged wild-type and TDP-43 mutants were generated by standard PCR methods. pF-wt was amplified with EcoRVpFTDPFW and EcoRVTDPRV and was then used as template to construct the various mutants. pF-F147/149L, pF-F229/231L, pF-F4L, and pF-Δ321-366 were generated with previously described primers (Buratti and Baralle, 2001; D'Ambrogio et al, 2009). The vectors to express the FLAG-tagged versions of TDP-43 were then generated by EcoR V cleavage of pcDNA5/FRT/TO (Invitrogen). To obtain the GFP-3′ UTR constructs, the sequence corresponding from the beginning of the fifth exon of TDP-43 to 53 nt after the fourth predicted polyadenylation signal (pA4) was amplified from human genomic DNA using the following primers XbaBglx5FW and Hind5000RV. The fragment was then cloned into the pEGFP-C2 vector (Clontech BD Biosciences) using the _Bgl_II and _Hind_III sites and the construction was verified by sequencing. The deletions Δ369 and Δ669 were made by PCR amplification using _Pfu_I polymerase with primers Δ369FW, Δ369RV, Δ669FW, and Δ669RV followed by _Dpn_I digestion (Promega). Primer sequences used for cloning, siRNA experiments, northern blot analysis, and EMSA are provided in the Supplementary data.
Cell culture and RNA interference
Stable expression of HA-Strep-TDP-43 (SH-TDP43) and the FLAG-tagged TDP-43 (F-TDP43) variants was achieved by Flp-recombinase-mediated recombination in Flp-In HEK293 cells (Invitrogen) followed by hygromycin B selection (InvivoGen). Each TDP-43-expressing vector and the pOG44 vector that expresses the Flp-recombinase (Invitrogen) were co-transfected with Effectene transfection reagent (Qiagen) and were gradually selected with Hygromycin B up to a concentration of 200 mg/ml. Individual clones were obtained by limited dilution. Induction of tagged TDP-43 (TG-TDP43) expression was achieved with 1 μg/ml tetracycline (Sigma). Cells were grown in DMEM-Glutamax-I (GIBCO) supplemented with 10% fetal bovine serum (EuroClone). Upf-1 silencing was performed as described (Matsuda et al, 2008) using Upf1siRNA (Supplementary Table I (Kim et al, 2005)). PM/Scl-100 and Dis3 silencing was carried out with Hiperfect (Qiagen) following manufacturer's instructions with the oligonucleotides listed in the Supplementary data.
Immunoblotting
Cells extracts were prepared in 15 mM HEPES (pH 7.5), 0.25 M NaCl, 0.5% NP-40, 10% glycerol, 1x protease inhibitor (Roche 1873580), 25 mM NaF, 10 mM β-glycerolphosphate, 0.2 mM Na3VO4, and 1 mM phenylmethylsulphonyl fluoride. Proteins were separated by SDS–PAGE and transferred to nitrocellulose (0.45 μM, Amersham Biosciences), and protein detection was carried out with standard western blotting techniques. Antibodies used: TDP-43 (ProteinTech Group 10782-2-AP and the previously described (Buratti et al, 2001)), p84 (Abcam ab487), tubulin (Sigma T5168), Upf1 (Santa Cruz Biotechnology sc-18260), PM/Scl-100 (Abcam ab50558), and DIS3 (Abcam ab68570).
Northern blotting
RNA was isolated with TRI REAGENT (Sigma) following manufacturer's instructions. RNA samples (10 μg, unless otherwise indicated) were loaded on 1% formaldehyde agarose gels, transferred to Hybond N+ nylon membranes (Amersham Biosciences) and probed with internally 32P-labelled sequences following pre-hybridization in ULTRAhyb® Ultrasensitive Hybridization Buffer (Ambion). Pre-hybridization and hybridization were carried out at 50 °C. The probes were generated by PCR using primers described in the Supplementary data and labelled with Rediprime II DNA Labeling System (GE Healthcare). The probe to detect GAPDH was generated as described in (Muro et al, 2003). Visualization of transcripts was carried out with a Cyclone Plus Storage Phosphor Scanner and the included OptiQuant Software (Perkin Elmer).
PCR analysis
qPCR was performed with iQ SYBR Green Supermix on a CFX96 Real-Time PCR Detection system (BIO-RAD). The primers to detect all TDP-43 variants are the following: RThx2FW, RThx3RV; RThV1x6FWd, RThV1x6RVd; and RThV2x7FW, RThV2x8RVb for the endogenous V1 and V2 transcripts, specifically; GAPDH: GAPDHFW, GAPDHRV. PCRs were performed for 15″ at 95 °C and 1′ at 60 °C for 45 cycles followed by the thermal denaturation protocol. Expression levels of the target gene relative to GAPDH RNA were determined using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Significant differences were determined by Student's _t_-test wherein P<0.05 was considered to be statistically significant.
RNA CLIP
CLIP cDNA libraries and high-throughput sequencing were performed as described before (Wang et al, 2009) with the following modifications. TDP-43 Antibody (Proteintech 10782-2-AP) was used for immunoprecipitation. The cDNA library was sequenced using paired-end approach on Illumina Genome Analyser, with 36 nt read-length from each direction. The 5′ and 3′ RNA linkers contained a three-nucleotide barcode AAA, which was removed from the sequences before mapping to the human genome sequence (version Hg18/NCBI36) allowing 1 mismatch using Bowtie version 0.10.1 (command line: -a -m 1 -v 1). 31438 unique sequences have mapped to the genome, one of which was GAGAGCGCGTGCAGAGACTTGGTGGTGCAT, which mapped to the 3′ UTR of TDP-43.
Binding assays
Production of recombinant GST-TDP-43 and EMSA was performed as described (Buratti and Baralle, 2001; Ayala et al, 2005).
Supplementary Material
Supplementary data
Review Process File
Acknowledgments
We thank M Gstaiger (Institute of Molecular Systems Biology, ETH Zurich, Switzerland) for the pcDNA5/FRT/TO/SH/GW vector, M Morgan (ICGEB, Trieste, Italy) for help with the EST analysis, T Curk (Artificial Intelligence Laboratory, Ljubljana, Slovenia) for the bioinformatic analysis of CLIP data, and S Dhir (ICGEB Trieste, Italy) for bioinformatics analysis of the TDP-43 gene sequences. This work was supported by the AriSLA and the European Union, EURASNET [grant number SHG-CT-2005-518238 to FEB].
Footnotes
The authors declare that they have no conflict of interest.
References
- Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, Graff-Radford NR, Hutton ML, Dickson DW (2007) TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer's disease. Ann Neurol 61: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351: 602–611 [DOI] [PubMed] [Google Scholar]
- Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD (2010) Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet 19: 3206–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE (2005) Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol 348: 575–588 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci 13: 867–878 [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. Embo J 20: 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS (2002) The yin and yang of the exosome. Trends Cell Biol 12: 90–96 [DOI] [PubMed] [Google Scholar]
- D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE (2009) Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res 37: 4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE (2009) Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett 583: 1586–1592 [DOI] [PubMed] [Google Scholar]
- Geser F, Winton MJ, Kwong LK, Xu Y, Xie SX, Igaz LM, Garruto RM, Perl DP, Galasko D, Lee VM, Trojanowski JQ (2008) Pathological TDP-43 in parkinsonism-dementia complex and amyotrophic lateral sclerosis of Guam. Acta Neuropathol 115: 133–145 [DOI] [PubMed] [Google Scholar]
- Gitcho MA, Bigio EH, Mishra M, Johnson N, Weintraub S, Mesulam M, Rademakers R, Chakraverty S, Cruchaga C, Morris JC, Goate AM, Cairns NJ (2009) TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol 118: 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatter T, Wepf A, Aebersold R, Gstaiger M (2009) An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol 5: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S, Iseki E, Yamamoto R, Minegishi M, Hino H, Fujisawa K, Togo T, Katsuse O, Uchikado H, Furukawa Y, Kosaka K, Arai H (2007) Concurrence of TDP-43, tau and alpha-synuclein pathology in brains of Alzheimer's disease and dementia with Lewy bodies. Brain Res 1184: 284–294 [DOI] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40: 572–574 [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE (2005) Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Kraemer BC, Schuck T, Wheeler JM, Robinson LC, Trojanowski JQ, Lee VM, Schellenberg GD (2010) Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol 119: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS (2009) Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res 37: 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE (2007) Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Lehner B, Sanderson CM (2004) A protein interaction framework for human mRNA degradation. Genome Res 14: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE (2003) Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA III, Fushimi K, Wu JY (2010) A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci USA 107: 3169–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci USA 107: 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Sato H, Maquat LE (2008) Chapter 9. Studying nonsense-mediated mRNA decay in mammalian cells. Methods Enzymol 449: 177–201 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Muro AF, Chauhan AK, Gajovic S, Iaconcig A, Porro F, Stanta G, Baralle FE (2003) Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J Cell Biol 162: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima-Yasuda H, Uryu K, Robinson J, Xie SX, Hurtig H, Duda JE, Arnold SE, Siderowf A, Grossman M, Leverenz JB, Woltjer R, Lopez OL, Hamilton R, Tsuang DW, Galasko D, Masliah E, Kaye J, Clark CM, Montine TJ, Lee VM et al. (2007) Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol 114: 221–229 [DOI] [PubMed] [Google Scholar]
- Neumann M, Mackenzie IR, Cairns NJ, Boyer PJ, Markesbery WR, Smith CD, Taylor JP, Kretzschmar HA, Kimonis VE, Forman MS (2007) TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol Exp Neurol 66: 152–157 [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133 [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesiridis GS, Lee VM, Trojanowski JQ (2009) Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet 18 (R2): R156–R162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A (2009) Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol 29: 1442–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ (2008) Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Good SK, Atkin S, Dewey CM, Mayer P III, Herz J, Yu G (2010) TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem 285: 6826–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Sinha R, Karni R, Krainer AR (2010) SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol 17: 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J (2001) SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. Embo J 20: 1785–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KJ, Yang CH, Fang YH, Cho KH, Chien WL, Wang WT, Wu TW, Lin CP, Fu WM, Shen CK (2010) Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med 207: 1661–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uryu K, Nakashima-Yasuda H, Forman MS, Kwong LK, Clark CM, Grossman M, Miller BL, Kretzschmar HA, Lee VM, Trojanowski JQ, Neumann M (2008) Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J Neuropathol Exp Neurol 67: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Parker R (1999) The exosome: a proteasome for RNA? Cell 99: 347–350 [DOI] [PubMed] [Google Scholar]
- Wang Z, Tollervey J, Briese M, Turner D, Ule J (2009) CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods 48: 287–293 [DOI] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH (2009) TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 106: 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, Smits V, Ceuterick-de Groote C, Van Broeckhoven C, Kumar-Singh S (2010) TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 107: 3858–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW (2004) Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13: 91–100 [DOI] [PubMed] [Google Scholar]
- Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK (2010) TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis 48: 56–62 [DOI] [PubMed] [Google Scholar]
- Xu YF, Gendron TF, Zhang YJ, Lin WL, D'Alton S, Sheng H, Casey MC, Tong J, Knight J, Yu X, Rademakers R, Boylan K, Hutton M, McGowan E, Dickson DW, Lewis J, Petrucelli L (2010) Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci 30: 10851–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Review Process File