Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence (original) (raw)
Abstract
Since the initial annotation of miRNAs from cloned short RNAs by the Ambros, Tuschl, and Bartel groups in 2001, more than a hundred studies have sought to identify additional miRNAs in various species. We report here a meta-analysis of short RNA data from Drosophila melanogaster, aggregating published libraries with 76 data sets that we generated for the modENCODE project. In total, we began with more than 1 billion raw reads from 187 libraries comprising diverse developmental stages, specific tissue- and cell-types, mutant conditions, and/or Argonaute immunoprecipitations. We elucidated several features of known miRNA loci, including multiple phased byproducts of cropping and dicing, abundant alternative 5′ termini of certain miRNAs, frequent 3′ untemplated additions, and potential editing events. We also identified 49 novel genomic locations of miRNA production, and 61 additional candidate loci with limited evidence for miRNA biogenesis. Although these loci broaden the Drosophila miRNA catalog, this work supports the notion that a restricted set of cellular transcripts is competent to be specifically processed by the Drosha/Dicer-1 pathway. Unexpectedly, we detected miRNA production from coding and untranslated regions of mRNAs and found the phenomenon of miRNA production from the antisense strand of known loci to be common. Altogether, this study lays a comprehensive foundation for the study of miRNA diversity and evolution in a complex animal model.
microRNAs (miRNAs) are ∼22 nucleotide (nt) regulatory RNAs that mediate broad post-transcriptional regulatory networks in most higher eukaryotes (Lai 2003; Flynt and Lai 2008). Although a variety of alternative biogenesis pathways exist (Yang and Lai 2010), most animal miRNAs are generated by the following canonical pathway. In the nucleus, a primary miRNA (pri-miRNA) transcript bearing a local inverted repeat is cleaved by the Drosha RNase III enzyme to yield the pre-miRNA hairpin (Kim et al. 2009). This is cleaved in the cytoplasm by a Dicer-class RNase III enzyme (Dicer-1 in insects) to yield a miRNA/miRNA* (star) duplex, of which one strand is predominantly transferred to an Argonaute (AGO) effector protein and guides it to target transcripts.
The founding miRNAs lin-4 and let-7 emerged from developmental genetic screens (Lee et al. 1993; Reinhart et al. 2000), but the vast majority of miRNAs were annotated from cloning of short RNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001) or from computational strategies (Grad et al. 2003; Lai et al. 2003; Lim et al. 2003a,b). The comparative approach has substantial power to discriminate miRNA genes as conserved hairpins exhibiting greater divergence in the terminal loop relative to the hairpin arms (Lai et al. 2003; Berezikov et al. 2005). However, only conserved miRNA genes are currently amenable to effective discovery by purely computational means. Instead, next-generation sequencing has lately become the method of choice for annotating new miRNAs, including species-restricted genes. As well, deeply sequenced small RNA data have yielded great insights into miRNA biogenesis, AGO sorting, and post-transcriptional modification.
In this study, we analyzed a diverse collection of small RNA libraries to provide the most comprehensive annotation of miRNAs in any species to date. In addition, the deep profiling of known miRNAs revealed alternative Drosha and/or Dicer-1 cleavages, frequent untemplated modifications, and candidate editing events of mature fly miRNAs Altogether, these findings provide a new foundation for studying miRNA biogenesis, modification, and emergence in Drosophila melanogaster.
Results
Small RNA data sets and processing
We combined 76 Illumina Drosophila small RNA data sets that we generated for the modENCODE project (48 of which were not previously reported) with 111 other published small RNA data sets; their accession IDs and library descriptions are provided in Supplemental Table S1. The 187 data sets range across developmental stages (i.e., different embryo timepoints, larval and pupal stages, male and female adults), tissues, and body parts (i.e., mass isolated imaginal discs/brains/salivary glands, heads, bodies, ovaries or testes); from cultured cell lines of diverse origins; from reads enriched in AGO1 or AGO2 effector complexes; from small RNA pathway mutants; and from a wide variety of combinations of these treatments. From 1.1 billion raw reads, just under 800 million (M) had linkers that we could identify and remove. The clipped reads were mapped to the dm3 genome assembly, yielding more than 488 M perfect mappers with at least 18 nt matching; an additional 51 M reads mapped perfectly to the genome following trimming of 3′ nucleotides.
Expression of known miRNA loci
The collected small RNA data included more than 214 M mature strand and more than 10 M star sequences from known miRNA loci (Supplemental Table S2). Four genes (bantam, mir-184, mir-8, and mir-2a-1/mir-2a-2) were sequenced more than 10 M times each, and these were present in each of the 187 libraries (except bantam, present in 186 libraries). In fact, the strong majority of miRNA loci were recorded in more than 100 data sets, despite known tissue-specific expression patterns of miRNAs (Aboobaker et al. 2005), the small size of certain data sets, and the fact that many libraries were specifically depleted of miRNAs (i.e., Piwi-family IP libraries or oxidized libraries). At the same time, the levels of such “omnipresent” miRNAs varied widely; for instance, bantam was sequenced from one time to 3.4 M times in different data sets.
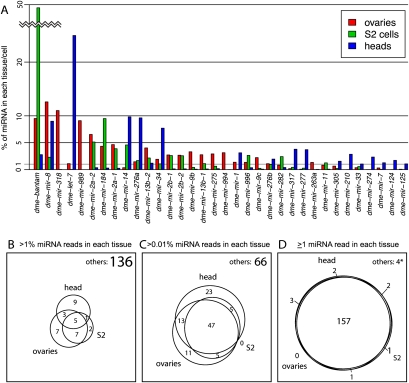
The majority of available Drosophila small RNA libraries were prepared from manipulations of ovaries, heads and S2 cells (see Methods), reflecting their adoption as major experimental systems for small RNA research. These contained in total about 73 M, 23.5 M, and 28 M reads mapped to miRBase 15 loci, respectively (Supplemental Table S3). mRNA expression in these three systems is quite distinct, and the same was true when considering their dominant miRNAs (Fig. 1A). The signatures of miRNAs contributing >1% of content in ovaries, heads, or S2 cells overlapped only moderately and in aggregate comprised only one-fifth of known miRNAs (Fig. 1B). However, the picture changed upon considering lower levels of expression. In particular, more than half of the miRNAs were common in the overlap of loci contributing >0.01% of reads in each tissue (Fig. 1C), and all but a few miRNAs were “coexpressed” in all three systems when considering levels down to single mature reads (Fig. 1D).
Figure 1.
Distinct and overlapping patterns of miRNA expression in different tissues and samples. (A) Graph shows those miRNAs that contribute more than 1% of miRNAs in aggregated sets of ovary, head, and S2 cell data totaling about 30–70 M reads specifically mapped to miRNAs. It is clear that many miRNAs are either strongly enriched or seemingly absent from one of the three sample types. (B–D) Venn diagrams that show the overlap in miRNAs detected in ovary, head, and S2 cells at various levels of expression. As the contribution of each miRNA decreases from 1% (B) to >0.01% (C), we observe increasing coexpression among these distinct tissue/cell types. When considering miRNA expression down to a single read in each library, we observe nearly complete coexpression. The few miRNAs that were not detected (4*) are either questionable as canonical miRNAs (miR-280 and miR-289) or were detected at only a few parts per million in the esoteric cell line OSS (miR-2280 and miR-2281).
We do not intend to suggest that extremely lowly expressed miRNAs are likely to influence gene expression. On the other hand, these data highlight that the concept of “coexpression” is fluid, and the cutoffs arbitrary. We infer that the depth of sequencing in these data sets provides the power to reveal even very weak miRNA expression, perhaps in cells with only spurious transcription across these loci.
Characteristics of miRNA loop and moRs
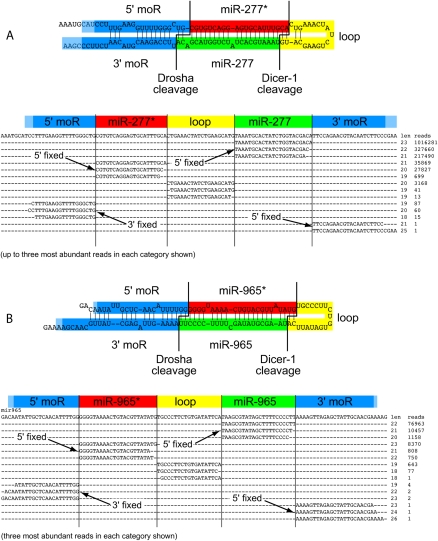
These deeply profiled data frequently included byproducts of miRNA processing, such as cleaved terminal loops as well as reads flanking the pre-miRNA (i.e., miRNA offset reads, or moRs) (Ruby et al. 2007; Shi et al. 2009). Note that the possibility of loop reads is constrained by the range of small RNA cloning; for example, the mir-1011 loop is only 9 nt, while the mir-989 loop is 99 nt. Figure 2 shows examples of loci with five phased species, whose characteristic dovetailing provides indisputable evidence for Drosha and Dicer-1 cleavage of their precursors. The 5′ ends of Drosophila miRNAs and miRNA* species are preferentially constrained relative to their 3′ ends (Ruby et al. 2007; Seitz et al. 2008). Indeed, both classes have _trans_-regulatory capacity (Okamura et al. 2008) and have been evolutionarily selected for particular loading and strand selection properties (Czech et al. 2009; Okamura et al. 2009; Ghildiyal et al. 2010). In contrast, the 3′ ends of 5′ moRs and the 5′ ends of 3′ moRs were preferentially fixed, likely reflecting their derivation from a single cleavage event by Drosha.
Figure 2.
Examples of miRNA loci exhibiting five phased species. (A) mir-277; (B) mir-965. The most abundant product is the miRNA (green) followed by its partner miRNA* species (red). The 5′ and 3′ ends of these RNAs dovetail with the abundant loop reads (yellow), as well as 5′ miRNA overlap (moR) and 3′ moR reads (blue). The convention of highlighting mature species green and star species red is continued in all subsequent figures.
These byproduct reads exhibited distinctive abundance, with 57,875 loop, 27,969 5′ moRs, and 970 3′ moRs in the aggregate data (Supplemental Table S2). 5′ moRs were consistently more abundant than 3′ moRs on a gene-by-gene basis, reminiscent of earlier observations that many 5′ pri-miRNA fragments, but rarely 3′ pri-miRNA fragments, were detected by tiling microarrays (Manak et al. 2006). This suggests that decay pathways act with distinct efficiency on the unprotected ends of Drosha-cleaved pri-miRNA flanks. Select loci had more balanced moRs, such as mir-3 with 891 5′ and 287 3′ moRs. moR levels also were not strictly correlated with miRNA abundance. For example, despite 35 M total reads for mature bantam, this locus had only four 5′ moRs and three 3′ moRs in the aggregate data. Such variable frequencies of moRs might reflect aspects of miRNA processing that are differentially regulated at certain loci. All of the mappings to miRBase loci, across the 187 data sets, can be accessed in the Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html.
5′ isomiRs of canonical miRNAs
Even though confident annotation of miRNAs relies upon the preferred production of specific small RNAs from a precursor hairpin, most miRNAs exhibit some heterogeneity in cloned species, referred to as isomiRs (Ruby et al. 2007; Wu et al. 2007; Morin et al. 2008; Seitz et al. 2008; Chiang et al. 2010). Indeed, highly expressed Drosophila loci such as bantam or mir-8 were associated with well over a hundred variant miRNA species, although these ranged in abundance over seven orders of magnitude (Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html).
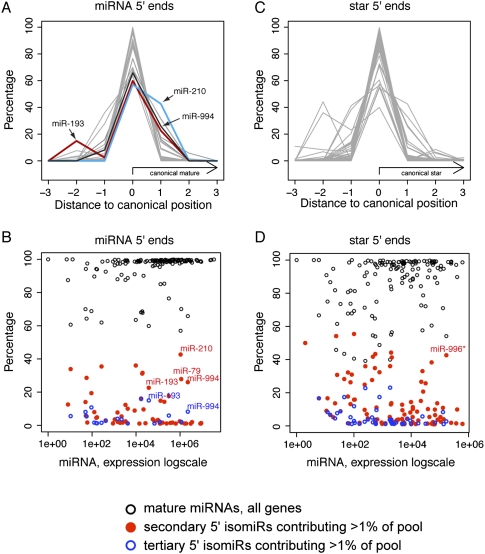
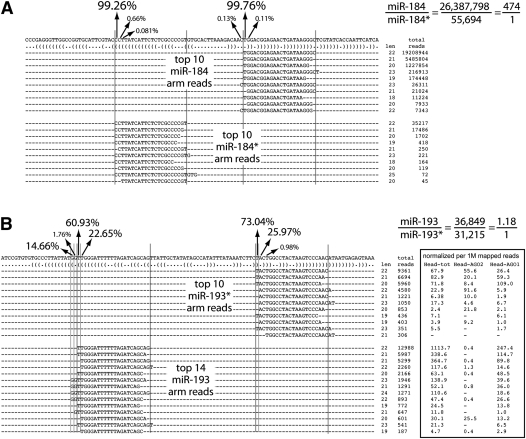
Since the regulatory spectrum of miRNAs is set by their 5′ ends (Lai 2002; Lewis et al. 2003; Brennecke et al. 2005), we were motivated to catalog their 5′ variation. The majority of loci exhibited high 5′ fidelity of both miRNA and star species (Fig. 3), as exemplified by mir-184 (Fig. 4A). This locus also illustrates that most loci have asymmetric accumulation of miRNA and star species, such that the dominant miRNA-type guide sequence generated by mir-184 is the miR-184 species. As reported from much smaller data sets (Ruby et al. 2007; Seitz et al. 2008), the 5′ ends of mature strands were detectably more precise than with star species (Fig. 3, cf. A,B with C,D). We also observed a general trend that the more highly expressed miRNAs and stars exhibited greater 5′ end fidelity than did lower expressed species (Fig. 3A).
Figure 3.
5′ variability of Drosophila canonical miRNAs. These charts summarize data for 135 canonical miRNAs that generated more than 1000 reads and had exclusively unique genomic mappings. (A,B) The 5′ end precision of mature miRNA species was generally high for well-expressed species; however, select loci generated abundant secondary and/or tertiary 5′ isomiRs. (C,D) The 5′ end precision of miRNA* (star) species was less than for mature miRNAs; still, only a relatively select group of highly expressed star species exhibited abundant 5′ isomiRs. The full analysis is available in Supplemental Table S4.
Figure 4.
Exemplary loci illustrating precision and variability in miRNA processing. (A) Most miRNAs, such as mir-184, exhibit precisely defined 5′ ends of both miRNA and star species. Since the mature strand of mir-184 is highly biased over its star species, there is one dominant miRNA-type regulatory species produced from this locus. (B) mir-193 is a locus exhibiting balanced accumulation of small RNAs from its hairpin arms. In addition, both 5p and 3p arms exhibit abundant secondary and even tertiary 5′ isomiR species. All of these accumulate in AGO1; therefore, mir-193 produces at least five substantial miRNA-type regulatory RNAs. Note that the 3p RNAs also accumulate in AGO2 as evidenced by their enrichment in a library prepared from small RNAs resistant to oxidization.
A subset of miRNAs and star species with highly imprecise 5′ ends were distinguished as clear outliers (Fig. 3A–D; Supplemental Table S4). Previously, the most striking case of a D. melanogaster 5′ isomiR was miR-210 (Fig. 3A,B), which exists as nearly equal populations of two different 5′ ends (Ruby et al. 2007). In this case, the mature miRNA is produced from the 3p arm, indicating heterogeneity at the Dicer-1 cleavage step. This trend held up with greater sequencing depth across the diversity of libraries, and inspection of AGO1 complexes from adult heads (Ghildiyal et al. 2010) confirmed the loading of both miR-210 5′ isomiRs into effector complexes (Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html). mir-79 was another locus with notable 5′ isomiR capacity on its mature (3p) strand (Fig. 3A,B). Its dominant reads assorted 70% and 27.8%, and a third 5′ class accounted for another 2.5% of miR-79 reads (Supplemental Table S4). All three of these 5′ isomiRs were recovered in similar proportions from ovary AGO1-IP complexes (GSE24310), indicating that they make substantial contributions to the miRNA target network controlled by mir-79.
mir-193 was even more remarkable in its extent of 5′ variation, which occurs on both miRNA (5p) and star (3p) strands (Fig. 4B). In fact, its mature (5p) species exists as a mixed population of RNAs with three distinct 5′ ends, comprising 60.9%, 22.7%, and 14.7% of miR-193-5p reads. All of these accumulated in relatively equal proportion in head AGO1 complexes (GSM466489) compared with total RNA (GSM466487) (Ghildiyal et al. 2010). Reciprocally, although its star (3p) species accumulated in AGO2 complexes (GSM466488), as is the case for many Drosophila miRNA* species (Czech et al. 2009; Okamura et al. 2009; Ghildiyal et al. 2010), both miR-193-3p 5′ isomiRs were also substantially incorporated into AGO1. Moreover, mir-193 exhibited reasonably balanced accumulation of its mature and star strands, with star species accounting for 30%–40% of total _mir-193_-derived reads in total RNA as well as in AGO1 complexes (Fig. 4B). Therefore, the combination of star utilization and alternative Drosha and Dicer processing strongly broadens the regulatory capacity of this locus for miRNA-type target regulation. All of the 5′ isomiR data are summarized in Supplemental Table S4.
Frequent antisense miRNA loci in Drosophila
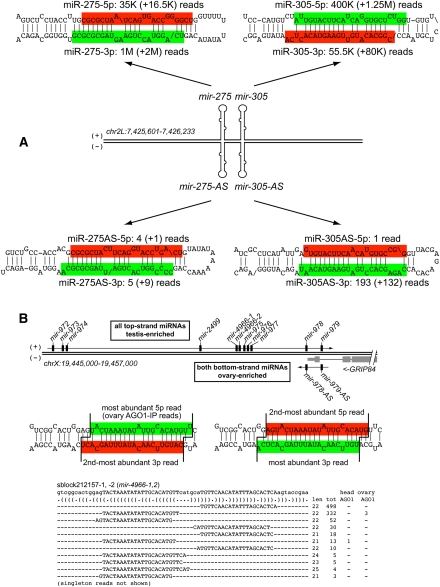
We, and others, reported that the Hox locus mir-iab-4 is transcribed and processed on its antisense strand, yielding mir-iab-8 (Ruby et al. 2007; Bender 2008; Stark et al. 2008; Tyler et al. 2008). In particular, mir-iab-8 is responsible for the sterility of mutants deleted for the locus (Bender 2008), and miR-iab-8-5p exhibits exceptional targeting capacity of the Hox genes abd-A and Ubx, distinct from miR-iab-4-5p (Stark et al. 2008; Tyler et al. 2008). We now recognized a dozen additional loci with confident patterns of antisense miRNA production, i.e., exhibiting a preferred small RNA duplex with 3′ overhangs and/or with reads in AGO1-IP libraries (Supplemental Table S5). These included mir-275/mir-305, for which we observed low abundance, but nonetheless specific, antisense miRNA/miRNA* duplexes for both members of the operon (Fig. 5A).
Figure 5.
Examples of antisense transcription and processing across miRNA operons. (A) The top genomic strand of the mir-275/mir-305 locus is abundantly converted into mature miRNAs, but the bottom genomic strand also exhibits confident evidence for miRNA production across both miRNA hairpins. Primary numbers indicate reads matching precisely to the highlighted species; numbers in parentheses sum all other isomiRs matching that hairpin arm. (B) The distal end of the mir-972-979 cluster on the X chromosome overlaps Grip84, transcribed on the other strand. We detected confident miRNA production from the antisense strands of mir-979 and mir-978. This locus also bears a perfect tandem hairpin (sblock212157/mir-4966) that is subject to alternate Drosha and Dicer-1 cleavage to produce multiple 5′ isomiRs on both hairpin arms; multiple species were also detected in AGO1-IP libraries. The entire hairpin is duplicated; thus, all reads could map to either location.
We also took note of mir-978 and mir-979, whose sense reads in animal libraries were by far most abundant in the testis (∼7000 and ∼500 reads recorded in GSM280085, respectively). Both genes exhibited antisense miRNA production (Fig. 5B) that was lower than sense production (mir-978-AS and mir-979-AS accumulated to 1/75 and 1/7 the level of their sense counterparts, respectively). Nevertheless, these were highly confident as antisense miRNA/miRNA* duplexes exhibiting 3′ overhangs and incorporation into AGO1 (Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html). Curiously, none of their antisense reads came from testis, and instead were found mostly in ovary data sets. This indicated the sexually dimorphic expression of sense and antisense strands of mir-978 and mir-979 in male and female gonads.
The distal end of the mir-972_→_979 cluster overlaps the 3′ end of Grip84, transcribed on the opposite strand (Ruby et al. 2007). In fact, mir-979 is contained within an intron of Grip84, while mir-978 is located just downstream of the annotated end of these gene (Fig. 5B). Grip84 is expressed by far at highest levels in ovaries among adult tissues (http://www.flyatlas.org/), consistent with the ovary-biased expression of these antisense miRNAs and supporting some functional connection between the expression mir-978-AS/mir-979-AS and Grip84.
In addition to 14 miRNA loci with confident evidence for mature antisense miRNAs (mir-iab-8, mir-307AS, and the 12 new annotations), six additional candidate antisense loci lacked star reads but had one to two reads in AGO1-IP libraries (Supplemental Table S5; Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html). In fact, one or more antisense reads were recorded for the majority of miRBase Drosophila miRNA loci (Supplemental Table S2). Although most of the latter are likely degradation products, it seems probable that some will eventually prove to be genuine Drosha/Dicer-1 products. These data support the notion that antisense processing may contribute substantially to the evolutionary emergence of novel miRNAs in Drosophila.
Novel genomic locations of confident miRNA genes include coding regions
Having analyzed reads mapping to sense or antisense to known miRNA loci, we were interested to annotate novel genomic locations of miRNA genes. Following considerable bioinformatics efforts to analyze the Drosophilid phylogeny for candidate miRNA genes (Lai et al. 2003; Ruby et al. 2007; Sandmann and Cohen 2007; Stark et al. 2007), it appears that few well-conserved miRNAs remain to be identified in this genus. Newly evolved, relatively species-specific miRNAs have been found (Lu et al. 2008; Berezikov et al. 2010), but these tend to accumulate to modest levels at best and require close inspection to distinguish them from a large background of RNA degradation fragments present in deep sequencing data. As with antisense miRNA loci, we identified novel genomic locations of miRNAs using stringent criteria, including the definition of specific 5′ ends and cloning of dominant miRNA/star species exhibiting 3′ overhangs (Chiang et al. 2010).
The vast majority of known miRNAs reside in intronic or intergenic space (Griffiths-Jones et al. 2008), and this remained the case with most novel miRNA loci that we annotated. Thirteen loci were intergenic, and 21 were located on the sense strands of introns (Supplemental Table S5). Inspection of the mir-972–mir-979 cluster revealed a cloned tandem hairpin just proximal to mir-975 (Fig. 5B). Pairing of the most abundant reads defines a duplex with atypical 3′ overhangs; however, these can be deconvolved into two alternate Drosha/Dicer-1 cleavages exhibiting 2-nt-3′ overhangs (Fig. 5B). One of the proposed cleavage registers places the Dicer-1 cut unusually far into the terminal loop, but its biogenesis was supported by the recovery of rare ovary AGO1-IP reads (GSE24310) whose small numbers were expected given low expression of this miRNA operon in ovary relative to testis. All told, this miRNA cluster is now the largest known in the D. melanogaster genome.
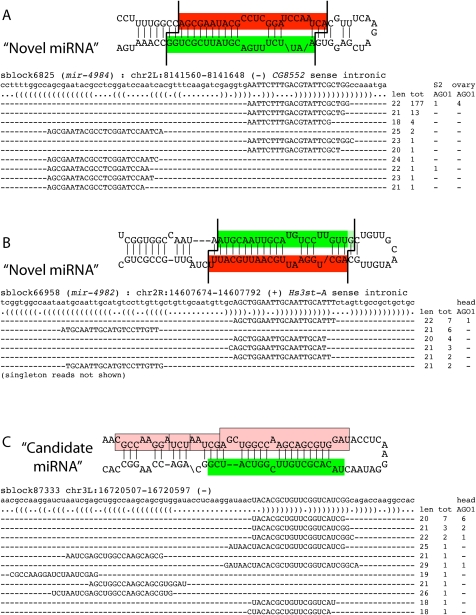
sblock6825/mir-4984 is an example of a novel confident miRNA annotated through several hundred reads conforming to a miRNA/miRNA* duplex and further supported by AGO1-IP reads in multiple tissues (Fig. 6A). sblock66958/mir-4982 is an example of a more modestly expressed locus, but one that still exhibited a confident miRNA cloning signature (Fig. 6B). Our annotations went to a lower limit of 12 mature strand reads in the case of sblock13008/mir-4946; however, its precise miRNA read was recorded in five libraries and it had star reads (Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html).
Figure 6.
Examples of novel miRNAs annotated in this study. (A) sblock6825/mir-4984 and (B) sblock66958/mir-4982 are novel miRNA loci that generate specific miRNA/miRNA* duplex species and had at least some reads in AGO1-IP libraries. mir-4982 approaches the lower limit for read accumulation needed for confident annotation. (C) sblock87333 is an example of a “candidate” miRNA locus that was not assigned a miRNA gene name at present. It exhibits heterogeneous 5p arm species (pink), and its dominant 3p arm is 20 nt in length, which is not typical for known miRNAs. Nevertheless, the 3p species clearly exhibit a preferred 5′ end, and several versions of the 3p species extending to 22 nt were present in head AGO1-IP data (GSM488489); one of the 5p reads would potentially pair with this duplex in an appropriate fashion. Therefore, this locus may eventually prove to be a genuine miRNA locus.
Potential mRNA-derived miRNAs must be evaluated especially carefully given the expectation that most mRNAs will generate at least some degradation reads. Only a handful of known miRNAs overlap untranslated regions of protein-coding genes (Rodriguez et al. 2004; Friedlander et al. 2008; Han et al. 2009); none have been confidently annotated from eukaryotic coding regions. We previously noted a few exonic hairpin candidates, but these did not have sufficient reproducibility, specificity, or star reads to reach confident annotation as genuine miRNAs (Ruby et al. 2007). Only recently did we annotate clear miRNA production from a Drosophila protein-coding transcript, mir-2280 within the 3′ untranslated region (UTR) of c-cup (Lau et al. 2009).
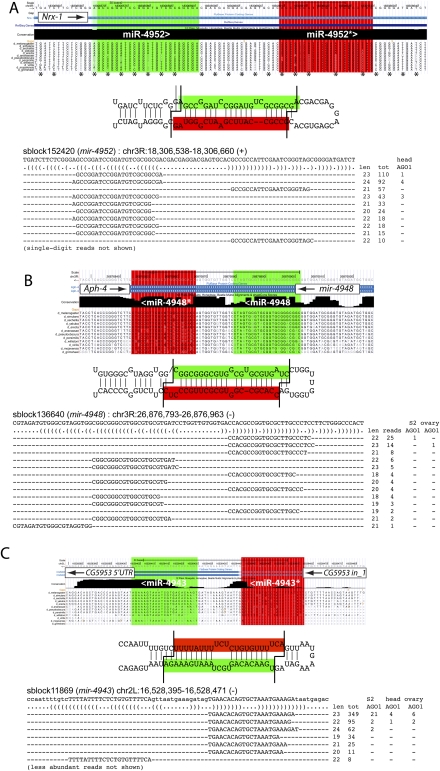
In this analysis, we included exonic loci in our pipeline of hairpin annotations and unexpectedly recovered a number of confident UTR- and CDS-resident miRNAs (Fig. 7A; Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html). Nine loci were located on CDS and two on UTRs (Supplemental Table S5). Their limited numbers confirmed that exons of protein-coding genes are not a major source of miRNA reads; nevertheless, Drosha/Dicer-1-mediated biogenesis of exonic miRNAs was reported by small RNA duplexes with appropriate 3′ overhangs, and usually also by reads in AGO1-IPs. Although the CDS miRNAs usually had conserved coding potential, they did not usually evolve in a way that suggested usage as _trans_-regulatory RNAs, that is, with loop divergence preferred over the hairpin arms (Lai et al. 2003). Instead, as illustrated by Nrx-1, the miRNA/star regions exhibited typical wobble position divergence similar to the terminal loop and flanking sequences (Fig. 7A).
Figure 7.
Examples of novel miRNAs generated from mRNAs. (A) A miRNA from the sense strand of the Nrx-1 coding region. This locus generates a specific miRNA/miRNA* duplex and exhibits some reads from head AGO1-IP data. Inspection of 12 species alignments indicates that the hairpin sequence evolves readily by codon wobbles, at a rate similar to the flanking nonhairpin codons. (B) A miRNA from the antisense strand of the Aph-4 coding region. In addition to specific miRNA/miRNA* duplex reads, this locus also generated a phased 5′ moR. (C) miRNA production from a primary-mRNA transcript in which the hairpin is produced from the pairing of intronic and exonic sequence of CG5953.
We identified three cases of miRNA production antisense to CDS regions, including aph-4 (Fig. 7B), and also from a hairpin spanning the sense strand of an exon-intron boundary in CG5953 (sblock11869/mir-4943) (Fig. 7C). While such arrangements might potentially serve regulatory functions, to target sense mRNAs or to disrupt mRNA splicing, it is also conceivable that these are simply neutrally evolving substrates. Further tests are needed to establish any _cis_- or _trans_-regulatory functions of these miRNA hairpins.
In total, we annotated at least 12 new antisense loci and 49 novel genomic locations of canonical miRNAs in D. melanogaster. Most of these are poorly conserved (excepting antisense and CDS loci) and modestly expressed (total counts from ∼2600 reads down to 12), but nonetheless judged confident for processing by Drosha/Dicer-1. Detailed summaries of the read evidence and structures supporting these miRNA annotations are provided in Supplemental Table S5 and the Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html.
Lower confidence cloned hairpins may comprise miRNA transitional intermediates
It seems unlikely that evolutionarily nascent miRNA genes would typically be “born” with all the necessary structural features for robust processing by miRNA biogenesis enzymes. Rather, many truly emergent miRNA hairpins might be processed inefficiently and/or imprecisely and probably do not deserve to be considered alongside miRNA loci that exhibit precise and efficient biogenesis. Nevertheless, we sought to segregate loci exhibiting partial evidence for processing by the Drosha/Dicer-1 pathway.
The aggregate list of initial hairpin loci with one or more mapped reads numbers over 200,000 and is not particularly informative. The vast proportion of these are clearly irrelevant as miRNA loci according to even loose criteria, but we segregated 61 compelling cloned loci that marginally failed confident classification, which we called “miRNA candidates” (Supplemental Table S5; Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html). Many of these exhibited putative miRNA/miRNA* duplexes, but these might not exhibit expected 3′ overhangs, or the reads might not have sufficiently precise termini (see Methods). These criteria are more stringent than those used for many previous miRNA annotations, and some loci deemed as candidates had evidence for putative star species as well as AGO1-IP reads (e.g., sblock87333 with nine AGO1-IP reads but also noncanonical sized mapped reads and several star reads with inconsistent overhangs; Fig. 6C). At least some of these miRNA candidates should gain confidence with additional small RNA data.
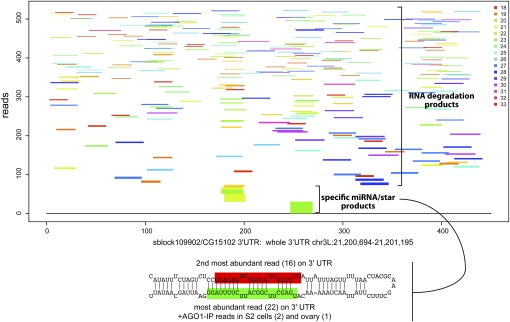
Some loci had remarkable features that placed them as compelling candidates for evolutionary transition intermediates toward miRNA birth. For example, the CG15102 transcript is broken down into heterogeneous RNA fragments that span the gamut of 18–30 nt (Supplemental Table S6). However, a majority of 21- to 22-nt reads mapped to a hairpin located in the 3′ UTR, comprising a duplex with 1- to 2-nt 3′ overhangs (Fig. 8). The putative miRNA species was recovered precisely in AGO1-IP libraries from S2 cells (GSM280088) and the ovary (GSE24310). Therefore, while this region clearly generates bulk reads via degradation, we infer that the CG15102 3′ UTR hairpin generates some short RNAs via Drosha/Dicer-1 cleavage. We hypothesize that such mixed evidence is the pattern expected for evolutionarily nascent miRNA substrates, and further study of such candidates may inform our understanding of the birth of miRNA genes. We provide detailed summaries of the read evidence and structures supporting these “candidate miRNA” annotations in the Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html.
Figure 8.
Example of a transitional miRNA locus, which exhibits signatures of both RNA degradation as well as Drosha/Dicer-1 processing across its precursor. Each read length has been plotted in a distinct color to emphasize the heterogeneity of cloned species mapping to the 3′ UTR of CG15102. The reads have been ordered on the _y_-axis with the most abundant individual species at the bottom. It can clearly be seen that a specific set of 21–22 nt reads are specifically made. These map to a typical pri-miRNA hairpin with a lower stem and a miRNA/miRNA* duplex region.
Absence of evidence for other previously annotated miRNA candidates
In our initial efforts at miRNA annotation, mir-280, mir-287, mir-288, and mir-289 emerged from comparative analysis of D. melanogaster and Drosophila pseudoobscura (Lai et al. 2003) but were not subsequently validated from small RNA sequencing (Ruby et al. 2007). The three latter genes were only tested because of their proximity to other clearly validated miRNA genes and otherwise did not score well on a genome-wide scan. Although these loci proved to be well conserved across the 12 flies (Berezikov et al. 2010), they lack classic patterns of miRNA evolution, namely, preferred nucleotide divergence in the terminal loop compared to the hairpin arms (Lai et al. 2003).
We obtained a few reads for these loci, and these were in the typical size range for miRNAs (21–23 nt). For example, precisely the same 21-nt read was recorded 13 times across seven different libraries from the annotated arm of mir-287. For mir-288, the dominant species of 23 nt corresponded exactly to the previously predicted product, and was sequenced six times in four libraries. These limited data were insufficient for confident miRNA validation, suggesting that they should be flagged in the miRBase registry. Nevertheless, their propensity to generate some specific small RNAs (Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html) suggests their possible function as conserved structured ncRNAs in flies.
We, and others, subsequently annotated D. melanogaster miRNA candidates using comparative studies of 12 sequenced fruitfly genomes (Ruby et al. 2007; Stark et al. 2007). We searched for short RNA production from several hundred conserved hairpin candidates not validated from the approximately 1 M mapped reads available at the time. Strikingly, the data from nearly three orders of magnitude greater sequencing failed to validate any of these loci as confident miRNA loci. Only a minor fraction of these lacked short RNA mappings, demonstrating that the aggregate data indeed sampled transcription across most of these loci. Nevertheless, these reads mapped haphazardly over the annotated hairpin and/or had heterogeneous sizes. Moreover, in contrast to the miRNA loci newly annotated in this study, almost all of which had some AGO1-IP reads, almost none of these conserved hairpins had AGO1-IP reads. The sole exceptions were a set of hairpins overlapping tRNAs and snRNA that each generated more than 10,000 total reads, whose six to nine AGO1-IP reads likely represented spurious association (Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html).
We conclude that there remain very few well-conserved Drosophila miRNA genes that have escaped discovery efforts. It remains plausible though, if not likely, that many of these conserved segments of the genome have been retained for functional or regulatory reasons other than miRNA production. We provide detailed analysis of the reads mapping to the previously described candidates in the Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html.
Untemplated modifications of miRNAs
The intermediate and mature products of miRNA loci can be modified at their 3′ ends, including by uridylation or adenylation (Kim et al. 2010). Mature miRNAs can be excised from either the 5′ or 3′ arms of different hairpins; thus, modifications to mature small RNAs are expected to occur collectively on both 5p and 3p species. In cases where the modification acts preferentially on the pre-miRNA hairpin, however, the untemplated nucleotides may exhibit a bias for 3p species.
We mapped each of the 187 libraries to the genome using prefix analysis, which we recently used to determine the nature of untemplated nucleotide matches to siRNAs and miRNAs subject to target-mediated tailing and degradation (Ameres et al. 2010). We binned the reads according to the nature of their 3′ untemplated nucleotides and pooled the data sets from each library normalized by sequencing depth (Supplemental Table S7). These analyses detected levels of 3′ uridylation and adenylation that were substantially higher than other types of modification, mirroring results obtained for mammalian miRNAs (Burroughs et al. 2010; Chiang et al. 2010). For uridylation and adenylation, we observed statistically significant twofold to 2.5-fold greater modification of 3p species compared with 5p species (Fig. 9), consistent with preferred additions onto pre-miRNA substrates.
Figure 9.
Patterns of 3′ untemplated additions in Drosophila miRNAs. (Left) Scenarios for 3′ untemplated additions to the pre-miRNA versus the mature miRNA/miRNA* species. Preferred addition to the pre-miRNA hairpin is expected to be reflected in a bias for modifications of 5p species relative to 3p species. (Right) The overall frequency of 3′ additions observed on Drosophila miRNAs are U > A > C > G. For U and A additions, _t_-test reveals a statistically significant preference for 3p additions, consistent with a preference for pre-miRNA modifications. C additions were much less frequent but also appeared to exhibit some 3p preference. Judging 5p U or A addition frequencies relative to G additions as background suggested that mature miRNA/miRNA* species are also subject to uridylation and adenylation. The full analysis is presented in Supplemental Table S7.
We also observed slightly more cytidylation on 3p species than 5p species, whose frequency was indistinguishable from the general rate of C addition for ncRNAs. While no Drosophila enzyme that would catalyze C addition is known, such an activity was detected in mammalian thymus (Edmonds 1965). The low rate (0.3%) of untemplated guanine addition did not differ between 5p and 3p species and was lower than that found among ncRNAs generally. Finally, we note that the slightly higher frequencies of U and A additions to 5p species, compared with other ncRNAs in general, indicated that uridylation and adenylation occurs detectably on mature miRNAs, in addition to pre-miRNAs.
Looking at addition patterns across all libraries, we found that miR-13a, miR-13b, miR-34*, miR-279, miR-312, and miR-92a were consistently adenylated, while miR-2c, miR-970, miR-988, miR-1003, miR-1008, miR-1010, and miR-1012 were consistently uridylated (Supplemental Table S7), indicating that the modifying enzymes exhibit preference for particular miRNA substrates. We further noted that 80% of all reads carrying 3′ additions specifically bore a single untemplated nucleotide. However, there was a strong correlation between mononucleotide and homo-polynucleotide additions of the same type (A: r = 0.63; T: r = 0.79), suggesting processivity of the modifying enzymes.
miRNA editing
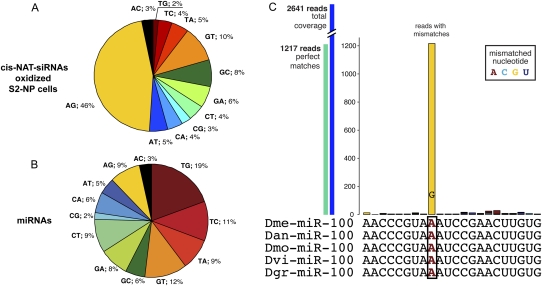
The Drosophila adenosine deaminase (dADAR) is relatively neural-specific, and consistent with this, most of its known mRNA targets are neural transcripts (Stapleton et al. 2006). However, RNA editing has also been suggested to occur in cultured S2 cells, based on the strong enrichment of A→G alterations in endo-siRNAs associated with AGO2 in S2 cells (Kawamura et al. 2008). We tested this notion using an independent data set of AGO2-associated reads from S2 cells (GSM280087). Most endo-siRNAs derive from TEs and have multiple mappings, thus confounding the genomic origin of reads that match imperfectly to TEs. We therefore chose to analyze putatively edited reads derived from uniquely mapping 3′ _cis_-natural antisense transcript siRNAs (3′-cis-NAT-siRNAs). We indeed observed strong enrichment for A→G alterations in these endo-siRNAs (Fig. 10A), confirming that dsRNA in S2 cells is subject to adenosine deamination. In contrast, analysis of miRNA species in S2 cells failed to provide similar evidence for preferred A→G alterations compared with other types of nucleotide changes (Fig. 10B). Surveys of mammalian miRNAs similarly suggested that there are relatively few instances of editing that can be detected in cloned short RNAs (Chiang et al. 2010), although these might be underestimated if pri-miRNA or pre-miRNA editing inhibits their biogenesis (Yang et al. 2006).
Figure 10.
RNA editing in Drosophila small RNAs. We collected S2 and head small RNA reads with one or two mismatches to 3′ _cis_-NATs or miRNAs and tabulated the nature of their nucleotide changes. (A) Endo-siRNAs from 3′ _cis_-NATs exhibit a preponderance of A→G changes indicative of adenosine deamination. (B) In contrast, miRNA reads do not collectively exhibit enrichment for A→G changes. (C) miR-100 is a highly conserved miRNA with abundant A→G transition reads present in multiple libraries. The full analysis is presented in Supplemental Table S8.
Nevertheless, individual occurrences of editing might have significant functional consequences, as shown for several mammalian miRNAs (Yang et al. 2006). We designed a computational pipeline to predict potential RNA editing candidates (see Methods) and focused our analysis on 36 S2 cell and head small RNA libraries. A number of edited miRNA candidates emerged (Supplemental Table S8), potentially affecting diverse aspects of miRNA biogenesis and/or function. We found particularly compelling those cases that satisfied additional criteria, such as the existence of a strong proportion of edited species in libraries generated by independent laboratories, the recovery of relatively large numbers (e.g., >100) of edited species, and/or cases in which the genomic identity of the edited nucleotide was highly conserved among Drosophilid genomes. Loci that satisfied all of these criteria included miR-100 (Fig. 10C), miR-971, and miR-33*. A full description of the candidate editing events and their levels of evidence are presented in Supplemental Table S8.
Conclusions
Deep sequencing yields many insights into known miRNA genes
Next-generation sequencing has revolutionized the collection of large-scale data and provided a foundation for recent stunning advances in small RNA research. Deep sequencing is now a standard technique to profile small RNA expression and continues to fuel the discovery of novel regulatory RNAs and biogenesis pathways. However, as small RNA samples are not typically normalized, there are now more than 100 M reads derived from a handful of fly miRNAs. In principle, it might be advantageous for pure discovery efforts to deplete highly expressed loci prior to sequencing. Nevertheless, valuable information has been gained from deep sequencing of known miRNA genes.
For example, in this study we performed careful annotations of 5′ isomiRs, which presumably broaden the regulatory capacity of miRNA genes given their frequent residence in AGO1 complexes. Simple inspection does not offer obvious structural clues as to why a subset of miRNA hairpins are susceptible to alternative Drosha and/or Dicer cleavage. Many of these alternative processing events occur within well-duplexed regions, which appear to present a well-defined cleavage surface. By analogy to other RNA binding proteins that modulate miRNA processing (Winter et al. 2009), we hypothesize that _trans_-acting factors could act upon specific miRNAs to adjust sites of RNase III processing.
We observed phasing of 5′moR/miR-5p/loop/miR-3p/3′ moR species for certain abundant canonical miRNA loci (Fig. 1). As small RNA data sets continue to accumulate and as broader windows of small RNA sizes are analyzed to capture more loop sequences, it may become commonplace to capture all five phased products of canonical miRNA biogenesis. In principle, alternative Drosha and/or Dicer-1 cleavages should be reflected in phased moR/loop reads. In the future, such data could help to distinguish miRNA variation that occurs as a consequence of alternative precursor cleavage, as opposed to subsequent exonuclease processing.
Deep sequencing of known loci also permitted untemplated additions and candidate editing events to be discerned. The current analyses extend our earlier observation of populations of miRNA reads with nongenome matching 3′ nucleotides in Drosophila (Ruby et al. 2007). It is now clear that 3′ uridylation (Hagan et al. 2009; Heo et al. 2009; Lehrbach et al. 2009) or adenylation (Katoh et al. 2009) of specific animal miRNAs can have profound effects on their processing and/or function. Uridylation of miRNAs is relatively common for mammalian pre-miRNAs as inferred from the preferred modification of 3p versus 5p hairpin reads (Chiang et al. 2010), and adenylation of mammalian miRNAs also appears common (Burroughs et al. 2010). Our studies provide broad evidence for both reactions on Drosophila miRNAs. In addition, we identified a limited set of high-confidence editing events in mature miRNAs. These comprise several classes of potential functional consequences, including changes in target specificity from altered seeds, and potentially altered processing and/or AGO sorting. These findings organize future experimental studies of miRNA modifications in Drosophila.
Evidence for a relatively limited number of miRNA substrates in Drosophila
It is of substantial interest to understand the dynamics of miRNA gene birth and death. This effort must rest upon a foundation of confident annotations of loci whose transcripts transit defined biogenesis pathways to yield genuine miRNA species. Careful annotation is necessary with next-generation sequence data sets, which can contain a large number and variety of short RNA reads generated by general RNA catabolism. In addition, in Drosophila, the endo-siRNA and piRNA pathways generate a tremendous diversity of short RNAs, whose incidental mapping to predicted hairpins cause many loci to masquerade as miRNA precursors.
To our knowledge, this study utilized the broadest sample diversity and largest read repository of any miRNA study to date. We annotated miRNAs on the basis of confident evidence, such as miRNA/star duplexes with appropriate overhangs and presence in AGO1-IP data, yielding a comprehensive view of canonical miRNA genes in this species. Despite our requirement for strict evidence, the depth of small RNAs analyzed permitted confident annotation of miRNAs expressed at vanishingly low levels. We do not expect such rare species to have substantial effects on endogenous gene regulation. Nevertheless, their defined processing characteristics are a testament to the depth of the underlying small RNA data and to their appropriate definition as “miRNAs.”
In theory, the more than 100,000 hairpins in the D. melanogaster genome, whose predicted structures are seemingly similar to those of confident miRNA genes (Lai et al. 2003), provide a vast set of potential substrates for entry into miRNA biogenesis. The pool of nascent miRNAs has been proposed to mediate a set of subtle regulatory interactions that may lead to their elimination if detrimental or possibly subject them to evolutionary retention if selected for beneficial activities (Bartel and Chen 2004; Chen and Rajewsky 2007). Our observations suggest that the pool of evolutionary nascent miRNAs is relatively limited and that at most only a couple hundred Drosophila hairpins are competent as miRNA substrate transcripts. The evidence for this viewpoint rests on our high genomic coverage of short RNA reads, the pervasive resequencing of the same set of miRNAs across a diverse cohort of unrelated tissues and cell types, and the recovery of a substantial set of neutrally evolving miRNA substrate transcripts. These data indicate that the cellular selection of miRNA substrates is much more restricted than we can envisage from current biochemical knowledge, and highlights the fact that substantial improvements in computational methods for the prediction of canonical miRNA genes remain to be had.
In concurrent work (Chung et al. 2011), we developed a computational model that effectively predicted mirtrons, which generate a subfamily of miRNA-class regulatory RNAs from splicing of short hairpin introns. Interestingly, we find that mirtrons and canonical miRNAs evolve and become fixed in Drosophila genomes according to distinct rates (Berezikov et al. 2010). This study extends the concept that the emergence and fixation of miRNA genes in different genomic locations may follow distinct and potentially independent evolutionary rules. For instance, even though we identified clear cases of canonical miRNA biogenesis from coding regions of mRNAs, the fact that there are no well-conserved cases of CDS miRNAs in Drosophila suggests that these are purged from genomes. On the other hand, the antisense strands of previously annotated miRNA loci stand out as a seemingly facile location for the expression of novel miRNAs, given their extremely limited genomic space (i.e., we annotated 12 novel miRNAs from a collective space of ∼15 kb antisense to known miRNA genes, compared with 49 miRNA hairpins annotated from the remaining 120 Mb of the genome). Altogether, our data suggest that there is no universal rate of “miRNA evolution.” Deep sequencing of small RNAs from across Drosophilid speciation should permit empirical tests of this notion.
Methods
Small RNA data sets
The complete listings of NCBI-GEO/SRA and modENCODE-DCC accession IDs for the 187 small RNA data sets analyzed are in Supplemental Table S1. Where possible, we began with raw sequences so that the data were processed uniformly. 3′ linker sequences were stripped using the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Except as noted, we used Bowtie (Langmead et al. 2009) to map to the dm3 genome assembly, using parameters to restrict to perfectly matching reads ≥18 nt and all genomic hits reported.
Analysis of miRNA 5′ variation
We selected 135 canonical Drosophila miRNAs that generated more than 1000 reads that were ≥18 nt and had exclusively unique genomic mappings. We tabulated the frequency of alternative 5′ ends and their position (nucleotide 5′ or 3′ to the base end) in Supplemental Table S4.
miRNA discovery
We used miR-Intess software tuned for performance on Drosophila (Lau et al. 2009; Berezikov et al. 2010). Nonrepetitive loci, including exonic locations, were assessed for hairpin structures using RNAshapes (Steffen et al. 2006) and for small RNA read patterns that reported confidently on Drosha/Dicer-1 cleavage. In general, we considered confident those loci with dominant mature/star reads exhibiting 3′ overhangs as duplexes, with <5-bp internal loops or asymmetric bulges. Bearing in mind that some confident loci exhibit alternative processing to generate an abundant isomiR (e.g., main Figs. 4, 5), we required 10 or more mature strand reads including up to one 5′ isomiR to constitute more than two-thirds of reads mapped to the hairpin arm, and two or more star reads.
Certain genuine miRNA loci might lack star reads if their duplexes were subject to strongly asymmetric strand selection. Since RNase III cleavage cannot confidently be inferred without star reads, we required in these cases that there be at least three reads in a wild-type AGO1-IP library, along with a proviso that the given species (along with up to one 5′ isomiR) constituted more than two-thirds of reads mapped to the hairpin arm. Without star reads, we considered hairpins with one or two reads in a wild-type AGO1-IP library, or exclusive AGO1-IP reads from mutant or knockdown samples (often signifying endo-siRNAs), to be insufficient evidence for miRNA annotation.
All of the loci were manually vetted to meet confident criteria, and the strong majority of annotated loci had both star reads as well as AGO1-IP reads. Compelling loci that met some, but not all confidence criteria were provisionally annotated as “candidates.” Further details of miRNA read characteristics are described in the Supplemental Text, and the complete mappings and structures of all the miRNA loci are available in the Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html.
Analysis of untemplated additions
We mapped all sequencing reads to the fly genome with a prefix matching algorithm (Ameres et al. 2010), which allowed a 3′ overhang of any number of mismatches on the reads. We binned the 3′ overhang according to their sequences: homo-A, -C, -G, -T, or X (X means mixed ACGT). We pooled multiple data sets, normalizing each data set by sequencing depth. We identified miRNAs that were consistently adenylated (or uridylated) as follows. For each miRNA, we identified the percentages of data sets in which it had higher than 1%, 5%, and 10% adenylation. Then we ranked all miRNAs by their percentages of data sets for each cutoff (1%, 5%, or 10%). The miRNAs that were in the top 20 for all three cutoffs were retained.
Identification of candidate RNA editing events
Reads were mapped using Bowtie allowing up to two mismatches (-v 2 --best), keeping only one alignment per read. The following filters were implemented to retain higher-confidence editing events, avoid SNPs, and minimize the impact of sequencing errors: (1) The average base sequencing quality score for a given position is greater than 20; (2) all candidate positions satisfy the neighborhood quality score criteria (NQS20/15); (3) modification is neither in the first nor last two bases of a read; (4) base coverage is greater than 15; and (5) frequency of the most abundant variant base is within 10%–85% interval of total coverage at mismatched position. In the absence of a prevalent variant, the candidate position is discarded. Candidates that passed this multilevel filtration procedure are listed in Supplemental Table S6.
Additional detailed descriptions of data generation and analysis can be found in the Supplemental Text and the eight Supplemental Tables. Finally, detailed information on reads mapping to miRBase loci, newly annotated miRNA genes, candidate miRNA hairpins, and unannotated loci from Ruby et al. (2007) and from Stark et al. (2007), can be browsed in the Supplemental analyses available online at http://www.macgenome.org/pub/lai_mirna/main.html.
Acknowledgments
We thank the Forstemann, Siomi, Wu, and Carthew laboratories for contributing to our studies by depositing their published small RNA data at NCBI-GEO/SRA. We thank Sue Celniker, the modENCODE transcriptome group at Indiana University (Peter Cherbas, Lucy Cherbas, Justen Andrews, Dayu Zhang, and Johnny Roberts), and Michael Brodsky for contributing samples from cultured cells and flies used for small RNA cloning. We also thank Zheng Zha and Lincoln Stein for helping with some of the GEO submissions and Peter Smibert for discussion. Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Alfred Bressler Scholars Fund, and the NIH (R01-GM083300 and U01-HG004261).
Footnotes
[Supplemental material is available for this article.]
References
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC 2005. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci 102: 18017–18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat Genet 5: 396–400 [DOI] [PubMed] [Google Scholar]
- Bender W 2008. MicroRNAs in the Drosophila bithorax complex. Genes Dev 22: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21–24 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC 2010. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet 42: 6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85 doi: 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N 2007. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8: 93–103 [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W-J, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC 2011. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res (this issue). doi: 10.1101/gr.113050.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ 2009. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 36: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds M 1965. A cytidine triphosphate polymerase from thymus nuclei. 1. Purification and properties of the enzyme and its polynucleotide primer. J Biol Chem 240: 4621–4628 [PubMed] [Google Scholar]
- Flynt AS, Lai EC 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 9: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD 2010. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y, Aach J, Hayes G, Reinhart BJ, Church G, Ruvkun G, Kim J 2003. Computational and experimental identification of C. elegans microRNAs. Mol Cell 11: 1253–1263 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kim YK, Heo I, Kim VN 2010. Modifications of small RNAs and their associated proteins. Cell 143: 703–709 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lai EC 2002. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30: 363–364 [DOI] [PubMed] [Google Scholar]
- Lai EC 2003. microRNAs: Runts of the genome assert themselves. Curr Biol 13: R925–R936 [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM 2003. Computational identification of Drosophila microRNA genes. Genome Biol 4: R42 doi: 10.1186/gb-2003-4-7-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N, Lim L, Weinstein E, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lau N, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC 2009. Abundant primary piRNAs, endo-siRNAs and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 16: 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP 2003a. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP 2003b. The microRNAs of Caenorhabditis elegans. Genes Dev 17: 991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI 2008. The birth and death of microRNA genes in Drosophila. Nat Genet 40: 351–355 [DOI] [PubMed] [Google Scholar]
- Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, Long J, Cheng J, Bell I, Ghosh S, Piccolboni A, et al. 2006. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet 38: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC 2008. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Cohen SM 2007. Identification of novel Drosophila melanogaster microRNAs. PLoS ONE 2: e1265 doi: 10.1371/journal.pone.0001265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD 2008. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr Biol 18: 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Hendrix D, Levine M, Haley B 2009. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat Struct Mol Biol 16: 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Carlson JW, Celniker SE 2006. RNA editing in Drosophila melanogaster: New targets and functional consequences. RNA 12: 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M 2007. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res 17: 1865–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M 2008. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev 22: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R 2006. RNAshapes: An integrated RNA analysis package based on abstract shapes. Bioinformatics 22: 500–503 [DOI] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC 2008. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev 22: 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N 2007. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE 2: e1020 doi: 10.1371/journal.pone.0001020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC 2010. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle 9: 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]