The WNKs: atypical protein kinases with pleiotropic actions (original) (raw)
. Author manuscript; available in PMC: 2011 Feb 8.
Abstract
WNKs are serine/threonine kinases that comprise a unique branch of the kinome. They are so-named owing to the unusual placement of an essential catalytic lysine. WNKs have now been identified in diverse organisms. In humans and other mammals, four genes encoding WNKs. WNKs are widely expressed at the message level, although data on protein expression is more limited. Soon after the WNKs were identified, mutations in genes encoding WNK 1 and 4 were determined to cause the human disease, Familial Hyperkalemic Hypertension (also known as pseudohypoaldosteronism II, or Gordon’s Syndrome). For this reason, a major focus of investigation has been to dissect the role of WNK kinases in renal regulation of ion transport. More recently, a different mutation in WNK1 was identified as the cause of hereditary sensory and autonomic neuropathy type II (HSANII), an early-onset autosomal disease of peripheral sensory nerves. Thus, the WNKs represent an important family of potential targets for the treatment of human disease, and further elucidation of their physiological actions outside of the kidney and brain is necessary. In this review, we describe the gene structure and mechanisms regulating expression and activity of the WNKs. Subsequently, we outline substrates and targets of WNKs, and effects of WNKs on cellular physiology, both in the kidney and elsewhere. Next, consequences of these effects on integrated physiological function are outlined. Finally, we discuss the known and putative pathophysiological relevance of the WNKs.
I. Introduction
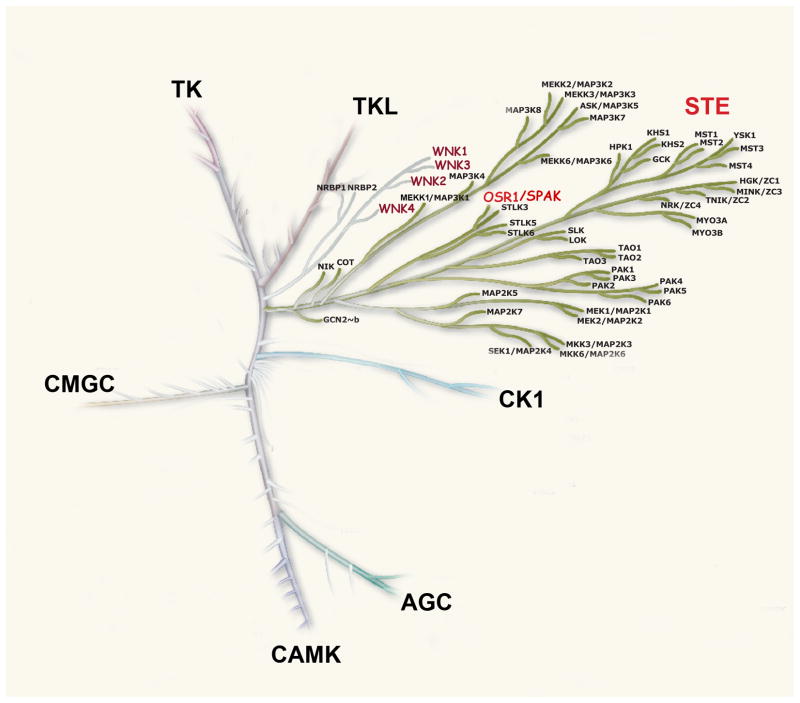
WNKs are serine/threonine kinases that comprise a unique branch of the kinome (Figure 1). The first member of the WNK (with no lysine [K]) family was cloned in 2000 by Cobb and colleagues from a rat brain cDNA library during a search for novel members of the MAP/extracellular signal-regulated protein kinase family (266). The WNKs were so-named owing to the unique placement of the lysine involved in binding ATP and catalyzing phosphoryl transfer in subdomain I, rather than subdomain II, where it is located in other serine/threonine kinases. WNKs have now been identified in multicellular organisms and unicellular organisms such as Giardia lamblia, but are absent from unicellular organisms such as S. cerevisiae (see Table 1).
Figure 1. Major families of the human kinome (adapted from (132)).
WNK kinases (in red) comprise a unique branch, which is most closely related to STE kinases. Note that OSR1 (and SPAK) are members of the STE protein kinase family.
Table 1. WNK kinases are expressed by a wide variety of organisms.
Accession numbers are provided only for WNK kinases for which a cDNA exists. Clones designated BC are partial cDNA clones, XM have incomplete cDNA ends. Arabadopsis and vertebrate isoform numbers are not equivalent.
| Species | Number of WNKs identified | Accessions for NCBI reference sequence |
|---|---|---|
| Arabidopsis thaliana | 9 | WNK1 NM_001035560.1, WNK2 NM_202625.1, WNK3 NM_148852.1, WNK4 NM_125220.4, WNK5 NM_115022.3, WNK6 NM_112761.3, WNK7 NM_103806.1, WNK8 NM_123564.3, WNK9 NM_122691.3 |
| Caenorhabditis elegans | 1 | NM_182275.3 |
| Giardia lambia | 1 | XM_001706061 |
| Oryza sp. | 2 | NM_001066596, NM_001054244 |
| Xenopus sp. | 2 | X. Tropicalis WNK1 NM_001097234, WNK3 NM_001005052X. Laevis WNK1 BC099030, WNK3 BC077899 |
| Mus musculus | 4 | WNK1 NM_198703, WNK2 NM_029361.3, WNK3 BC060731, WNK4 NM_175638 |
| Rattus rattus | 4 | WNK1 NM_053794, WNK3 NM_001163607, WNK4 NM_175579.3 |
| Homo sapiens | 4 | WNK1 NM_018979, WNK2 NM_006648, WNK3 NM_020922, WNK4 NM_032387 |
Arabidopsis thaliana has at least 9 WNKs, the largest number identified, whereas mammals have 4 WNKs (Figure 2), excluding splice variants or isoforms derived from alternative promoter usage [section II.A.]. In humans, the four genes encoding WNKs 1–4 have been localized to chromosomes 12p13.33 (WNK1), 9q22.31 (WNK2), Xp11.22 (WNK3) and 17q21.31 (WNK4); these isoforms have also been identified in multiple other mammalian species. Other species in which WNK kinases have been identified or predicted include Caenorhabditis elegans, Oryza, Phycomyces, Drosophila melanogaster, Xenopus sp., Danio rerio and chicken.
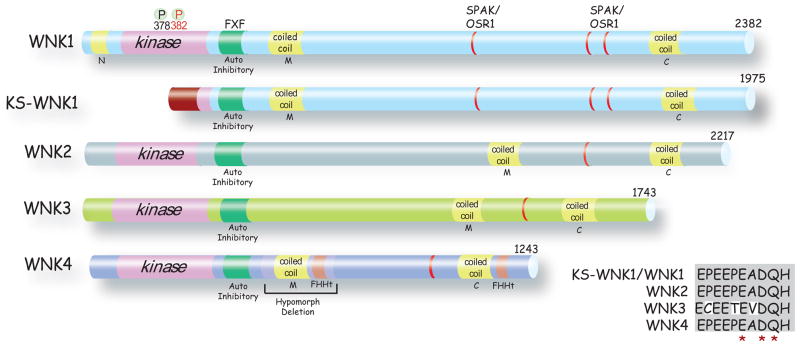
Figure 2. Structures of WNK Kinases.
WNKs 1–4 and KS-WNK1 are shown, with the length of each, indicated. All, except KS-WNK1, contain homologous kinase domains (pink), autoinhibitory domains (green) with essential phenylalanine residues (F) separated by a single amino acid (X). All WNKs have a coiled-coil domain (yellow) near the carboxyl terminus (C) and within the middle portion of the protein (M). WNK1 also contains an amino terminal (N) coiled-coil domain. Two phosphorylated (P) serine residues that are essential for WNK1 activation are shown (378 and 382). The alternative first exon in KS-WNK1 is shown in dark red. SPAK/OSR1 binding regions are shown in red. Sites of FHHt mutations are shown in orange. The portion of the molecule deleted to generate WNK4 hypomorphic mice (167) is indicated. Inset shows an alignment of WNK kinase acidic motifs. The acidic motif is highly conserved between WNK family members. The residues mutated in WNK4 that lead to FHHt are marked *. The functional consequences of disrupting this motif in other members of the WNK family are unknown.
In mammals, the WNK kinases are widely expressed, at the message level, as determined by RT-PCR and Northern blotting. Data on protein expression, however, is more limited owing to the poor availability of well-validated antibodies and the sometimes-contradictory results. WNK1 is widely expressed, with highest levels in the testis, heart, kidney and skeletal muscle and lower expression in brain (163). In chloride-transporting epithelia, WNK1 displays tissue-specific cell distribution. In the kidney, colon and gall bladder it is cytoplasmic; in the ducts of the liver and pancreas it is mainly localized on the lateral membranes (30). WNK2 is predominantly expressed in heart, brain and colon (245). WNK3 is expressed at low levels in brain, lung, kidney, liver and pancreas, and in fetal tissues including placenta, fetal brain, lung and kidney; very low levels are present in fetal heart, thymus, liver and spleen (80, 245). WNK4 is expressed in tissues containing secretory epithelia including kidney, pancreas, bile duct, colon, brain (blood-brain barrier), epididymis and skin (95, 245).
Based on their expression pattern, the WNKs are likely to have multiple physiological functions in diverse tissues. Soon after the WNKs were identified, mutations in genes encoding two member of the WNK family (WNKs 1 and 4) were identified by positional cloning as causing the human disease Familial Hyperkalemic Hypertension (FHHt, also known as pseudohypoaldosteronism II, or Gordon’s Syndrome) (258). For this reason, a major focus to dissect their function has been with regard to renal regulation of ion transport. The symptoms of this disease are reversed by treatment with thiazide diuretics, which specifically inhibit the Sodium Chloride cotransporter SLC12a3 (NCC), suggesting a primarily renal defect. More recently, a mutation in WNK1 was identified as the cause of Hereditary sensory and autonomic neuropathy type II (HSANII), an early-onset autosomal recessive disorder characterized by loss of perception to pain, touch, and heat due to a loss of peripheral sensory nerves (211). Thus, the WNKs represent an important family of potential targets for the treatment of human disease, and further elucidation of their physiological actions outside of the kidney and brain is necessary. Knowledge on the physiological significance of WNK2 and WNK3 is particularly lacking.
In this review, we first describe the gene structure [section I] and mechanisms regulating expression and activity of the WNKs [section II]. Subsequently, we outline substrates and targets of WNKs, and their effects on cellular physiology [section III]. Next, consequences of these effects on integrated physiological function are outlined [section IV]. Finally, we discuss the known and putative pathophysiological relevance of the WNKs [section V].
II. WNK kinase gene structure, expression and regulation
A. Genomic structure
Databases, such as AceView, indicate that variation of WNK kinase transcripts is complex, but here we will focus on transcripts for which there is published experimental evidence.
1. WNK1
The human WNK1 gene spans 160kb, contains 28 exons, and its transcriptional regulation is complex, with the existence of both an isoform derived from an alternative promoter and splice variants. The existence of variant WNK1 transcripts was suggested by the work of Lifton and colleagues, which showed transcripts of varying length on multiple-tissue Northern blots (258). The WNK1 promoter region contains a CpG island that extends from −1453 to +166 of the ATG start codon, and in common with other CpG island promoters, lacks a TATA-box. Comparison with the human expressed sequence tag database revealed multiple potential transcription initiation sites (245).
Primer extension and 5′ RACE-PCR identified two transcription start sites in the CpG island, 219 and 179 base pairs upstream of the ATG start codon (41). Both of these sites were confirmed to be functional using Northern Blot analysis and reporter assays. Two additional transcription initiation sites were identified within exon 1 (downstream of the CpG island), resulting in truncated WNK1 transcripts lacking the first 639 nucleotides of the coding sequence, but containing the entire kinase domain. It is unknown whether there is any functional difference between the full length and truncated form.
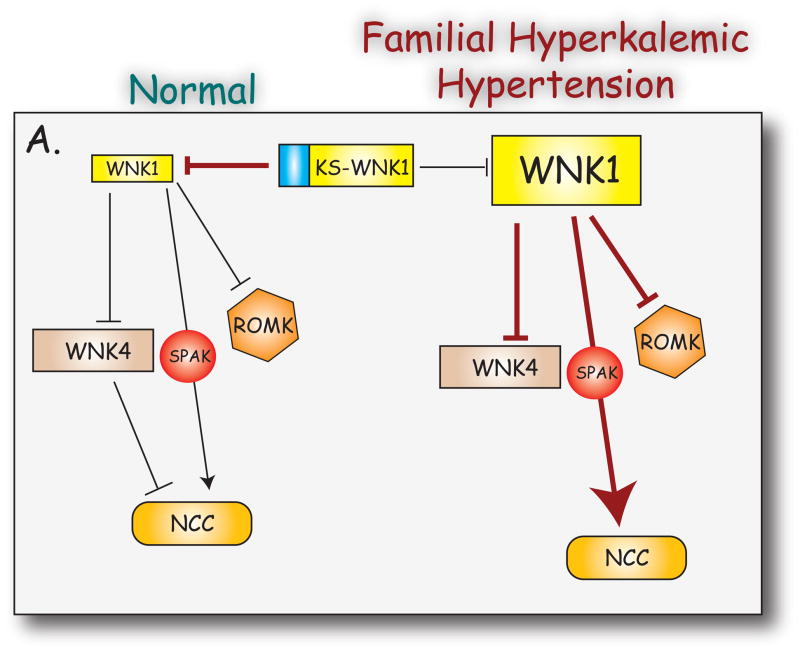
Xu and colleagues reported the existence of a novel truncated isoform of WNK1(272), now typically called kidney-specific WNK1 (KS-WNK1). This isoform was detected in human (41) and mouse (163), and is likely to play a role in the regulation of blood pressure [section III.E.]. Importantly, KS-WNK1 appears to be expressed specifically in the distal convoluted tubule (DCT) of the kidney (41, 163). The transcript encoding this isoform is generated from an alternative promoter upstream of exon 4, and its first exon, 4a, differs from exon 4. The protein encoded lacks the first 437 amino acids of the full length WNK1 (including almost the entire kinase domain), but instead begins with 30 amino acids that include a cysteine-rich region; the sequence of the KS-WNK1 transcript from exon 5 onwards is identical to that of full length WNK1 (Figure 2 and 3). The human and mouse versions of KS-WNK1 share a high degree of identity (87% identical).
Figure 3. Kidney-specific N terminal WNK1 exon generates a kidney-specific isoform (KS-WNK1).
This isoform lacks the entire kinase domain of full length WNK1, and exon 4 is replaced by exon 4a (shown). The nucleotide and amino acid sequences of mouse and human KS-WNK1 are highly similar, (nucleotide identities indicated by *, amino acid differences indicated by red letters; amino acids indicated by single letter code). A cysteine-rich region within exon 4a is indicated by the box.
Additional complexity arises from the presence of two 3′ polyadenylation sites, which may alter mRNA stability and translational efficiency (41). Transcripts containing these polyadenylation sites are expressed ubiquitously, with the longer variant being more abundant. One of these variants is 10.5 kilobases in length, while the other is 9 kilobases, and indistinguishable in size from the 9 kilobase KS-WNK1 transcript. Both of these polyadenylation sequences were also identified in the mouse WNK1 gene (163).
Alternative splicing of exons 9, 11 and 12 also occurs, leading to altered coding (41, 163, 245, 272), though exons 11 and 12 appear to be used less frequently (41). For example, in adult mice, exon 11 is largely absent from both WNK1 and KS-WNK1 in kidney, but abundant in testis (163). In the developing embryo, exon 11 is absent in WNK1 transcripts in placenta, but abundant in neural tissues (163). The functional significance of these variant forms of WNK1 is unknown, but exons 11 and 12 encode an amino acid sequence that is proline rich, suggesting a potential transmembrane domain flanked by a flexible conformation (163). This region also shares homology with several extracellular matrix proteins, which may be significant in regulation of tight junctions.
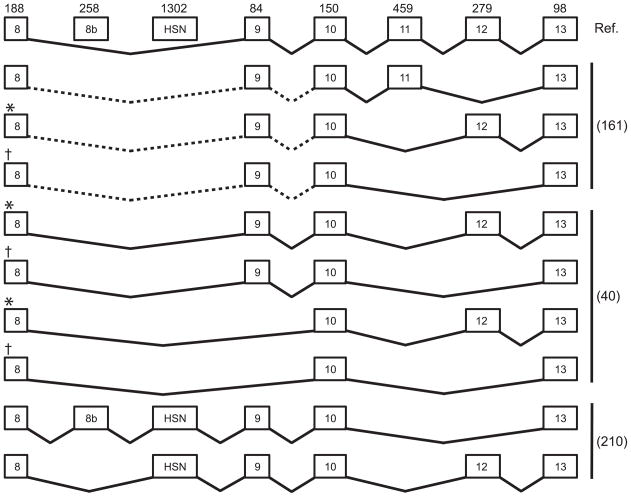
The identification of mutations in WNK1 as the causative defect of Hereditary Sensory and Autonomic Neuropathy Type II (HSANII) [section V.C], resulted in the identification two additional WNK1 splice variants, containing two novel exons (211). These variants were only identified in neuronal tissue, and contain exon HSN2, present in mRNA from brain, spinal cord, dorsal root ganglia and sciatic nerve, as well as exon 8b, present only in brain and spinal cord (Figure 4). Alternative splicing was observed in these variants downstream of exon 10.
Figure 4. WNK1 splice variants.
WNK1 displays significant variability in splicing between exons 8 and 13. The numbers above the exon boxes indicates length of each exon in nucleotides. Dashed line connecting exon indicates splicing between these exons was not determined. For variants described in O’Reilly (162), * and † indicate that these maybe identical to the variants described by Delaloy (41) since splicing 5′ of exon 10 was not determined. The renal transcripts identified O’Reilly are distinct from those identified by Shekarabi (211) since HSN-containing transcripts are neuron-specific. The functional consequences of alternative splicing are unknown. Note the additional splice variant that results in hereditary sensory and autonomic neuropathy II.
Other transcripts have also been identified, including a rare transcript arising from an alternative splice acceptor site in intron 3 (41). This transcript is predicted to encode a short (394 amino acids) isoform predicted to be catalytically inactive. As with all the other splice variants described, the functional and/or pathophysiological roles of these putative isoforms remain to be elucidated. The importance of knowledge of the full complement of splice variants is illustrated by studies using a transgenic approach, in which lacZ was expressed from a WNK1 BAC reporter (40). WNK1 mRNA is detected at high levels in heart by Northern blot, but the major WNK1 transcript in the heart lacks exon 12 (41). However another study did not detect WNK1 protein on heart sections (30); the epitope used to generate the antibody used in this study was encoded by exon 12, which explains the lack of detection.
2. WNK2
The human WNK2 gene spans 136kb and contains 30 exons. A CpG island spans a putative transcription start site, extending from −1063 to +623 (82), and inappropriate silencing of this CpG island by methylation may play a role in glial tumorigenesis [section V.E.]. Promoter analysis using luciferase reporter constructs showed that the 5′ region of the CpG island displays strong promoter activity in HEK293 cells, and the 3′ portion within exon 1 strongly inhibits this promoter activity (82). A similar pattern of activity was observed in a glioma cell line.
While, based on analysis of the genomic sequence and cDNA clones, there are numerous putative WNK2 transcript variants, there is little experimental evidence in this respect. A study examining the molecular basis of T cell-mediated recognition of pancreatic cancer cells identified a WNK2 isoform as a tumor epitope (87). This isoform is only 779 amino acids long, versus the 2297 amino acids of the canonical WNK2 isoform, and is truncated at the C-terminus. It also lacks the first 14 amino acids, has a further 51 amino acid deletion, and a 3 amino acid change. Apart from its ability to act as a pancreatic tumor epitope, the functional roles of this isoform, or whether it is more widely expressed, are unknown.
3. WNK3
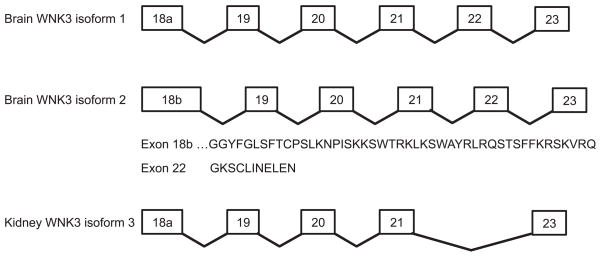
The human WNK3 gene spans 165kb and contains 24 exons, and like the WNK1 and WNK2 genes, has a predicted 1454 base pair CpG island spanning the first exon (80). Two exons display alternative splicing, both of which maintain the open reading frame (Figure 5). Exon 18 contains two splice donor sites, with resulting exon 18 lengths of either 163 base pairs (exon 18a) or 304 base pairs (exon 18b). Expression of the 18a transcript was detected by RT-PCR at low levels in adult brain, lung, kidney and pancreas, with higher levels in liver; in fetal tissues, levels were high in kidney, low in placenta, brain and lung, and very low in heart, thymus, liver and spleen (80). In contrast, the longer 18b transcript was restricted to adult and fetal brain only. The resulting additional 47 amino acids in the brain-specific isoform are likely to have functional consequences (65) and [section III.E.2.]. Transcript variants with or without exon 22 have also been identified in almost all tissues, with transcripts lacking exon 22 being more abundant (80).
Figure 5. WNK3 splice variants.
Alternative splicing generates two distinct variants in brain, and one in kidney. Exon 18a is present in both brain and kidney, but exon 18b is brain-specific and adds an additional 47 amino acids to the protein encoded by exon 18a. Exon 22 is skipped in kidney, resulting in an isoform lacking an 11 amino acid stretch present in brain (65). The renal isoform has been shown to stimulate activity of NCC (65). In contrast brain isoform 2 exhibits an inhibitory effect on NCC, but since NCC expression has not been detected in the brain, this is probably not physiologically relevant.
4. WNK4
In contrast to the other human WNK kinases, the human WNK4 gene is fairly small, spanning 16kb and containing 19 exons. In common with them, it has a predicted CpG island, spanning from −380 to +425, relative to the ATG codon (McCormick, unpublished observations). A putative initiator element, believed to be required for transcription initiation from TATA-less promoters is located at −27. A 216 base pair region upstream of this transcription initiation site is sufficient to confer strong promoter activity in Cos-7 cells (123). Although a variant has been identified in mouse brain (58), the only other WNK4 splice variant results from improper splicing of exon 22, leading to replacement of the 46 amino terminal acid residues with 37 alternative residues (J.A. McCormick, unpublished observations). Interestingly, this deletes a serine residue that can be phosphorylated by serum- and glucocorticoid-induced kinase 1 (SGK1) (194), as well as the negative signal regulatory domain identified by Yang and colleagues (281).
5. Evolutionary relationships of WNK kinases
Comparison of genomic sequences indicates that C. elegans and Drosophila carry only a single gene homologous to the WNK kinases, indicating that the ancestral WNK gene underwent duplication in higher animals. The chromosomal regions containing WNK2 and WNK3 are syntenic, sharing three other homologous gene pairs matched in orientation besides the WNKs (80). This suggests that the syntenic regions arose from a genomic duplication, and diverged in sequence, with several additional small genes either inserted or lost. In contrast, the genomic regions containing WNK1 and WNK4 do not demonstrate conserved synteny with each other, or with the regions containing WNK2 and WNK3, suggesting they diverged from the ancestral WNK gene independently.
B. Regulation of WNK kinase expression and abundance
Little is known about the transcriptional regulation of the WNK kinases, with most studies examining regulation of WNK1, KS-WNK1 and WNK4 by the mineralocorticoid hormone aldosterone, and manipulation of dietary electrolytes, both of which are relevant to renal electrolyte balance and control of blood pressure. The physiological significance of the effects of dietary and hormonal manipulation on WNK kinase levels are discussed later [section IV].
1. WNK1 and KS-WNK1
Given the role of WNK1 in the regulation of ion transport, and ultimately blood pressure, O’Reilly and colleagues performed extensive work to examine the effects of aldosterone and dietary manipulation of sodium and potassium on the expression levels of WNK kinase transcripts in mice (162). Chronic (6 day) manipulation of dietary sodium intake in either direction (high sodium or low sodium) had no significant effects on the expression of WNK1, although there was a trend towards lower expression on a low sodium diet. Mice on a low sodium diet displayed a significantly lower expression level of KS-WNK1 relative to mice on a high sodium diet.
Several groups have compared the basal abundance of WNK1 to KS-WNK1 in kidney tissue, at the message level. O’Reilly and colleagues (162) noted that KS-WNK1 was highly expressed along the DCT, but reported that expression of WNK1was at levels that were ‘near background’. Lazrak and colleagues (115) also determined that the vast majority (91%) of renal WNK1 transcripts in rat are KS-WNK1. The only data with respect to protein abundance suggest that both KS-WNK1 and WNK1 can be detected by immunoblot of kidney tissue (249). Three groups showed that the ratio of KS-WNK1 to WNK1 is reduced by dietary potassium deprivation, whereas the ratio is increased by dietary potassium loading (Figure 6) (115, 162, 249). In situ hybridization on kidney sections from mice fed varying dietary potassium, confirmed the increase in KS-WNK1 expression on a high potassium diet, though the reduction on a low potassium diet was not observed (162).
Figure 6. Effects of dietary potassium intake on renal WNK kinase expression.
O’Reilly and colleagues (162), Lazrak and colleagues (115), and Wade and colleagues (249) all detected significant increases in the ratio of KS-WNK1/WNK1 during high potassium diet. O’Reilly and colleagues reported that high potassium intake increased renal WNK4 mRNA significantly.
The mineralocorticoid hormone aldosterone regulates blood pressure by acting on the distal nephron to increase the reabsorption of sodium (with concomitant reabsorption of water through aquaporin channels), and also regulates serum potassium levels by enhancing renal potassium secretion (139, 232). In mice, adrenalectomy did not alter expression levels of either WNK1 or KS-WNK1 (162). Chronic aldosterone excess resulted in a significant increase in KS-WNK1 expression, but had no effect on WNK1 levels. This finding appears paradoxical, given that dietary sodium restriction, which increases aldosterone secretion, reduced KS-WNK1 expression (162), but suggests that the response of KS-WNK1 to aldosterone may be altered by extracellular fluid (ECF) volume or by serum potassium concentration. Dietary sodium restriction reduces ECF volume, stimulating secondary hyperaldosteronism without changes in serum electrolytes, whereas chronic aldosterone infusion (mimicking primary hyperaldosteronism) raises ECF volume and causes hypokalemia. Thus, the reduction in KS-WNK1 under low sodium conditions may be a response to conserve extracellular volume (162). KS-WNK1, but not WNK1, mRNA expression has also been shown to be induced by aldosterone in a mouse cortical collecting duct cell line that stably expresses the mineralocorticoid receptor (MR). Aldosterone was shown to induce KS-WNK1 mRNA expression in mpkCCD cells, derived from cortical collecting duct (CCD), with no effects on WNK1 or WNK4 levels (156). One limitation of these studies is that in vivo, KS-WNK1 does not appear to be expressed in the cortical collecting duct (162). Analysis of the KS-WNK1 promoter region identified a putative glucocorticoid response element (GRE), to which activated mineralocorticoid receptors can bind; none were identified in the WNK1 promoter (41). It is possible that this element mediates the transcriptional activation of the KS-WNK1 promoter by aldosterone.
As described in [section II.A.1], two WNK1 transcripts with initiation sites within the CpG island (regulated by proximal promoter 1, P1, see Figure 7), and downstream of the CpG island, within exon 1 (regulated by proximal promoter 2, P2) were identified by 5′RACE PCR in humans (41). Reporter studies, using luciferase constructs to test various regions of P1 showed that the region from −1 to −1200 of the ATG codon displays promoter activity (41). At the molecular level, a 153 base pair region of P1 contains putative binding sites for the transcriptional activators Sp1, Oct-1, HNF-1 and HES-1. It also contains a CUP element, which acts as a repressor by connecting with a downstream C/EBP binding site, one of which is also present. However, the highly homologous mouse P1 region lacks the HES-1, CUP, C/EBP, and Sp1 sites, but has two Sp1 binding sites absent in the human promoter region. In the mouse promoter region, deletion of a region between −700 and −977, which contains five putative Sp1 binding sites, reduced reporter activity by 50% (287). Finally, the region from −1200 to −2500 appears to contain both repressor and activator motifs (41).
Figure 7. Human WNK1 and KS-WNK1 promoter region structures.
Delaloy and colleagues mapped multiple transcription initiation sites for WNK1 by 5′ RACE-PCR, indicated by bent arrows (41). The renal-specific promoter, PKS-WNK1, initiates expression of KS-WNK1, which lacks the WNK1 kinase domain. Horizontal lines indicate consensus transcription factor binding sites identified with the TESS program. The translation start site for P2 transcripts is indicated by *.
A fragment spanning P2 (from +13 to +626) also displayed promoter activity, which was strongly inhibited by deletion of a putative GATA-1 binding site at +607. This promoter was more active, and deletion of the GATA-1 site had a more pronounced effect in HEK293 and MDCK cells (both kidney-derived) than in CHO cells, which may have implications for regulation of expression of this truncated form of WNK1 in vivo (39). The P2 region contains numerous consensus Sp1 binding sites and several putative binding sites for other transcription factors including the CCAAT binding factor. It also contains putative binding sites for transcriptional repressors such as WT1-KTS and CUP, and for transcriptional activators produced in the brain, heart, and testis (NF-ATp) or that may be involved in regulating kidney development (AP-2alphaA).
The kidney-specific isoform of WNK1, KS-WNK1, is transcribed from an alternative promoter, which lies within intron 4 (see Figure 7). Reporter studies identified promoter activity with a fragment extending from just −70 to +14 in CHO, HEK and MDCK cells(41), which contains consensus binding sites for PU-1 and C/EBPα, as well as the GRE which may mediate aldosterone-dependent transcription (156). Further analysis identified a strong distal enhancer, which conferred high promoter activity specifically in MDCK cells. The location of this enhancer region was narrowed down to a 157 base pair fragment that displays high homology to sequences upstream of NCC and Kallikrein, which are also restricted to the DCT in the kidney. Further analysis of this region may shed light on the mechanisms by which the DCT differentiates during development.
In the human disease FHHt, two large (22 kilobases and 41 kilobases), overlapping deletions within intron 1 of the WNK1 gene have been identified as a causative defect (258). Wilson and colleagues showed that this deletion leads to increased expression of WNK1, although this was only assessed in lymphocytes (258). Using a transgenic mouse approach, Delaloy and colleagues examined the effects of intron 1 deletion on expression of WNK1 and KS-WNK1 (39). When intron 1 was deleted, ectopic expression of a KS-WNK1 reporter was observed, particularly in skeletal muscle, heart and cerebellum; there was no effect on extra-renal expression of WNK1. In the kidney, both WNK1 and KS-WNK1 showed a significant increase in expression levels in the DCT. Furthermore, KS-WNK displayed ectopic expression in other renal segments, and the expression of WNK1 was increased in segments normally expressing it. These data suggest that one or more repressors constitutively prevent expression of KS-WNK1 outside the DCT, and represses WNK1 expression in this segment in particular. Alternatively, intron 1 could contain an insulator that prevents interaction between elements regulating the WNK1 and KS-WNK1 promoters. Indeed, both repressors and insulators were identified in vitro (39). While the lengths of the human and mouse first introns differ substantially (60 kilobases and 30 kilobases respectively), sequence comparison suggests that the repressors and insulators identified in mouse are likely to reflect the intronic regulation of the human WNK1 gene. The relevance of dysregulation of WNK1 and KS-WNK1 expression by deletion of intron 1 is discussed in [section IV.].
While numerous putative transcriptional activators of WNK1 and KS-WNK1 have been identified in silico, none have been confirmed as regulators in cells or animals. In mouse, WNK1 shows a reduction in expression levels as embryonic development proceeds (287), but little else is known about extra-renal regulation during development or by experimental manipulation in vivo. Finally, a 3308 base pair region of the WNK1 gene lies antisense to the spliced gene RAD52, raising the possibility of regulated alternate expression.
2. Developmental regulation of WNK1 expression
Transgenic mice expressing a lacZ reporter under the control of the WNK1 promoter have provided insight into developmental regulation of WNK1 expression (40). Reporter activity is detected as early as embryonic day 8,5 (E8.5) in the developing vasculature and primitive heart, with peak expression during the early stages of cardiac development. In the embryonic central nervous system, reporter activity was almost absent in embryos, being restricted to the capillaries surrounding the neural tube and some neurons of the floor plate (40), a group of cells that directs differentiation and axonal trajectories (175). In adults, lacZ expression was detected in the cerebellum, especially in the granular layer and cerebellar Purkinje cells. These data indicate that during development, WNK1 expression is most likely regulated at the transcriptional level, though the signals triggering changes in expression remain to be determined.
3. WNK2 and WNK3
Very little is known about regulation of WNK2 expression levels, although methylation-dependent silencing across 1.3 kilobases of the CpG island spanning the putative transcription initiation site has been observed in gliomas (82). Analysis of the human 5′ flanking sequence from −1128 to −263, the smallest fragment shown to confer promoter activity reveals multiple putative transcription factor binding sites, including several Sp-1 binding sites, typically found in CpG islands (J.A. McCormick unpublished observations).
WNK3 expression levels are highly developmentally regulated in the CNS (97). In the hippocampus and cerebellum, it is virtually absent at postnatal day 10, but becomes highly expressed by postnatal day 21.
4. WNK4
In rats, O’Reilly and colleagues found that manipulation of dietary sodium levels had no significant effect on renal WNK4 mRNA levels, though there was a trend to decrease on a low sodium diet (162). In contrast, increased dietary potassium leads to increased expression of WNK4; decreased potassium had no effect, as determined by Real Time PCR and in situ hybridization (162).
While adrenalectomy or chronic aldosterone infusion had no effect on WNK4 mRNA expression levels (162), two putative negative glucocorticoid response elements (nGREs) have been identified in the 5′ upstream region of the WNK4 gene. Administration of the synthetic glucocorticoid dexamethasone to HEK293 cells resulted in a 28–35% reduction in expression levels of endogenous WNK4 (123). Significantly, in a transgenic mouse model in which expression of the glucocorticoid receptor (GR) is induced by doxycycline, overexpression of GR led to a significant decrease in WNK4 mRNA levels in the CCD (159). While expression of WNK4 was also trending to lower levels in the DCT of these mice after 2 days of GR overexpression, this trend had disappeared after 15 days, suggesting adaptation. Analysis of 337 and 285 base pair fragment of the WNK4 promoter using reporter assays confirmed the presence of the two nGREs functionally. Electrophoretic mobility shift assays showed that the two putative nGREs interacted strongly with nuclear extracts, and these interactions could be blocked by an anti-GR antibody; Chromatin Immunoprecipitation assays confirmed these findings. Since signaling through the mineralocorticoid receptor requires binding to GREs, the possibility remains that aldosterone regulates WNK4 expression. However, it may be that the role of the nGREs is to allow suppression of WNK4 expression levels by glucocorticoids in non-mineralocorticoid target tissues. Further complexity regarding steroid hormone regulation of WNK4 expression arises from the observation that dexamethasone increases WNK4 mRNA levels 3-fold, in Reissner’s membrane of the inner ear (104). Glucocorticoids thus appear to either increase or decrease WNK4 expression levels, depending on the cell type studied.
As in the WNK1 promoter region, a putative GATA-1 transcription factor binding site has been identified in the WNK4 promoter region (see Figure 8) (124). Protein acetylation is an important epigenetic mediator of gene expression (124). Treatment of HEK293 cells with trichostatin A (TSA), a histone deacetylase inhibitor, resulted in increased WNK4 expression, and deletion analysis indicated that the GATA-1 may play an important role in mediating this response. TSA treatment led to an increase in GATA-1 acetylation, which correlated with its ability to bind the WNK4 promoter region and stimulate WNK4 promoter activity. Interestingly, GR may also contribute to acetylation-mediated control of WNK4 promoter activity, since a fragment containing the nGRE at −285 was responsive to TSA, whereas a fragment containing the nGRE at −337 was not (124). Opposing responses to TSA have been observed in other GR-mediated transactivation processes, including regulation of the MMTV gene, a classic GR-regulated gene (12, 113, 191).
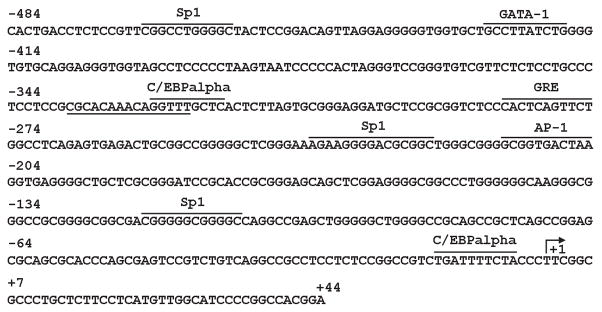
Figure 8. Human WNK4 promoter region structure.
Li and colleagues analyzed the hWNK4 promoter region using TRANSFAC-TESS and Match™ online software (124). Bent arrows indicate transcription initiation sites; horizontal lines indicate putative transcription factor binding sites. Note the presence of two glucocorticoid responsive elements (GRE), which probably function as negative GREs to inhibit WNK4 expression (123), and the GATA-1 binding site whose acetylation upregulates WNK4 expression levels (124).
5. Circadian regulation of WNK kinase expression
Nine members of the WNK kinase family have been identified in Arabidopsis thaliana (155). Interestingly, several of these WNK kinase isoforms have been shown to be under circadian regulation (152, 155), and regulate flowering time (253). While the Arabidopsis thaliana WNK kinases and circadian pathways differ significantly to those in mammals, it is tempting to speculate that the WNK kinase maybe under circadian regulation in mammals. For example, perhaps WNK4 is regulated through its two nGREs by glucocorticoids, which in humans, peak in the early morning and reach their nadir at night.
C. Protein Structure
1. Kinase Domain
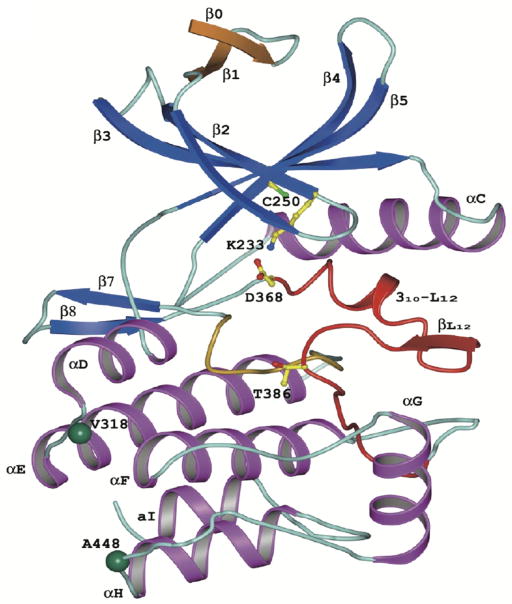
The WNK kinases display a wide range of sizes (1243 amino acids for WNK4, up to 2382 amino acids for WNK1) but share several highly conserved regions (Figure 2). The most highly conserved region is the kinase domain, with the four homologues sharing 84% or more sequence identity. The crystal structure of the kinase domain of WNK1 has been solved (see Figure 9) for a low activity conformation in which the activation loop phosphorylation site was mutated to an alanine (S382A) (143). While the conserved catalytic residues in the catalytic loop adopt standard positions and peptide conformations, they adopt different side chain conformations, resulting in the absence of hydrogen-bonds found in other kinase structures. As predicted from the primary amino acid structure, a cysteine residue (C250) occupies the position in subdomain II occupied by the catalytic lysine residue in other protein kinases. However, the side chain of this cysteine is unlikely to play a role in catalysis due to its distance from the active site. Instead, a lysine in subdomain I (K233) provides the catalytic side chain, and results in a large cavity in the back of the ATP-binding site.
Figure 9. Topology of the WNK1 kinase domain (taken from (143)).
The five conserved β strands are blue and α helices are magenta. The extra N-terminal β strand is shown in gold. The activation loop is shown in red and the catalytic loop in yellow. Lys-233, Cys-250, Asp-368, and Thr-386 are shown in ball-and-stick representation. Cys-250 occupies the position in subdomain II occupied by the catalytic lysine residue in other protein kinases. Instead, a lysine in subdomain I (Lys-233) provides the catalytic side chain, and results in a large cavity in the back of the ATP-binding site. The green balls represent two substrate-specificity determinant residue Val-318 and Ala-418.
Protein kinases with very high amino acid identity in the kinase domain, such as the SGKs, can display differences in substrate specificity (108). In the case of the WNK kinases, WNK1 is able to bind and phosphorylate synaptotagmin 2; WNK4 phosphorylates synaptotagmin 2 much less effectively (117, 269); similarly, WNK4 phosphorylates the target kinases SPAK (Ste20-related proline alanine-rich kinase) and OSR1 (oxidative stress response 1) much less efficiently than WNK1 does (see Section IIID4). Using homology-based structural modeling, Min and colleagues identified two residues (V318 and A448) in the substrate binding groove that might be involved in substrate recognition (143). Mutation of these residues prevented interactions between WNK1 and synaptotagmin 2, and reduced phosphorylation by 40–50%. In WNK4 these two residues are replaced by a glutamate and a lysine; in WNKs 2 and 3, the valine is conserved, but the alanine is not, raising the possibility that there may be substrate overlap between WNKs 1–3.
2. Autoinhibitory domain and autophosphorylation site
Activity of protein kinases can be regulated by the presence of an autoinhibitory domain that lies outside the catalytic domain. This domain suppresses kinase activity until an activating signal releases it from the inhibitory site, providing additional modifications are not required for activation. One such additional modification is autophosphorylation of residues in the activation loop. These appear to be two major mechanisms by which WNK kinase activity is regulated. In the WNK kinases, the autoinhibitory domains lies immediately C-terminal to the kinase domain, are approximately 70 amino acids in length and share at least 46% identity (254, 267) (green in Figure 2). Mutation analysis of WNK1 (267), identified conserved two phenylalanine residues, F524 and F526, that appear to play an important role in autoinhibition and are conserved in all WNK kinases. In addition to autoinhibition, it appears that WNK kinases can inhibit the kinase activity other WNK kinases (exhibiting ‘cross’ inhibition) suggesting interactions between, and cross-regulation of, the WNK kinases. For example, both WNK1 and WNK3 are able to phosphorylate WNK4 in vitro (278). WNK4 autoinhibitory domain inhibits the catalytic activity of both WNK1 (120, 254), WNK2 (120), and WNK3 (278), in vitro. The physiological implications of this cross inhibition are not clear, but do correspond to inhibitory functional effects observed in overexpression models (224, 278, 279, 281)
For full activation of WNK1, phosphorylation of S382 is required, and phosphorylation of serine 378 enhances activity (267). The residues mediating this phosphorylation are located outside the activation loop, probably in the serine-rich region N-terminal to the kinase domain. At the structural level WNK1 features a large unique cavity adjacent to the catalytic site, formed by an outward displacement of the C-terminus of the activation loop. Unusually, the activation loop also displays remodeling of the activation loop at the N-terminus. This unique structure of the activation loop may allow the development of specific WNK1 inhibitors or activators.
3. Coiled-coil domains
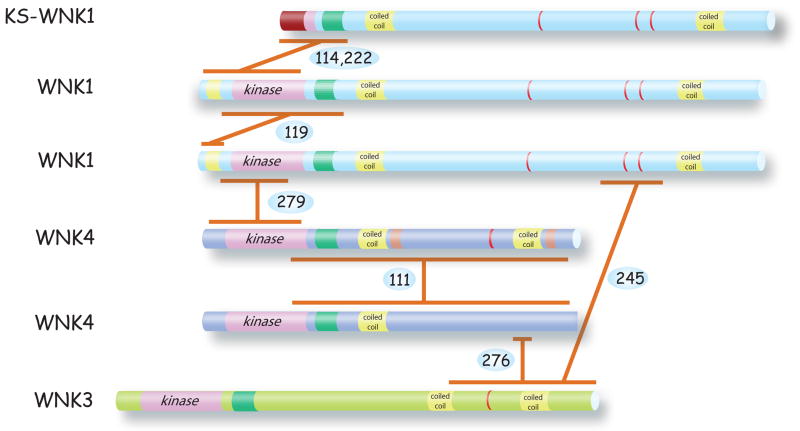
Coiled-coil domains are characterized by a heptad repeat pattern in which residues in the first and fourth position are hydrophobic, and residues in the fifth and seventh position are predominantly charged or polar. Since they can interact with each other, coiled-coil domains play a major role in protein-protein interactions in the dynamic assembly and disassembly of protein complexes (133). All members of the WNK kinase family are predicted to contain coiled-coil domains (see yellow domains in Figure 2) as determined using the programs COILS or N-COILS. Experimental confirmation of these possible interactions is generally lacking, but several WNKs have been reported to associate on protein complexes, both in vivo and in cells and oocytes (see Figure 10). Yeast two-hybrid data revealed that the N-terminus of WNK1 (residues 1–222) interacts with a fragment including the WNK1 autoinhibitory domain and the second putative coiled-coil domain (120). However, the N-terminus, which contains a putative coiled-coil domain at residues 190–217, was unable to interact with itself. Other interactions that might involve the coiled-coil domains include those between WNK1 and KS-WNK1 (115, 224), which interact via their N-termini, and WNK3 and WNK4, which interact through the coiled-coil domains at their C-termini (278). Gel filtration experiments suggest that WNK1 may exist as a tetramer (120), and interactions through the coiled-coil domains may enable the formation of complexes through scaffolding.
Figure 10. Protein-protein interactions.
Shows sites of interactions between WNK kinases, detected either in vitro, in cells, or in vivo.
References are indicated by blue circles. A
4. Proline-rich sequences
All members of the WNK kinase family contain putative binding sites for proteins that bind proline-rich motifs. Protein domains that bind proline-rich motifs are often involved in the modulation of signaling. The unique properties of proline provide a mechanism for highly discriminatory recognition of proline-rich motifs without requiring high affinities (Kd values are typically in the μM range) (102).
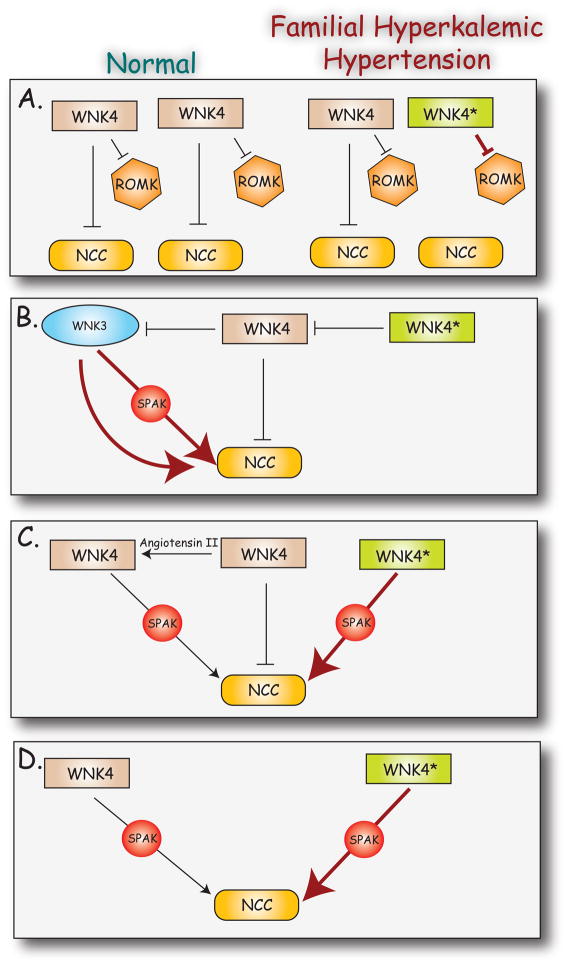
The Src homology domain 3 (SH3) is the most common protein interaction module in mammals, with 300 protein members in the human genome (101). SH3 domains play keys roles in many processes, including regulation of cell growth and the immune response, and also form structural components in a variety of systems. Typically, SH3 domains bind to canonical proline-rich domains, including a “core” PXXP motif in the target protein. Amino acid sequence analysis reveals that all members of the WNK kinase family contain putative SH3-binding motifs, and several have been confirmed experimentally. He and colleagues examined the role of three PXXP motifs near the N-terminus of WNK1 in the inhibition of potassium transport through ROMK1 and showed that mutation of all three motifs completely prevented inhibition (77). WNK1 was then shown to interact specifically with the endocytic scaffold protein, intersectin, and this interaction was required for internalization of ROMK1. WNK4 was also shown to mediate endocytosis of ROMK1 via interaction with intersectin inhibition (77). Interestingly, three mutations associated with FHHt (E559K, D561A, and Q562E, mouse numbering) lie adjacent to three conserved PXXP motifs (which begin at P545, P552, and P555). Introduction of the FHHt mutations resulted in an increased interaction between WNK4 and intersectin, resulting in increased endocytosis of ROMK1. A reduction in surface ROMK abundance might contribute to the hyperkalemia observed in FHHt (section IV.C.).
5. Other features
The FHHt-causing mutations in WNK4 may lie within another conserved region within the WNK kinases, termed the acidic motif due to the presence of 5 acidic residues in a 10 amino acid stretch. This motif is only present in the WNK kinases and is conserved between chordates, but absent from other phyla (80). The functional significance of this motif is unclear, since analysis of its functional role has involved removal of the entire domain, rather than point mutagenesis (66, 153), and it is possible that the presence of so many acidic amino acids is merely coincidental. It should be noted that the residues in WNK4 that cause FHHt are conserved in all WNK kinases (see Figure 2). It is likely that mutations within this region cause structural alterations that impair interaction of the PXXP motifs with intersectin, rather than the acidic amino acids performing a specific role inWNK4 function.
The N-terminus of KS-WNK1, which differs from that of WNK due to alternative promoter usage, is cysteine-rich, containing 6 cysteine residues in the first 50 amino acids. The functional relevance of these cysteine residues is unclear, but the unique exon 4A binds to WNK1 and inhibits its actions on WNK4 and on ROMK (115, 127, 224, 249).
There are large regions of the WNK kinases that share little homology with each other. It is possible that motifs within these regions may play a role in determining isoform-specific intereactions and functions of the WNK kinases. Generation of chimeric proteins in which these regions have been switched between different isoforms might give insight regarding their functions.
III. WNK kinases and regulation of molecular and cellular processes
A. Regulation of WNK activity
1. Tonicity
To identify regulators of WNK1, Xu and colleagues tested a number of agents and stimuli on HEK 293 cells, including growth factors, heat shock and lysophosphatidic acid, to determine whether they could increase WNK1 activity (120, 266). The only stimulus tested that led to a reproducible increase in WNK1 activity was hypertonicity (0.5M NaCl or 0.5M sorbitol), which is likely to be relevant to the role of WNK1 in renal physiology [section IV.A.]. In addition to a direct effect on WNK1 kinase activity, hypertonicity alters intracellular localization of both WNK1 and WNK4 (285) [section III.A.4.]. Phosphopeptide mapping identified multiple residues on WNK1 phosphorylated following exposure to hypertonicity. Two ofthese sites are located N-terminal to the kinase domain (S15 and S167), one within the activation loop of the kinase domain (S382), previously identified as being required for full activation of WNK1 (267). The other three sites are located downstream of the kinase domain (S1261, T1848, and S2372) (285). While stimulation of WNK1 phosphorylation by sorbitol increased its kinase activity, phosphorylation at S1261 may inhibit the interaction of WNK1 with two of its substrates [section III.D.4.], SPAK and OSR1, which mediate responses to hypertonicity. Three constitutively phosphorylated sites within the C-terminal region of WNK1 (Ser2012, Ser2029, and Ser2032) were also identified.
Hypotonic stress also activates WNK1 kinase activity (120, 149). In most experiments, hypotonicity has been generated using a low chloride, and often low potassium, incubation medium. Recently, Uchida and colleagues tested whether tonicity, Na, Cl, or K was key to activation. They reported in preliminary form that low Cl and K, but not low Na, was sufficient to stimulate WNK1 kinase activity (Uchida abstract ASN 09). As hypotonic, low chloride conditions activate NKCC and NCC, and inhibit KCC, these effects on kinase activity may be physiologically relevant [section IV].
2. Insulin and insulin-like growth factor 1 (IGF-1)
Insulin and IGF-1 regulate many physiological processes through the activation of phosphoinositide 3-kinase (PI3-kinase), which phosphorylates phosphatidyl inositol(4,5)_P_2 to generate the second messenger phosphatidyl inositol(3,4,5)_P_3 (PI(3,4,5)P3). PI(3,4,5)P3 acts as a cofactor for 3-phosphoinositide-dependent protein kinase-1 (PDK1) which activates members of the AGC family of kinases, including protein kinase B/Akt1, p70 ribosomal S6 kinase (S6K), and SGK, via phosphorylation of their activation loops. These kinases have been shown to play roles in cell growth, proliferation and survival, control of cellular glucose uptake, and ion transport (136).
Sequence analysis revealed that WNK1 contains a putative Akt1/SGK phosphorylation site (Arg-Xaa-Arg-Xaa-Xaa-Ser/Thr, T58/60; T60 in human, T58 in rat); only WNK4 also contains a putative phosphorylation site in the same vicinity (248). In vitro studies showed that Akt1 strongly phosphorylated WNK1 at T58/60. IGF-1 induced phosphorylation at this site in HEK 293 cells, and this phosphorylation was mediated by Akt1. The functional significance of phosphorylation of WNK1 at T58/60 is unclear, since it did not affect its catalytic activity on a substrate (247). Identification of physiological substrates of WNK1 may resolve this issue. Subsequent studies have confirmed that WNK1 is a substrate for Akt1 (92, 197, 269), and that activation of the PI3-kinase/Akt1 pathway is required for insulin to stimulate WNK1 phosphorylation (92, 197).
SGK1 is a widely expressed protein kinase, that has been most extensively characterized with regard to its role as a mediator of aldosterone action in the kidney (136). Similarly to Akt1, SGK1 phosphorylates WNK1 at T58/60 (269).
3. WNK4 and serum and glucocorticoid-induced kinase 1
Ring and colleagues identified a highly conserved Akt1/SGK phosphorylation site at S1169 of WNK4 (190), which was confirmed to be an SGK1 phosphorylation site. Functionally, a WNK4 mutant, S1169D, which mimics phosphorylation at S1169, reversed the inhibitory effect of WNK on activity of the epithelial sodium channel (ENaC) and the renal outer medullary potassium channel 1 (ROMK11), but not on NCC (198) (see below). Src family protein tyrosine kinase modulates these SGK1-mediated effects of WNK4 on ENaC and ROMK11, but it is unknown whether Src directly phosphorylates WNK4, or acts by inhibiting SGK1-dependent phosphorylation of S1169 (284).
SGK1 also phosphorylates WNK4 at a second site, S1196 (194); phosphorylation at this site appeared to be more intense that at S1169. Interestingly, this serine is not located in a classic Akt1/SGK phosphorylation site consensus sequence, and is also phosphorylated by WNK1. Similarly to the effects of phosphorylation of WNK4 at S1169, mutations of WNK4 mimicking phosphorylation at both of these sites (S1169D/S1196D) resulted in the inability of WNK4 to exert an inhibitory effect on NCC [section IV.A.].
4. Intracellular localization
Alteration of intracellular localization is an important mechanism by which the actions of proteins can be modified. For example, in order to be activated by PDK1, Akt1 must translocate to the plasma membrane, an event that involves interaction of the N-terminal Pleckstrin homology domain of Akt1 with PI(3,4,5)P3 (47). PDK1 is also recruited to the plasma membrane by interaction with PI(3,4,5)P3. It is not known whether WNK kinases interact with phosphoinositides, but sequence analysis does not reveal the presence of any putative phosphoinositide interaction domains (McCormick, J.A., unpublished observations). Another example of control of intracellular localization is illustrated by aldosterone signaling through its receptor, MR. In the absence of ligand, MR resides both in the cytoplasm and in the nucleus. Binding of aldosterone to MR increases the number of MR molecules in the nucleus, where it acts as a transcription factor. Translocation of MR from the cytoplasm to the nucleus is mediated through the interaction of a nuclear localization signal within the MR sequence, with importin-α at the nuclear pore (230).
The intracellular location of WNK1 under basal conditions is primarily cytoplasmic, and it is excluded from the nucleus (247, 285). Phosphorylation of WNK1 does not alter its intracellular localization itself (247, 285), but WNK1 rapidly redistributes to small vesicles in response to hypertonicity (285). In HEK 293 cells treated with sorbitol, WNK1 colocalized with the vesicle coat protein, clathrin, and partially with AP-1, which is recruited to budding vesicles at the trans-golgi network (285). Redistribution of WNK1 is mediated though the non-catalytic C-terminus. Overexpression of catalytically inactive WNK1 or WNK4 reduces the membrane expression of ROMK1 through a clathrin-dependent endocytosis mechanism (32, 98), so the ability of WNK4 to relocalize to clathrin-coated vesicles is likely to be functionally relevant. In INS-1 cells, a pancreatic β-cell line, WNK1 localizes to the plasma membrane, and also to insulin-containing vesicles (117), where it might play a role in mediating insulin secretion.
In vivo, WNK4 has been reported to colocalize with ZO-1, at tight junctions in multiple epithelia (95, 258), as well as in lateral membranes and cytoplasm (95). In vitro, similarly to WNK1, exposure of HEK 293 cells to hypertonicity induces a rapid redistribution of WNK4 from the cytoplasm to an unidentified membrane compartment (210). This redistribution is reversible, i.e. WNK4 is not trafficked to a lysosomal compartment for degradation. Only hypertonicity that causes cell shrinkage led to WNK4 relocalization, which may be relevant with regard to regulation of the Na+-K+-2Cl− contransporter (NKCC1). NKCC1 is activated by cell shrinkage, and WNK4 has been shown to regulate its activity, through interaction with SPAK kinase (58). Despite WNK4 being implicated in EGF signaling [section III.B.4.], treatment of cells with EGF did not induce redistribution of WNK4, suggesting that different stimuli lead to distinct mechanisms of WNK4 action.
Immunofluorescence microscopy using an anti-WNK2 antibody revealed that it is localized to the plasma membrane, as well as to the cytoplasm (146). Knockdown of WNK2 expression with siRNA appeared to preferentially reduce plasma membrane WNK2 levels. Further analysis revealed that localization of WNK2 to the plasma membrane is dependent on the presence of C-terminal residues 1922–2156, within which the WNK coiled-coil domain is located.
WNK3 is localized diffusely in HeLa cells, and translocates to the nucleus on induction of apoptosis (246).
5. Circadian regulation
As described in [section II.B.4.], the expression of WNK kinases is under circadian regulation in Arabidopsis. Phosphorylation of downstream targets involved in circadian regulation is increased at the peak period of expression, and this is likely to be a consequence of both the increased WNK levels, as well as an effect on intrinsic activity. It is not known if WNK kinase expression displays circadian rhythm in vertebrates, or whether its activity is regulated, but the closely-related MAP Kinases display rhythmicity in their phosphorylation, which peaks at night (74, 107, 200). Furthermore, WNK3, presumably the brain isoform, is expressed at high levels in brain regions involved in circadian regulation, including the supraoptic nucleus, suprachiasmatic nucleus, and other areas of the reticular activating system that control the sleep/wakecycle (97).
B. Roles of WNKs in MAP kinase signaling
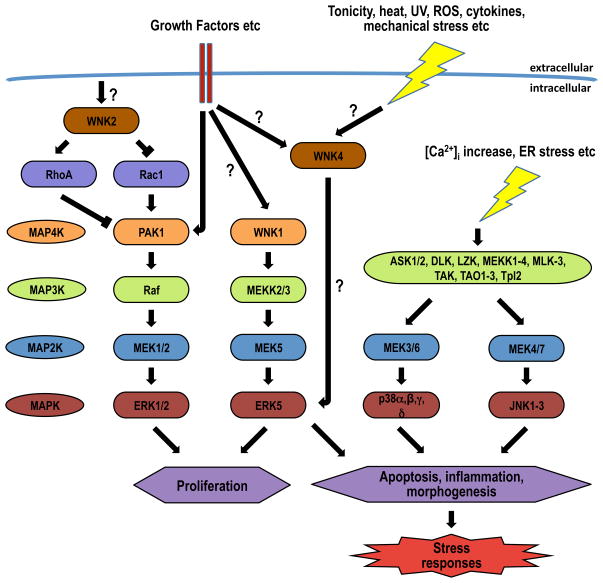
Mitogen-activated protein kinases (MAPKs) mediate signal transduction activated by a wide variety of extracellular stimuli (110). The MAPK family can be subdivided into six subgroups in mammals, including extracellular signal-regulated kinase 1/2 (ERK 1/2), c-Jun NH2-terminal kinase (JNK), and p38 MAP kinases (110). Activation of the MAPKs occurs through a cascade involving the sequential phosphorylation of protein kinases (starting with a MAP4K following receptor activation), culminating in phosphorylation and activation of the MAPK by a MAPK kinase (MAP2K). MAPKs then regulate the activity of transcription factors, the ultimate effectors of the MAPK pathway. Each MAPK is activated by a specific MAPKK, which is in turn regulated by relatively specific extracellular stimuli. The ERK pathway is stimulated primarily by growth factors and tumor promoters, whereas the JNK and p38 MAPK pathways are activated by pro-inflammatory cytokines and environmental stress, including hyperosmotic stress. The kinase domains of the WNK kinases are most similar to the MAP2K family, but initial studies in HEK293 cells showed that WNK1 does not exhibit MAP2K or MAP3K activity in several MAPK pathways, suggesting that it belongs to a distinct pathway (266). Subsequently, however, members of the WNK kinase family have been shown to modulate MAPK signaling (see Figure 11).
Figure 11. Overview of MAPK signaling pathways, with known positions of WNK kinase action.
The evidence used to generate this figure is discussed in [section III.B.]. The MAPK pathway is activated by growth factors and both environmental and intracellular stress, resulting in proliferative or stress responses through regulation of gene transcription. The ERK pathway is stimulated primarily by growth factors and tumor promoters, whereas the JNK and p38 MAPK pathways are activated by pro-inflammatory cytokines and environmental stress, including hyperosmotic stress. Members of the WNK kinase family influence MAP kinase signaling pathways as shown. EGF receptor activation stimulates WNK1 which in turn activates of ERK5. Stimulation of ERK5 by WNK1 requires MEK5 and MEKK2/3 activity, and WNK1 physically interacted MEKK2/3. WNK2 inhibits ERK1/2 activation by controlling the balance of upstream regulators of PAK1 activity, RhoA and RacI. WNK4 stimulates both ERK1/2 and p38 MAPKs in response to both hypertonic stress and stimulation with EGF. ? indicate where intermediates in the signaling pathways are unknown.
1. WNK1 and ERK5 signaling
Following identification of the autoinhibitory domain of WNK1 (267), Xu and colleagues re-examined the effects of WNK1 on MAPK signaling, since this domain may have masked WNK1 actions in their initial studies (266, 270). Removal of the autoinhibitory domain leads to much greater kinase activity in vitro (267), so WNK1 lacking this domain was used. ERK5 is a MAPK that is activated by both proliferative and stress stimuli (252); its upstream regulators are the MAP3K MEKK2/3, and the MAP2K MEK5 (252). Transient transfection studies revealed that WNK1 overexpression resulted in activation of ERK5, but not the closely related MAPK ERK2, in HEK293 cells (270). Further studies showed that the effect of WNK1 on ERK5 activity required both intact MEK5 and MEKK2/3 activity, and that WNK1 physically interacted with, and activates, MEKK2/3. Finally, WNK1 is also required for EGF-mediated activation of ERK5 in both Hela (270) and in neural progenitor cells (225). These data suggest that WNK1 functions as a MAP4K in this pathway, but the activation of MEKK2/3 by WNK1 appeared to be independent of its kinase activity, and other regulatory mechanisms may be involved e.g. complex formation.
2. WNK1 and SPAK/OSR1 signaling
The MAP4Ks STE20 (sterile 20)-like kinases SPAK and OSR1 play important roles in ion homoeostasis and cell volume control (43). STE20p was originally identified in budding yeast, and acts as a MAP4K in the pheromone-response pathway (43). However, STE20p is also able to directly phosphorylate non-MAPK pathway proteins, such as myosin I (261), suggesting that SPAK/OSR1 may also transduce signals through non-MAPK pathways. In mammalian cells, under hyperosmotic or hypotonic low Cl− conditions SPAK/OSR1 have been shown to activate NKCC1, NKCC2 and NCC by direct phosphorylation (46, 149, 178, 185), with WNKs 1, 3 or 4 acting upstream of SPAK/OSR1. WNK1 phosphorylates SPAK and OSR1 at an equivalent residue located within the T-loop of the catalytic domain (T233 in SPAK, T185 in OSR1) and a serine residue located within a C-terminal non-catalytic region (S373 in SPAK, S325 in OSR1) (248). Interactions of WNK kinases with SPAK/OSR1, and their functional consequences are described in more detail below [section III.E.].
3. WNK2 is a negative regulator of MAPK signaling
Knockdown of WNK2 expression levels using siRNA was shown to have no effect on ERK5 activation, but surprisingly led to an increase in phosphorylation of the MAPK ERK1/2 (147). Furthermore, reducing levels of WNK2 resulted in an enhancement of EGF activation of ERK1/2. Analysis of the signaling pathway from binding of EGF to its receptor down to activation of MEK1/2, the MAP2K that phosphorylates ERK1/2, showed that WNK2 affects ERK1/2 activity by modulating activity of MEK1. This modulation appears to pivot around S298 of MEK1, which can be phosphorylated by the protein kinase PAK1, and plays a role in priming MEK1 for activation by Raf-1, or promotes its incorporation into a protein scaffold containing ERK1/2 (31, 48). WNK2 does not interact with MEK1, or phosphorylate it at S298, but rather acts through interference with activity of PAK1 (146, 147). Knockdown of WNK2 activates PAK1, but WNK2 does not phosphorylate it directly (146). WNK2 regulates PAK1 activity by controlling the balance of the activity of upstream regulators of PAK1 activity, RhoA and Rac1, which display reciprocal activity (53). WNK2 was found to physically interact with both RhoA and Rac1 (146). WNK2 knockdown decreases RhoA activation, but promotes Rac1 activation, resulting in PAK1 activation. The effects of WNK2 on the ERK1/2 pathway may be significant in the etiology of cancer [section V.E.].
4. WNK4 modulates phosphorylation of ERK1/2 and p38
In vitro, overexpression of WNK4 increases phosphorylation of both ERK1/2 and p38 MAPKs in response to both hypertonic stress and stimulation with EGF (146). A similar effect on ERK1/2 phosphorylation following EFG stimulation was also observed for WNK1 (225). Mutation of WNK4 to either a kinase-dead form (D312A) or an FHHt-mutant form (Q565E) did not affect the ability of WNK4 to phosphorylate these MAPKs. The upstream and downstream mediators of the WNK4 effects have not been determined, but as discussed in [section III.A.4.], differences in intracellular trafficking of WNK4 in response to hypertonicity and EGF stimulation are likely to play an important role in transducing downstream effects.
5. PI3-kinase signaling
As described in [section III.A.], WNK1 is a phosphorylation target of the PI3-kinase-dependent kinase Akt1, and WNK4 is a target for the related kinase SGK1. Conversely, WNK1 has been shown to modulate signaling through the PI3-kinase pathway, independently of its kinase activity. In vitro kinase assays performed on HEK293 cell lysates transfected with WNK1 and SGK1 showed that WNK1 increased specific phosphorylation of the synthetic substrate Crosstide, and of the physiological substrate Nedd4-2, by SGK1 (268). Activation of SGK1 by WNK1 was ERK-5 independent, but appears to be mediated through phosphorylation of T256, the PDK1 target site in the activation loop of SGK1. WNK1, however, does not directly phosphorylate SGK1 (268, 269), and the mechanism of regulation of SGK1 activity by WNK1 may require Akt1, since a kinase-dead mutant of Akt1 reduced activation of SGK1 by WNK1. However, caution should be taken in extrapolating this finding to a physiological setting, since the effect was observed in an overexpression system, and other kinase-dead members of the Akt1 family, or members of the closely-related SGK family itself, might exert a similar effect in vitro. For example, the inhibition by kinase-dead Akt1 may simply be due to competition with SGK1 for binding to WNK1 (which does not directly phosphorylate SGK1) or another required binding partner, similar to transcriptional squelching (22).
Both Akt1 and SGK1 phosphorylate WNK1 at T58/60 (247, 269), but this does not increase WNK1 kinase activity. Instead, phosphorylation of WNK1 at T58/60 may enhance the formation of a protein complex which somehow activates SGK1. This activation is independent of the WNK1 kinase domain, as demonstrated by stimulation by amino acid residues 1–220 of WNK1 alone (269). Finally, knockdown of WNK1 with siRNA prevents both IGF-1 (269) and H2O2 (25) from stimulating SGK1 activity.
Taken together, these data suggest an interesting interplay between Akt1, SGK1 and WNK1 in mediating IGF-1 signaling. IGF-1 treatment of cells leads to Akt1-dependent phosphorylation of T58/60 of WNK1, which then activates SGK1, which in turn phosphorylates WNK1 at T58/60. This positive feedback system may act as a mechanism to amplify IGF-1 signaling, which may be relevant, in vivo, to body growth. Mice lacking Akt1 display a mild growth defect (26, 29) while mice lacking both Akt1 and SGK3 have a severe growth defect (J.A. McCormick, unpublished observations), indicating the Akt1s and SGKs interact in the regulation of growth. Significantly, WNK1 is also able to activate SGK3 in vitro (269), as well as SGK2, but the physiological roles of SGK2 are currently unknown.
Akt2 has been shown to be the primary mediator of PI3-K signaling in insulin-dependent glucose homeostasis (28). Activation of the PI3-kinase/Akt1 pathway is required for insulin to stimulate WNK1 phosphorylation at T58/60 (92, 197), and insulin also activates SGK3 (292). Mice lacking both Akt2 and SGK3 display a more severe diabetic phenotype than mice lacking Akt2 alone ((James A. McCormick and David Pearce, unpublished observations (28)), while mice lacking SGK3 alone display normal glucose homeostasis (137). Perhaps a similar interplay between the Akts, SGKs and WNKs is important in the pathophysiology of diabetes?
6. Other pathways
The D. melanogaster WNK homologue was identified in a genome-wide RNAi screen to identify regulators of the WNT-Wingless pathway, using a transcriptional reporter-based assay (35). WNK negatively regulates the WNT pathway, in an induction-independent manner.
The D. melanogaster WNK homologue was also shown to interact with the protein CG8368 in a large scale yeast two-hybrid screen to generate a protein interaction map in D. melanogaster (64). The interacting partner is an RNase H-like protein that may play a role in RNA maturation, and nothing is known about the role of WNK kinases in its regulation.
D. Direct phosphorylation targets of WNK kinases
In addition to phosphorylation of components of the signaling pathways or inter-WNK phosphorylation described above, the WNK kinases have been shown to directly phosphorylate several additional downstream substrates.
1. WNK1 and synaptotagmins
The synaptotagmins act as calcium sensors in neurons and neuroendocrine cells, and regulate both endocytosis and exocytosis (196, 239). Synaptotagmin 2 was identified as interacting with the kinase domain of WNK1 by yeast two-hybrid screening; synaptotagmins 1, 3 and 9 were also shown to interact, but their interactions with WNK1 were not further characterized (117). Pull-down assays and coimmunoprecipitation confirmed the interaction between WNK1 and synaptotagmin 2. Interestingly, the kinase domain of WNK4 did not interact with any of the synaptotagmins despite being 85% identical to the WNK1 kinase domain, suggesting that specific residues within the kinase domain of WNK1 confer binding specificity. For example, changing the surface charge by introducing the mutation V318E prevented WNK1 from binding to synaptotagmin 2 without grossly changing its structure.
Synaptotagmin 2 contains two C2 domains, which bind Ca2+ and allow it to act as a Ca2+ sensor by stimulating its interaction with phospholipids (182, 207). Introducing mutations into synaptotagmin 2 that impair Ca2+ binding prevent the interaction between synaptotagmin and WNK1; increasing Ca2+ enhances their interaction (117). WNK1 phosphorylates synaptotagmin 2 within the C2 domains on T202 and T386. However, the effects of WNK1 on synaptotagmin 2 are more complex than simple phosphorylation, as shown by studies examining its phospholipid vesicle interactions. At low Ca2+ levels, synaptotagmin 2 will interact with WNK1, and if WNK1 is activated, be phosphorylated. Increasing Ca2+ levels push synaptotagmin 2 away from interacting with WNK1 and towards interacting with membranes, but if synaptotagmin 2 has already been phosphorylated by WNK1, a higher level of Ca2+ will be required to induce its membrane association. Phosphorylation by WNK1 thus appears to regulate synaptotagmin 2 by modulating the dynamics of its Ca2+–dependent membrane interactions.
2. SMAD2
Transforming growth factor beta (TGF-β) regulates cell proliferation, migration, differentiation, and apoptosis, and has broad (patho)physiological actions. TGF-β binds to a type II serine/threonine kinase receptor which recruits and phosphorylates a type I receptor. Activated receptors then phosphorylate and activate receptor-activated SMADs (R-SMADs), which interact with SMAD4 in the cytoplasm. The R-SMAD/SMAD4 heterodimer then translocates to the nucleus where it acts as a transcriptional regulator. Using yeast two-hybrid screening, Lee and colleagues determined that the kinase domains of WNK1 and WNK4 directly interact with R-SMADs 2 and 3 (116), which specifically transduce signals from TGF-β. Both WNK1 and WNK4 phosphorylate SMAD2 in vitro, at serines 110, 260 and 465, with S465 phosphorylation, a key phosphorylation site for SMAD2 activation, being the major phosphorylation site. Knockdown of WNK1 using siRNA led to a decrease in SMAD2 protein levels, but surprisingly increased levels of phosphorylated SMAD2 and its effects on transcription, suggesting dual roles in SMAD signaling.
In addition to transducing TGF-β signals, the SMADs also act via non-canonical means, for example by direct regulation of β–catenin (183, 289). Of more relevance to the WNK kinases, there is abundant evidence of crosstalk between the TGF-β and MAPK pathways (83). The identification of serines (110 and 260) phosphorylated by WNK1 provides evidence for a possible role for the WNK kinases in regulating SMAD signaling, though the upstream pathways (TGF-β-dependent or otherwise) are unknown. These two serines have also been identified as sites phosphorylated by calmodulin-dependent kinase II (serines 110 and 260) (257) and protein kinase C (serine 110) (274), and are not phosphorylated by the TGF-β receptor in the canonical TGF-β pathway. The physiological relevance of the effects of the WNK kinases on SMAD signaling remains to be determined.
3. Vacuolar H+-ATPase (V-ATPase)
The Arabidopsis thaliana WNK8 was found during a yeast two-hybrid screen to identify potential regulators of the Arabidopsis thaliana V-ATPase subunit C (81). This interaction was shown to be specific, and involved the C-terminus of WNK8, which contains a coiled-coil motif. WNK8 was then shown to phosphorylate V-ATPase subunit C at four sites, as determined by MALDI-TOF MS analysis, and could phosphorylate subunit C from other eukaryotes. WNK8 was also able to phosphorylate the cytoplasmic V1 domain protein complex, and could also phosphorylate subunits A, G and B/H. This study was the first to identify a kinase that directly interacts with and phosphorylates the V-ATPase, but it was not determined whether the phosphorylation events had any effect on V-ATPase complex structure, or activity.
In mammals, V-ATPases play numerous roles in physiology and disease (89). Most notably, they play an important role in protein trafficking by acidifying endosomes and lysosomes, which may be relevant to the regulation of ion channel activity. In neuronal cells, they participate in the loading of neurotransmitters into vesicles. V-ATPases are also localized to the plasma membrane in a wide variety of cells, including the proximal tubule, and the intercalated cells of the renal connecting segment and collecting duct, where they participate in acid-base homeostasis. Regulation of the C subunit in particular may play an important role in squamous cell carcinoma (173), in which mutations of WNK3 have been identified (Table 5).
Table 5.
WNK polymorphisms associated with blood pressure variability
| Polymorphism | Association | Reference |
|---|---|---|
| WNK1 | ||
| Intron 3 | No BP Change | 109 |
| T1056P | No BP Change | 109 |
| T1056P | Dose BP Effect | 169 |
| M1808I | Dose BP Effect | 169 |
| Intron 10 | BP Effect | 237 |
| Intron 23 | BP Effect | 237 |
| Exon 4 | Children BP Effect with age | 238 |
| Exon 10 | Children Dose BP Effect with age | 238 |
| Exon 11 | Children BP Effect with age | 238 |
| Intron 7 | BP response to thiazides | 240 |
| Intron 8 | BP response to thiazides | 240 |
| Intron 10 | Thiazide-sensitive, Low urinary Na, when combined with alpha adducing and Nedd-4 polymorphisms | 171 |
| Intron 1 | Blood pressure, essential hypertension, urinary K excretion | 157 |
| WNK4 | ||
| Intron 10 | BP Effect | 51 |
| Intron 10 | No BP Effect | 14,219 |
| Intron 14 | BP Effect | 109 |
| M546V | 100 | |
| P556T | 100 | |
| P1173T | 100 | |
| A589S | High Htn Population | 226 |
4. SPAK and OSR1 kinase
One important physiological substrate for WNK kinases is OSR1/SPAK, although the ability of the several WNK kinases to activate OSR1/SPAK has been reported to differ between members (8). In vitro, WNK1 (149, 248) phosphorylates SPAK and the related kinase OSR1 at two sites. The first conserved phosphorylation site (T233 in SPAK, T185 in OSRI) is located within the activation loop of the catalytic domain. The second is located within the C-termini (S373 in SPAK, S325 in OSR1), and its phosphorylation results in stimulation of SPAK/OSR1 activity. Although WNK4 also phosphorylates SPAK/OSR1 (248), Anselmo and colleagues (8) reported that WNK1 and WNK4 had strikingly different effects on OSR1 and SPAK. These investigators found that the activity of WNK1 towards OSR1/SPAK is much higher than the activity of WNK4, and importantly phosphorylation of OSR1 causes a shift gel migration; in contrast, phosphorylation by WNK4 does not cause such a shift and does not lead to activation of either OSR1 or SPAK.
Ahlstrom and colleagues (6) recently succeeded in synthesizing a full length WNK4. They found that the protein was capable of phosphorylating SPAK, but also noted that it was strongly associated with a second, unidentified kinase, which was also capable of phosphorylating SPAK. Thus, although the WNK1 and WNK4 kinase domains are 85% homologous, they appear to exert differential substrate specificity, including differential activity with respect to OSR1/SPAK (8). Finally, WNK3 also phosphorylates the serine residue in SPAK/OSR1, though the significance of this is unknown (149)
E. Ion channel regulation in vitro
The identification of mutations in WNK kinases as being causative of FHHt led to a flurry of research into the roles of the WNK kinases in the regulation of ion transport by the distal nephron. There have also been several studies examining effects of WNK kinases on neuronal channels and transporters. In this section, we will describe the wealth of in vitro data that have given insight into potential physiological roles of the WNK kinases, and these data will be placed into a physiological context in [section IV.]. The effects of WNK kinases on ion transport proteins are summarized in Tables 2, 3, and 4.
Table 2.
Summary of WNK1 effects on ion transport in vitro.
Table 3.
Summary of WNK3 effects on ion transport in vitro.
Table 4.
Summary of WNK4 effects on ion transport in vitro.
1. Structure of the distal nephron
The renal distal tubule is defined as the region of the nephron between the macula densa and the confluence with another tubule to form the collecting duct. This region comprises a short segment of thick ascending limb, the DCT, the connecting tubule (CNT) and the initial segment of cortical collecting tubule (ICT) (reviewed in (184)). NCC has been localized exclusively to the DCT at the mRNA level using in situ hybridization(9, 18, 164) and single nephron PCR(244), as well as at the protein level (1, 177). Expression of NCC is therefore considered a marker of the DCT. Further expression analysis has revealed that the DCT can be subdivided into an “early” DCT (DCT1) and a “late” DCT (DCT2)(164). Both DCT1 and DCT2 express NCC, but the DCT1 does not express the sodium-calcium exchanger (Na/Ca)(164) or ENaC(128), which are both expressed at the DCT2(209). The collecting duct, while expressing ENaC, does not express Na/Ca(164). The K excreting channel, ROMK1, and the Na-K-ATPase are also expressed all along the distal tubule(141). Activity of the Na-K-ATPase at the basolateral membrane of tubule cells is the driving force for sodium reabsorption along the entire nephron.
2. WNK regulation of NCC trafficking
NCC was an obvious candidate target for regulation by the WNK kinases since the thiazide diuretics, which specifically inhibit NCC, reverse all of the symptoms of FHHt (68). Initial studies to examine the effects of WNK kinases on NCC activity have relied on heterologous expression in Xenopus oocytes, due to the lack of a mammalian cell system that robustly displays NCC activity. Mammalian cell systems, on the other hand, provide data regarding the intracellular trafficking of NCC, which is relevant to regulation of its activity.
In Xenopus oocytes, WNK4 exerts an inhibitory effect on NCC activity. Rather than affecting total cellular NCC protein abundance, WNK4 reduces NCC abundance at the plasma membrane (23, 66, 67, 259, 279). WNK4 and NCC associate in a protein complex involving the C-termini of both proteins (23, 259, 281). The role of the kinase domain in NCC regulation is unclear, having been shown to be dependent (67, 259) and independent (281) on kinase activity, depending on the study. A truncated form of WNK4 lacking the entire kinase domain is still able to inhibit NCC activity (224, 281), and a short region near the carboxyl terminus of WNK4 that is required for NCC inhibition has been identified. A study using chimeras of WNK3 and WNK4 suggests that the N-terminus of WNK4 mediated NCC inhibition (199), and the truncated forms of WNK4 used in other studies (281) may be exerting a dominant-negative effect on endogenous WNK kinases.
The mechanism by which WNK4 reduces surface NCC expression appears to be through insertion of NCC into the plasma membrane, rather than endocytosis. Studies in both Xenopus oocytes (66) and mammalian cells (23) showed that the ability of WNK4 to reduce NCC surface expression is not affected by expression of a dominant-negative dynamin, suggesting that clathrin-dependent processes are not involved. Evidence that WNK4 inhibits forward trafficking of NCC to the plasma membrane was provided by the observation that Brefeldin A, an inhibitor of forward trafficking, reduces NCC surface expression (223). Following washout of the Brefeldin A, NCC surface expression increased, but overexpression of WNK4 inhibited this process. The inhibitory effect of WNK4 on NCC is sensitive to inhibition of lysosomal proton pumps, suggesting that WNK4 reduces trafficking of NCC to the plasma membrane, ultimately leading to enhanced lysosomal degradation (23). In HEK 293H cells, WNK4 reduced NCC surface expression and redirected it to a lysosomal compartment as shown by increased co-localization with LAMP-2, and an increased association of NCC with the lysosomal adaptor protein, AP-3 (223). More recently, in COS-7 cells overexpressing WNK4, accumulation of NCC in the lysosomal compartment was observed (291). Overexpression of a dominant-negative mutant of sortilin, a lysosomal targeting receptor, was able to reverse the inhibitory effect of WNK4 on NCC. NCC and sortilin directly interact, raising the possibility that together with WNK4, they comprise a complex that targets NCC to the lysosome for degradation. It is not known whether sortilin and AP-3 also interact with each other. Thus, the primary mechanism of WNK4 inhibition of NCC is through increased trafficking of NCC for lysosomal degradation.
WNK1 does not regulate NCC activity directly, but rather exerts an indirect effect through suppression of WNK4 inhibition of NCC (66, 279). Functional studies of WNK1 have revealed that its actions to modulate WNK4 require physical association with WNK4 and intact kinase activity (281). KS-WNK1, which lacks intrinsic kinase activity, interacts with and inhibits WNK1 kinase activity, inhibiting its ability to suppress WNK4 inhibition of NCC, presumably through a dominant-negative mechanism (224).
WNK3 is a potent activator of NCC activity in oocytes, and expression of a kinase-dead WNK3 mutant strongly inhibits NCC activity (187). Phosphorylation of NCC is increased by co-expression with WNK3 (187), but WNK3 does not appear to phosphorylate NCC directly (278). Furthermore, WNK3 and WNK4 interact physically, and compete to regulate NCC activity (278). The ability to inhibit WNK3 stimulation of NCC activity was not displayed by WNK1 or KS-WNK1. In vivo, this competition might allow a greater dynamic range of NCC regulation. Interestingly, an FHHt mutant WNK4 (Q562E) was not able to inhibit WNK3, which may be relevant to the disease [section V.A.]. Chimera studies suggested that the stimulatory effect of WNK3 is conferred by its N-terminus (199). However, as with WNK4, there is controversy regarding whether the C-terminus plays a key role, since the C-terminus of WNK3 is sufficient to activate NCC activity (278).
As discussed in [section II.B.2.], brain and kidney express alternative WNK3 isoforms which differ by small peptide insertions into the C-terminal encoded by exon18b and exon 22 in the brain isoform. Glover and colleagues examined the effects of both of these isoforms on NCC activity, and showed that in contrast to the renal isoform, which activates NCC, the brain isoform inhibits NCC activity (65). Increased or decreased NCC activity was associated with corresponding alterations in NCC surface expression, and the effect of the renal isoform was not dependent on SPAK kinase. These data suggest that the insertion of amino acid in the C-terminus of the brain isoform play a key role in preventing this isoform from activating NCC.
Similarly to WNK1, SGK1 suppresses WNK4 inhibition of NCC activity (194), and induction of SGK1 expression by aldosterone may play a role in mediating aldosterone-dependent stimulation of NCC activity. SGK1 and WNK4 directly interact, and SGK1 phosphorylates WNK4 at a non-canonical serine (S1169), which is also phosphorylated by WNK1. Phosphorylation of WNK4 at S1169 prevents WNK4 inhibition of NCC activity (194).
3. WNK regulation of NCC phosphorylation
SPAK and OSR1 kinase were identified as WNK1-interacting partners by yeast two-hybrid screen (149); WNK1 phosphorylates SPAK (149, 248), which in turn phosphorylates NCC at four N-terminal residues, T49 (185), T53 (149, 248), T58 (149, 248), and S71 (149) (note that amino acid numbering is based on rodent sequence). Interaction with SPAK is mediated through an arginine at position 19 in NCC, as illustrated by a loss of interaction when this site is mutated (185). Mutation of T53, T58 and S71 to alanines significantly reduces NCC activity in Xenopus oocytes, with T58A and S71A being greatly inhibitory (65, 170). The effect of these mutations on NCC activity is not dependent on a change in NCC surface expression (65, 170) indicating that phosphorylation of NCC at the N-terminal stimulates the cotransporter’s intrinsic activity. In HEK293 cells, T53A significantly reduced NCC activity under basal conditions, with no apparent effect of T49A or T58A (185), but T58A prevented stimulation of NCC activity by hypotonicity. These data, however, are limited by the high background in the system, and the data do not appear to have been normalized for specific activity. The mutant T58D NCC, in which phoshorylation at this site is mimicked, has higher basal activity than wild type NCC (65). Interestingly, coexpression with a constitutively active WNK3 mutant is able to further increase activity of this mutant NCC (65). This may involve phosphorylation at the other residues in NCC, but it may represent an alternative mechanism of activation by WNK3. In Xenopus oocytes, angiotensin II prevents WNK4 inhibition of NCC (198), an effect that specifically requires WNK4 and is probably mediated through SPAK.
4. WNK regulation of ENaC
WNK1 increases ENaC activity in both Xenopus oocytes and CHO cells by activating phosphotidylinositol 3-kinase, stimulating the ENaC regulator SGK1 (268, 269). This effect is dependent on an intact WNK1 kinase domain, but also requires an intact N-terminal domain. KS-WNK1, which lacks both the kinase domain and the amino terminal domain, has also been reported to stimulate ENaC (156), though the significance of this is unclear since KS-WNK1 is not expressed at high levels in the CCD in vivo (162). Similar to its effect on NCC, WNK4 inhibits ENaC activity, an effect that was suppressed by SGK1 but apparently did not require kinase activity (189). The same group, however, subsequently identified a serine residue in WNK4, S1169, which is phosphorylated by SGK1 (190). A phosphomimetic mutant of WNK4, S1169D, showed an almost complete loss of the ability to inhibit ENaC activity, suggesting that this site is crucial for effects on ENaC. It is possible that phosphorylation at the non-canonical SGK1 phosphorylation site in WNK4, S1196, which reduces WNK4 inhibition of NCC (194), also plays a role in ENaC regulation. The E3 ubiquitin ligase Nedd4-2 interacts through its three WW domains with the C-terminal PY motifs in each ENaC subunit, regulating the plasma membrane expression of ENaC (217, 221). Mutations in ENaC that eliminate the PY motifs result in increased ENaC activity and Liddle syndrome, a Mendelian form of hypertension (208). Deletion of the PY motifs from ENaC prevented WNK4 from inhibiting its activity, hinting at a possible mechanism by which WNK4 regulates ENaC (189). An FHHt mutant WNK4 (Q562E) does not inhibit ENaC activity when expressed in Xenopus oocytes(189). Taken together, these data suggest that dysregulation of ENaC activity by mutations in the WNK kinases may play a role in FHHt. WNK3 did not have an effect on ENaC activity in Xenopus ooctes (121)
5. Regulation of ROMK1
WNK4 strongly inhibits ROMK1 activity in vitro, through a kinase-independent mechanism(98, 115). WNK4 reduces ROMK1 abundance at the plasma membrane, but in contrast to its inhibition of NCC, the effect is dynamin-dependent and involves clathrin-mediated endocytosis (98). WNK4 and ROMK1 interact, but interaction is not sufficient to result in ROMK1 inhibition (77). FHHt-causing mutations in WNK4 (E559K, D561A and Q562E) increase this interaction and inhibit ROMK1 to a greater extent than wild type WNK4 (77, 98). Furthermore, deletion of the region containing these three amino acids prevents WNK4 inhibition of ROMK1, and a synthetic peptide of this region blocks the interaction between WNK4 and ROMK1, indicating it plays a key role in the WNK4-ROMK interaction (153). Taken together, these data suggest a possible mechanism for the hyperkalemia observed in FHHt, whereby potassium secretion is reduced by lower apical membrane expression of ROMK1. Direct phosphorylation of WNK4 by SGK1 reverses WNK4 inhibition of ROMK1 (190), with further complexity added by attenuation of the SGK1 effect by the protein kinase c-Src (284). In contrast to its effects on inhibition of NCC by WNK4, angiotensin II does not affect inhibition of ROMK1 by WNK4 (198).
Studies in Xenopus oocytes and HEK-293 cells have shown that WNK1 also inhibits ROMK1 activity. The effect is either dependent on intact kinase activity (115, 218), or independent on it, depending on the study(32). Three proline-rich motifs (PXXP) within amino acids 1–119 of WNK1 are sufficient to inhibit ROMK1 (77, 251). Time course studies of ROMK1 plasma membrane expression suggest that WNK1 increases endocytosis of ROMK1 in a dynamin-dependent manner (32), an effect involving interactions with the scaffolding protein intersectin (77). Interaction with intersectin occurs via the N-terminal proline-rich motifs of WNK1, and triple mutation of these motifs ablates this interaction, as well as preventing ROMK1 inhibition (77). WNK4 inhibition of ROMK1 also seems to require its interaction with intersectin, and interestingly, FHHt-causing mutations in WNK increase the interaction (77). Recently, direct evidence was provided that WNK1 stimulates ROMK1 endocytosis, and the clathrin adaptor molecule ARH is required (55). It remains to be determined whether ARH is a WNK1 substrate, and whether other WNK kinases modulate WNK1-ARH interaction.
KS-WNK1 alone does not affect ROMK1 activity, but reverses WNK1 inhibition of the channel (127). Two regions within KS-WNK1 have been identified that mediate its antagonistic effect on WNK1 regulation of ROMK1 (127). Not surprisingly, one region, including amino acids 31–253, contains the autoinhibitory domain; the other, which spans amino acids 1–77, contains the unique cysteine-rich sequence encoded by exon 4a. Both of these regions coimmunoprecipitate with amino acids 1–491 of WNK1, and mutational analysis revealed that the two essential phenylalanine residues in the autoinhibitory domain are required for region 31–253 to inhibit WNK1. In order for KS-WNK1 to inhibit WNK1, amino acids 120–491 of WNK1 are required, rather than the N-terminal proline-rich motif (127).
WNK3 has also been shown to inhibit ROMK1 activity in Xenopus oocytes, through an effect on plasma membrane abundance, rather than conductance or open probability (121). Interestingly, introduction of a mutation homologous to the FHHt-causing WNK mutation Q562E enhanced the inhibitory effect of WNK3, suggesting that this region may play an important role in the function of the WNKs in general.
6. WNK regulation of K-Cl cotransporters
There are four mammalian isoforms of K-Cl cotransporter (KCC), KCC1 to KCC4, which differ in their tissue distribution and physiological roles (5). KCC1 is ubiquitously expressed and is involved in the regulation of cell volume. KCC2 is restricted to neurons, where it plays a critical role in regulating intracellular chloride concentration. KCC3 and KCC4 are expressed in several tissues, including the kidney and central nervous system, and their diverse physiological functions include blood pressure control through vascular smooth muscle relaxation (4), transepithelial ammonium transport (7), renal K+ secretion (49), and myocardial K+ loss during ischemia (277). There is emerging evidence that WNK kinases regulate KCC function, and given the existence of multiple isoforms of both the WNKs and KCCs, regulation and physiological consequences are likely to be complex. Regulation of KCC activity is likely to be coupled to regulation of NKCC activity, since conditions that activate KCCs inhibit the NKCCs [section III.E.6.]. For example, cell swelling, high intracellular Cl−, and protein phosphatases stimulate KCCs, but inhibit NKCCs, and the reciprocal maneuvers inhibit KCCs and activate NKCCs (60). WNK kinases are good candidates for regulators of KCC and NKCC activity, since phosphorylation status controls their activities.
The first study examining regulation of KCC by WNK kinases showed that in Xenopus oocytes, wild type WNK3 strongly inhibits hypotonic activation of all 4 KCC isoforms, and activity of KCC2 under isotonic conditions (38). Coexpression of a kinase-dead mutant of WNK3 (WNK3 D294A) with the KCCs resulted in activation of all four KCCs, with the level of stimulation only slightly lower than that induced by hypotonicity in the absence of wild type WNK3. Under hypotonic conditions, only KCC1 and KCC2 could be further activated by coexpression of WNK3 D294A, probably since KCC3 and KCC4 were already functioning at their _V_max (38). WNK3 D294A exerts its stimulatory effect on KCC via activation of protein phosphatases, since treatment with phosphatase inhibitors prevented WNK3 D294A-mediated activation. WNK3 inhibition of KCC4 probably does not require interaction with SPAK, as illustrated by the ability of a WNK3 mutant unable to bind SPAK to retain its inhibitory effect (178).
Similarly, WNK4 inhibits hypotonic activation of KCC2 (58), KCC1, KCC3 and KCC4 (61). The requirement for hypotonicity for KCC activation could also be bypassed by kinase-dead WNK4, but only for KCC2 and KCC3 (61). Coexpression of wild type SPAK had no effect on WNK4 inhibition, and inhibition was kinase-dependent. However, coexpression of a kinase-dead SPAK and a constitutively active WNK4 mutant resulted in significant activation of KCC2 activity under isotonic conditions, suggesting that SPAK is unable to phosphorylate and inhibit KCC2 (possibly via protein phosphatase activation) and lies in the same regulatory pathway as WNK4 (58).
Recently, Rinehart and colleagues identified two phosphorylation sites, T991 and T1048, which play a role in regulation of KCC3 activity. Mutation of these sites to alanines, which prevents their phosphorylation, strongly activated KCC, confirming that KCC is active in the dephosphorylated state (188). Knockdown of WNK1 expression in HEK293 cells expressing KCC3 reduced phosphorylation at both of these threonines, though it was not determined whether this stimulated KCC3 activity (188). Since these two phosphorylation sites are conserved between KCC isoforms and between species, it is likely that these residues play a key role in regulation of KCC activity in general.
7. NKCC1 and NKCC2
Two genes encode the bumetanide-sensitive Na+-K+-2Cl− cotransporters NKCC1 and NKCC2. NKCC1 is ubiquitously expressed, and in polarized epithelial cells it is localized to the basolateral membrane, except in the choroid plexus where it is apical (176). Physiologically, it plays a role in cell volume regulation, cell proliferation and survival, epithelial transport, and neuronal excitability (195). NKCC2 is expressed exclusively at the apical membrane of the thick ascending loop of Henle (TALH), and plays an important role in salt reabsorption at this segment. This is best illustrated by Bartter’s disease Type I, in which inactivating mutations in NKKC2 result in a severe salt-wasting nephropathy accompanied by polyuria, hypokalemic metabolic alkalosis, and hypertrophy of the juxtaglomerular complex (154).
NKCC1 activity is strongly inhibited under hypotonic conditions. An initial study showed that in Xenopus oocytes, WNK4 inhibited NKCC1 under mildly hypotonic conditions by reducing its surface expression (95). A more recent study found no effect of WNK4 alone under isotonic conditions, but instead found significant activation of NKCC1 activity when SPAK was coexpressed (58). These changes in NKCC1 activity were unrelated to changes in surface expression levels (58). Activation under these conditions required the kinase activity of both SPAK and WNK4; mutation of the kinase domain of either kinase prevented activation by the wild type form of the other kinase. A putative SPAK-binding motif initially identified in cation-Cl− cotransporters (RFXV) was found within WNK4; mutation of this motif (WNK4 F997A) led to inhibition of NKCC1 activity when SPAK was coexpressed, suggesting this interaction is required for activation under isotonic conditions. Activation was also observed in hypotonic conditions when SPAK was coexpressed with WNK4, which might explain the opposite effects initially observed when SPAK was not coexpressed (95). No effects of WNK4 or SPAK alone were seen under hypertonic conditions, but when co-expressed there was a small reduction in hypertonic activation of NKCC1 (58). In this situation, the kinase of activity of SPAK appeared to be more important than that of WNK4 (58). Further regulation of WNK4/SPAK-dependent activation of NKCC1 may be achieved by interplay with apoptosis-associated tyrosine kinase (AATYK), an inhibitor of NKCC1 activity (59). AATYK directly interacts with SPAK through two RFXV motifs (174), and protein phosphatase 1 (PPI) is recruited to form a complex. It has been proposed that dephosphorylation of SPAK by PPI leads to inhibition of WNK4/SPAK activation of NKCC1 (59).
NKCC2, which is mutated in Bartter’s disease Type I, displays increased phosphorylation and activity under low Cl− hypotonic conditions (178). Coexpression with WNK3 significantly increases NKCC2 activity in Xenopus oocytes, an effect associated with increased phosphorylation of NKCC2 (178, 187). SPAK alone has no effect on NKCC2 activity, but enhances WNK3 stimulation of the cotransporter (178). Kinase-dead mutants of both WNK3 and SPAK suppressed NKCC2 activity, suggesting that both kinases are required to maintain basal NKCC2 activity. However, the kinase-dead mutants did not have an additive inhibitory effect on NKCC2, suggesting WNK3 lies upstream of SPAK in this pathway (178). Similarly to WNK4, WNK3 contains a putative SPAK-binding RFXV motif; mutation of this motif (WNK3 F1337A) prevented WNK3 stimulation of NKCC2, and also ablated interaction between WNK3 and SPAK. Mutation of T96, T101 and T111 in NKCC2, the residues whose phosphorylation is associated with increased activity, also prevented WNK3 activation of NKCC2 (178). Furthermore, consistent with an essential role for phosphorylation at these sites, a kinase-dead WNK3 mutant was unable to stimulate NKCC2 activity. These findings suggest that WNK3 acts as an intracellular Cl− sensor, and through SPAK increases NKCC2 activity in response to hypotonicity.
8. Regulation of TRP channels
The mammalian transient receptor potential (TRP) superfamily consists of six protein subfamilies, TRPC, TRPM, TRPV, TRPA, TRPML and TRPP. TRPs are non-voltage-gated cation channels involved in diverse physiological processes, and in the pathogenesis of various diseases. For example, multiple TRPs have been implicated in kidney disease (260) and cardiovascular disease (86), and may provide targets for the treatment of pain (33). Activity of TRPs is regulated by environmental stimuli including stretch, tonicity, and temperature (86). There is some evidence from in vitro studies that the WNK kinases play a role in regulation of TRP channel activity.
TRPV4 plays an important role in the regulation of system water balance, as illustrated by abnormal osmotic regulation in mice lacking TRPV4 (126, 144) and the recent identification of a loss-of-function polymorphism in humans that leads to hyponatremia via a reduction of hypotonicity-responsiveness of the channel (235). In addition to being expressed in the osmosensing nuclei of the hypothalamus (125), TRPV4 mRNA is highly expressed in the kidney (42, 125), and at the protein level, is abundant from the TALH onwards (236). In the kidney, TRPV4 may play a role in Ca2+ reabsorption. In HEK293 cells, transiently transfected TRPV4 stimulates Ca2+ entry in response to hypotonicity or the TRPV4 activator 4α-PDD; coexpression with WNK1 or WNK4 inhibited responses to both stimuli (57). For both kinases, the reduction in TRPV4 activity was associated with a reduction in surface expression of TRPV4, and for WNK4, was shown to require kinase activity. Similarly to regulation of NCC by WNK4, deletion of the C-terminus (WNK4 Δ1086–1211) of WNK4 abrogated the ability of WNK4 to inhibit TRPV4 (57). The extreme C-terminus of WNK4 contains a putative coiled-coil domain, as well as an SGK1 phosphorylation site (194). FHHt mutants of WNK4 (WNK4 E559K and WNK4 Q562E) were less effective at inhibiting TRPV4 activity, which may be relevant with regard to the presentation of hypercalciuria observed in patients with FHHt arising from WNK4 mutations (134, 135). Mice lacking TRPV4 display a trend towards hypercalcemia, but are not hypercalciuric on a standard diet (144); perhaps, similarly to mice transgenic for FHHt WNK4 mutations (112), a high Na+ diet would elicit a hypercalciuric phenotype. In contrast to WNK4, mutation of the kinase domain of WNK1 did not interfere with its ability to inhibit TRPV4 activity, but mutation of the putative WNK1 phosphorylation site S382 did.
TRPV5 may be a more important player in the regulation of calcium homeostasis by the kidney than TRPV4, since mice lacking TRPV5 display a 6-fold increase in urinary Ca2+ excretion on a standard diet which persists on a Ca2+-restricted diet (79). Data concerning effects of WNKs on TRPV5 are contradictory. In Xenopus oocytes, coexpression of wild type WNK4 significantly activated TRPV5 (91), but had no effect on TRPV6 activity; WNK1 had no effect on either channel. Surface biotinylation assays, and expression of a fluorescently tagged TRPV5 showed that WNK4 stimulates TRPV5 activity by increasing its surface abundance (91), possibly through a positive effect on N-glycosylation of the channel (90). Stimulation of TRPV5 by WNK4 requires an intact kinase domain, but FHHt mutants of WNK4 retained their activating ability (91). Interestingly, coexpression of NCC inhibited WNK4 stimulation of TRPV5, which may be relevant in cases of FHHt where the primary defect is in WNK4, and NCC expression is likely to be increased (112). Increased NCC expression could inhibit TRPV5 activity, leading to a reduction in Ca2+ reabsorption, and hypercalciuria. In HEK293 cells, WNK4 inhibited activity by stimulating calveolae-mediated endocytosis (Chou Long)
In Xenopus oocytes, WNK3 stimulates Ca2+ transport by TRPV5, and in contrast to WNK4, by TRPV6 (288). WNK3 increases surface expression of TRPV5, and similarly to WNK4, the mechanism appears to involve an effect on N-glycosylation and the secretory pathway. The stimulatory effect on TRPV6 may also occur through an effect on surface expression (288). For both channels, activation requires an intact kinase domain.
9. Other chloride transporters and channels
Members of the SLC26 family of exchangers transport a broad array of monovalent and divalent anions, including OH−, SO42−, Cl−, I−, HCO3−, formate, and oxalate, with differences in ion specificity between family members (150). Tissue expression patterns of SLC26 exchangers is also highly variable; SLC26A2, for example is expressed in most tissues (70, 75), whereas SLC26A9 appears to be restricted to lung (130) and gastric epithelial cells (271).
SLC26A6, a Cl−/base exchanger (CFEX), is expressed in the renal proximal tubule (105) where it physiological roles have been best described (214), as well as in the intestine (265), pancreatic ducts (130), testis tubule (105) and liver tubule (105). Similar to its effects on NKCC1 and NCC, coexpression of WNK4 with CFEX in Xenopus oocytes resulted in strong inhibition of CFEX activity (95). In contrast, WNK4 had no effect on the activity of SLC26A4 (Pendrin), which is mutated in Pendred syndrome, an inherited form of sensorineural hearing loss and goitre (52).
SLC26A9 is likely to function as a Cl− channel with minimal OH−/HCO3− permeability (45). In Xenopus oocytes, WNKs 1, 3 and 4 all inhibited SLC26A9-mediated Cl− transport (45), and kinase-dead mutants of WNKs 3 and 4 were still able to inhibit the channel (kinase-dead WNK1 was not tested). The kinase domain of WNK3 (WNK3 1–410) had no effect on SLC26A9 activity. Taken together, these data suggest that with regard to SC26A9 regulation, motifs C-terminal to the kinase domains of the WNKS may act as protein scaffolds. Similarly to WNK-mediated regulation of other channels and transporters, surface abundance of SLC26A9 is reduced by co-expression with WNK kinases. The physiological significance of WNK regulation of SLC26A9 is currently unknown, but together with WNK4 regulation of CFEX, these data hint at possible physiological roles for WNK regulation of SLC26 exchangers in extrarenal tissues.
A genetic strategy using Caenorhabditis elegans revealed a role for WNK1 in regulation of Cl− channel activity in a more physiological context. The SPAK and OSR1 homologue C. elegans germinal centre kinase 3 (GCK-3) (44) physically interacts with WNK1, through the WNK1 RFXV motif (78). WNK1 phosphorylates GCK-3 at S419 in vitro, although the functional relevance of this is unclear, since mutation of S419 had no effect on basal GCK-3 activity. Mutant C. elegans, lacking functional WNK1, displayed growth arrest at the early L2 larval stage, as well as defective development of the excretory canal (78). A similar defect in excretory tube formation was also observed in C. elegans in which GCK-3 had been disrupted. A constitutively active GCK-3 mutant (T280E) rescued the WNK1-deficient phenotype, indicating that WNK1 lies upstream of GCK-3 in the signaling pathway. Further genetic analysis revealed that WNK1-GCK3 act through modulation of CLH-3, a CLC ion channel. Downregulation of CLH-3 suppressed the effect of GCK-3 mutation, suggesting that CLH-3 negatively regulates excretory tube formation, and WNK1-GCK-3 inhibits this effect.
The cystic fibrosis transmembrane conductance regulator (CFTR) is a cAMP-gated chloride channel located on the apical surface of epithelia including the lung, intestine and pancreatic ducts; these are the tissues primarily affected by defects in CFTR in cystic fibrosis(122). WNK1 and WNK4 may regulate CFTR activity since they are likely to be co-expressed in several tissues (95, 280). In Xenopus oocytes, co-expression with WNK1 reduced conductance of activated CFTR by 40% (280); this effect appeared to be partially kinase-dependent. WNK4 inhibited CFTR conductance to a similar degree as WNK1, but in the case of WNK4, the effect was kinase-independent. Furthermore, surface biotinylation studies revealed that WNK4, but not WNK1, reduced CFTR surface abundance. WNK3 had no effect on CFTR activity in this system.
10. Paracellular Chloride permeability
Immunohistochemical studies suggest that WNK kinases may be expressed near or within tight junctions (95, 258). Thus, it was logical to test whether these kinases alter the properties of tight junctional complexes. Several studies indicated that WNK kinases alter paracellular Cl− permeability in vitro, which may be relevant in the pathogenesis of FHHt. In the earliest set of studies, two groups generated MDCKII cell lines in which expression of wild type or mutant WNK4s is tetracycline-inducible (96, 276). Wild type WNK4 significantly (96) or trended towards (276) increasing paracellular chloride permeability, while a kinase-dead WNK4 mutant had no effect on chloride permeability (96). Both studies identified no changes in the abundance or localization of proteins involved in regulation of tight junction permeability (claudins, occludins, or ZO-1), and Kahle and colleagues could not detect changes in tight junction structure by freeze-fracture EM (96). Since overexpression of WNK4 was associated with increased phosphorylation of claudins 1–4 (276), and kinase-dead WNK4 had no effect on paracellular chloride permeability (96), the effect of WNK4 seems to be kinase-dependent. A more recent study has confirmed the finding that wild type WNK4 increases paracellular chloride permeability, and identified claudin 7 as another WNK4 phosphorylation target (231).
Similar to overexpression of WNK4, MDCKII cells overexpressing WNK1 display an increase in paracellular permeability, associated with increased phosphorylation of Claudin 4 (168). However, an in vitro kinase assay suggests that WNK1 does not directly phosphorylate claudin 4 (278). WNK3 appears to have no effect on paracellular chloride permeability (121).
F. Control of cellular processes
Given the effects of the WNK kinases on signaling through growth factor pathways, including the MAPK and PI3-Kinase pathways, it is not surprising that the WNK kinases modulate cell proliferation, migration and differentiation. In addition, WNKs have also been implicated in the regulation of exocytosis. It is likely that the role of the WNKs in influencing these processes will become a much greater area of focus in the future.
1. WNK1 and cell proliferation and migration
Two studies that examined the effect on WNK1 on cell proliferation revealed opposing effects, depending on the cell type studied. These findings provide further evidence that WNK1 can influence signaling pathways in multiple ways.
As discussed in [section III.A.], insulin signaling is mediated through activation of PI3-kinase and subsequent activation of the Akt and SGK kinases. A key endpoint for this insulin-dependent signaling cascade is the stimulation of cell proliferation (203), and WNK1 is also activated by it [section III.A.2.]. Knockdown of WNK1 expression levels in 3T3-L1 preadipocytes using siRNA led to a significant increase in [3H]thymidine in the absence of insulin, and an even greater increase in the presence of insulin (92). Furthermore, knockdown of WNK1 enhanced cell proliferation in the presence of serum (the dependence of this effect on insulin directly was not assessed), as determined by increased cell numbers; this increase was not due to an effect on apoptosis. These data indicate that in this in vitro system, WNK1 acts as a negative regulator of cell proliferation.
In contrast, WNK1 acts as a positive regulator of proliferation, as well as of differentiation and migration, in C17.2 neural progenitor cells (225). Stable knockdown of WNK1 in C17.2 cells results in is a striking reduction (approximately 80%) in their proliferation rate, and an impairment of EGF-stimulated proliferation. Knockdown of WNK1 also inhibited migration of C17.2 cells, a process known to be mediated by EGF signaling (17).
WNK1 was also identified in an RNAi-based large-scale screen to identify proteins involved in cell growth and viability in the D. Melanogaster Kc167 and S2R+ cell lines (19). Knockdown of WNK1 reduced S2R+ cell proliferation significantly.
2. WNK1 and cell differentiation
C17.2 cells in which WNK1 has been knocked down do not differentiate to the same extent as normal C17.2 cells, as shown by alterations in gross morphology and expression of various differentiation markers (225).
A more extensive characterization of the role of WNK1 in mediating neuronal cell function was performed after WNK1 was identified as a binding partner for LINGO-1 in a yeast two-hybrid screen (290). LINGO-1 is a transmembrane protein that forms part of a complex with the Nogo66 receptor (142), this complex playing a role in mediating inhibition of neurite extension in neuronal cells, oligodendrocyte differentiation and axon myelination (11). The interaction between WNK1 and LINGO-1 was confirmed in vivo, and was mediated through amino acid residues 123 to 510 of WNK1, which contain the kinase domain. Knockdown of WNK1 expression in both PC12 cells and primary cortical neuron cells using siRNA inhibited the ability of Nogo66 to impair cell differentiation. Nogo66 signaling is mediated through activation of RhoA (160), and knockdown of WNK1 impairs this activation. The mechanism underlying may involve an effect on the RhoA inhibitor, Rho-GDI1, since WNK1 and Rho-GD1 could be coimmunoprecipitated from rat brain. The Nogo66-LINGO1-RhoA signaling pathway is known to interact with MAPK signaling pathways (275), and the MAPK is likely to play a role in the regulation of neurite extension, given its involvement in cytoskeletal remodeling (180).
3. WNK1 and exocytosis
As described in [section III.D.1.], WNK1 interacts with and phosphorylates synaptotagmin 2 (117), which plays a role in the control of exocytosis (63). Furthermore, WNK1 and synaptotagmin 2 were co-localized to secretory granules in a pancreatic β-cell line (117), and synaptotagmins play a role in the control of insulin secretion (62). While these data suggest a possible role for WNK1 in the regulation of exocytosis, more direct evidence comes from a study examining the interaction between WNK1 and Munc18c, another binding WNK1 binding partner identified in a yeast two-hybrid screen (165). Munc18c also plays a role in insulin signaling, as illustrated by its effects on insulin-stimulated GLUT4 vesicle translocation in vitro (233, 234), and the defective glucose-stimulated insulin secretion observed in mice heterozygous for Munc18c (166). Munc18c is a cytoplasmic protein that acts as a scaffold to regulate recruitment of the membrane protein syntaxin 4 to the SNARE complex (85, 103, 186); this role may provide the mechanism by which Munc18c modulates exocytosis. WNK1 interacts with Munc18c at the plasma membrane and in the cytoplasm, and its effects on Munc18c appear to be completely kinase-independent (165). Competitive inhibition of endogenous WNK1-Munc18c complexes with an N-terminal (amino acids 1–172) fragment of Munc18c significantly inhibited glucose-stimulated insulin secretion in vitro (165). This inhibition appeared to be due to an inhibition of SNARE complex formation, as shown by a reduction in syntaxin 4-VAMP2 association in secretory granules (165).
4. WNK2 and cell proliferation
Similarly to the effects of WNK1 in 3T3-L1 preadipocytes, WNK2 inhibits DNA synthesis, as determined by bromodeoxyuridine pulse-labeling, and cell proliferation in HeLa cells. More general effects of WNK2 on cell proliferation may be of significance in cancer, since WNK2 inhibits colony formation by glioma cells in a kinase-independent manner [section V.E.].
5. WNK3 and cell survival
The observation that endogenous WNK3 translocates from the cytoplasm to the nucleus of HeLa cells following induction of apoptosis with actinomycin D (246) stimulated further analysis of its role in cell survival. Overexpression of WNK3 increased survival of actinomycin D-treated cells, while knockdown of endogenous WNK3 levels by RNA interference increased apoptosis. WNK3 was shown to physically interact with two proteins known to play a role in apoptotic pathways, procaspase-3 and Heat-shock protein 70 (Hsp70) (246). Knockdown of WNK3 resulted in an increase in capsase-3 activity, suggesting that WNK3 inhibits apoptosis through an inhibitory effect on caspase-3 activity. WNK3 possibly acts as an adaptor or scaffold protein in a procaspase-3 regulatory complex, since it does not directly affect procaspase-3 phosphorylation (246). Regulation of apoptotic pathways by WNK3 may be relevant in cancer [section V.E.].
IV Physiological roles of WNK Kinases
WNK kinases have become a major focus of investigation because they comprise a previously unrecognized signaling pathway. Importantly, this pathway appears to be essential for normal development, for regulation of arterial pressure, for normal electrolyte balance, and for sensory nerve function. Mechanisms by which WNK kinases regulate these processes are beginning to be understood, but major knowledge gaps, and continuing controversies, render understanding incomplete. Integrative and pathophysiological roles of WNK kinases will be discussed here.
A. Effects of WNK4 on NaCl Transport
Physiological actions of WNK kinases were first recognized when WNK mutations were found to cause familial hyperkalemic hypertension. This autosomal dominant syndrome, described by Paver and Pauline (172), is characterized by hyperkalemia and hypertension, implying disordered renal Na, K, and Cl homeostasis (for a review see (179)). Infusion experiments comparing effects of several sodium salts on potassium excretion showed that intrinsic potassium secretory capacity remains intact in FHHt (206, 227), implying that the hyperkalemia is functionally derived, and does not result from an inability to secrete K. Schambelan and colleagues (206) postulated that the primary abnormality is an increased rate of chloride reabsorption along the distal nephron. This was projected to reduce the lumen-negative transepithelial voltage (they inferred that the Cl movement is electrogenic), which would facilitate Na reabsorption. They postulated that increased NaCl reabsorption would expand ECF volume, generating hypertension, and reduce potassium secretion. They named this defect a ‘chloride shunt’, but noted that they did not have enough information to speculate further on its nature. Take and colleagues (227) corroborated effects of different sodium salts in patients with FHHt, and they confirmed that the defect was corrected by administering a thiazide diuretic. They postulated, therefore, that the ‘chloride shunt’ reflected increased NaCl reabsorption along the thiazide-sensitive nephron; owing to inaccurate information about sites of thiazide action at that time (213), they postulated that the defect lay in the connecting tubule; it has become clear subsequently that the thiazide-sensitive segment is the DCT instead (9, 164). These authors speculated that increased activity of the thiazide-sensitive Na-Cl cotransporter would reduce Na delivery to potassium secretory segments of the nephron, thereby limiting potassium secretion. Potential mechanisms for the defect in K secretion will be discussed further, below.
More recently, additional evidence has accumulated supporting the role of NCC in FHHt. Farfel and colleagues (135) showed that the magnitude of blood pressure lowering that occurs with thiazide diuretics is much larger in patients with FHHt from WNK4 mutations, than in unaffected individuals. Further, the same group showed that individuals with FHHt caused by mutations in WNK4, waste calcium and develop osteopenia (135); these features are opposite to those observed in Gitelman syndrome (106), a syndrome in which dysfunction of the NCC leads to hypokalemic alkalosis and hypocalciuria. Thus, physiological information from humans with FHHt suggests that WNK4 mutations cause FHHt, at least in part, by activating NCC.
NCC activity is regulated by several physiological factors, including dietary salt deprivation (50), luminal salt delivery (99), angiotensin (201, 202), and aldosterone (243). Regulation occurs at several levels, including message abundance (164), protein abundance (1), and apical membrane trafficking (201, 202). NCC is also activated when it is phosphorylated along its amino terminal cytoplasmic domain (as discussed above). Perturbations that enhance NCC activity, such as dietary NaCl deprivation, short-term angiotensin infusion, and aldosterone administration all increase the abundance of phosphorylated NCC (27, 228, 241).
Lalioti and colleagues tested effects of WNK4 in vivo by introducing two copies of wild type or FHHt-mutant WNK4 into mice (112). Blood pressure was lower in mice with two extra copies of wild type WNK4; when stressed with a low potassium diet, the serum potassium was also reduced. Morphological studies showed that extra WNK4 copies reduced NCC abundance in kidney cortex. These results were interpreted as suggesting that wild type WNK4 functions as an inhibitor of NCC activity. Ohta and colleagues used a different approach to study physiological effects of WNK4 in mice (167). They generated WNK4 ‘hypomorphs’, in which exon 7 of WNK4 is deleted (see Figure 1); this construct was designed to delete the carboxyl terminus of WNK4, but when animals were generated, they instead expressed a modified WNK4 in which exons 7 and 8 were deleted, but the carboxyl terminus was intact; the resulting protein was expressed. The deleted segment does not include the kinase domain, but does include a coiled-coil domain (labeled M in Figure 2) and the acidic region mutated in some patients with FHHt. The truncated WNK4 demonstrated moderately reduced kinase activity toward SPAK, when tested in vitro. Animals expressing this mutant WNK4 exhibited reduced SPAK and OSR1 phosphorylation, and reduced NCC phosphorylation, although total NCC abundance was unaffected. The blood pressure was somewhat reduced in the hypomorphic animals, although this was evident only during the awake period (night time). Dietary salt restriction did not have an additional effect in either wild type or hypomorphic animals. The authors conclude that the results indicate that the physiological role of WNK4 is to activate NCC, not to inhibit it. Thus, physiological results reported to date are contradictory with respect to the predominant effects of WNK4 on NCC, some suggest that its net effect is to inhibit NCC; other suggest that its net effect is to stimulate NCC.
In contrast to the contradictory information concerning effects of wild type WNK4 on NCC, there is wide agreement that mutant WNK4 increases arterial pressure and serum potassium largely by activating NCC. In addition to the clinical data discussed above, two groups have studied mice expressing mutant WNK4. Lalioti generated animals transgenic for two copies of FHHt-mutant WNK4 Q562E on a wild type background (112). These mice demonstrated increased serum K concentration, compared with wild type animals, on low, moderate and high K intakes, and elevated arterial pressure. The abundance of NCC was increased and DCT segments appeared hypertrophic; the defects were corrected by thiazide diuretics or by crossing the mice with NCC knockouts. These results show clearly that FHHt does not simply result from a loss of WNK4 function, as the animals retained two copies of the wild type WNK4 gene. Similar results were obtained by Yang et al. (282) who knocked FHHt-mutant WNK4 D561A into animals expressing a single endogenous WNK4. This model mimics human disease, with one copy of wild type and one copy of mutant WNK4. The resulting mice were hypertensive and hyperkalemic, relative to wild type controls. The abundance of phosphorylated of NCC was increased substantially, and the functional defects could be corrected by infusing sodium sulfate or administering a thiazide diuretic. Since the Q562E WNK4 mutant is unable to inhibit WNK3 activation of NCC in vitro, and suppresses the actions of the wild type WNK4 on NCC (278) it is possible that unrestrained WNK3 activity plays a role in the etiology of FHHt.
Two groups have postulated mechanisms by which WNK4 may switch from an inhibitory to a stimulatory mode (198, 278). The first model, proposed by Gamba and colleagues, high circulating angiotensin II levels turn WNK4 from being inhibitory to being stimulatory, with respect to NCC. The second model, postulated by Yang and colleagues (278), suggests that FHHt-causing mutant WNK4 acts to stimulate WNK3 activity; it does so by acting as a dominant-negative regulator of wild type WNK4, which normally inhibits NCC. These models are discussed further, below (Section VA, and Figure 12).
Figure 12. Models by which mutations in WNK4 cause FHHt.
In this figure, an arrow indicates stimulation, a large arrow indicates increased stimulation, and a T shape indicates inhibition. Model A (112) shows that mutation of one of two WNK4 genes leads to a loss of inhibition of NCC and increased suppression of ROMK. Model B (278) shows that mutant WNK4 exerts a dominant-negative effect on wild type WNK4’s inhibition of WNK3 (second copy of WNK4 is omitted for clarity). The uninhibited WNK3 is shown activating NCC via both SPAK-dependent and SPAK-independent processes. Model C (198) shows that angiotensin II converts WNK4 from an inhibitory mode to a stimulatory one. Mutant WNK4 is shown acting in the stimulatory mode, even in the absence of angiotensin II. Model D (282) shows that wild type WNK4 acts to stimulate NCC. According to this model, mutant WNK4 has a simple gain-of-function effect.
One other concern about experiments performed using physiological model systems arises because of homology between components of the signaling and transport systems. First, the amino terminal domains of the various cation chloride cotransporters are homologous, including several of the key phosphorylation sites. Several ‘phospho-NCC’ antibodies recognize not only NCC, but also NKCC2 (and perhaps NKCC1) (unpublished observations). Further, there have been variable reports of the expression and importance of OSR1 in kidney tissues, versus SPAK, which appears to be the dominant renal member of the family. SPAK is expressed predominantly along the TAL and can phosphorylate NKCC2, as well as NCC (46, 149, 178, 185). Thus, studies showing changes in the abundance of ‘phospho-NCC’ may sometimes represent changes in the abundance of phosphorylated NCC and NKCC2.
B. Effects of WNK1 on NaCl transport
There is less information concerning effects of WNK1 on NaCl transport than there is concerning effects of WNK4. Unlike patients with FHHt from WNK4 mutations, patients with FHHt from WNK1 mutations do not appear to waste calcium (3); in as much as hypocalciuria is a hallmark of NCC inactivity, this indicates either that the defect in WNK1 patients does not involve NCC, or that there are additional mechanisms that stimulate calcium retention. Some have suggested that hypertension in individuals with WNK1 mutations is not very sensitive to thiazide diuretics (263), but few data support this contention, and countervailing observations have been made (Jeunmaitre, personal communication).
Technical difficulties have frustrated attempts to study effects of WNK1 in vivo. Deletion of WNK1 is embryonic lethal; deletion of a single copy, however, reduces blood pressure (286), but the effects of WNK1 heterozygosity on NCC, phosphorylated NCC or SPAK abundance were not reported, so the mechanisms hypotension induced by WNK1 deletion remain speculative. Based on in vitro effects of KS-WNK1 to antagonize WNK1, and on WNK1 to regulate NCC (discussed above), two groups provided preliminary evidence suggesting that WNK1 and KS-WNK1 do regulate NCC in vivo. Hadchouel and colleagues generated mice with inactivation of KS-WNK1, to test the hypothesis that KS-WNK1 acts as a natural WNK1 antagonist. NCC abundance was increased 2-fold in affected mice, which also showed enlargement of DCT profiles; while there was some evidence of ECF volume expansion, with suppressed aldosterone levels and a slight increase in diastolic blood pressure, hyperkalemia and hypertension were not present (Hadchouel et al, ASN 2008). Liu and colleagues compared mice transgenic for the amino terminal KS-WNK1 1–253 fragment, driven by a kidney-specific cadherin promoter, and wild type mice (127). NCC abundance and blood pressure were reduced in mice over-expressing KS-WNK1 1–253, at baseline, suggesting that KS-WNK1 does act to inhibit NCC in vivo. In contrast, KS-WNK1 knockout mice had increased NCC abundance, but like the mice discussed above, they had normal blood pressure on a control diet. When stressed with a high salt diet, they did become hypertensive; when stressed with a high K diet, they did become hyperkalemic. Thus, these data strongly support the model first postulated by Subramanya and colleagues (224), that KS-WNK1 acts as an endogenous dominant-negative WNK1 regulator, thereby inhibiting NCC activity in vivo. One caveat of these studies is that the kidney-specific cadherin promoter is widely active in the kidney, and transgenes using it typically display a mosaic expression pattern. Therefore, KS-WNK1 may be exerting an inhibitory effect on WNK1 in nephron segments where it is not normally expressed. These results, however, suggest that other effects of WNK1 must be involved in generating the full FHHt phenotype, as the derived models, unlike the FHHt WNK4 models, do not fully recapitulate the human disease.
C. Effects of WNK4 on potassium transport
In mammals, aldosterone secretion is driven by two distinct stimuli, serum potassium and ECF volume (through rennin and angiotensin II). Surprisingly, aldosterone has different effects on kidney transport pathways, depending on whether it is secreted in response to hyperkalemia or ECF volume depletion. This ability of aldosterone to transduce different effects depending on the physiologic condition has been termed the ‘_aldosterone paradox_’ (73). In vitro studies discussed above suggested that WNK4 inhibits ROMK by reducing its membrane abundance, and that mutant WNK4 is more active in this regard; if disordered ROMK activity is responsible for hyperkalemia in the setting of FHHt, then such individuals should be unable to excrete K unless high distal flow rates are induced. In contrast, as noted above, the potassium secretory capacity of individuals with FHHt, is normal when stimulated by non-chloride sodium salts (227). Further, the renal abundance of ROMK has been reported to be normal in both WNK4 Q562E transgenic and WNK4 D561A knockin animals, and microperfused collecting ducts from D561A knockin animals exhibit normal ROMK activity (112, 282). These data however should not be taken as excluding a role for WNK4 in regulating ROMK physiologically. The fact that ROMK is not activated more, despite the presence of hyperkalemia, suggests that some ROMK dysfunction may be present in the disease; quantitation of ROMK abundance by western of whole kidneys may also be insensitive to physiologically relevant changes. In addition, infusing non-chloride sodium salts may activate maxi-K channels, which drive flow-induced potassium secretion (204) and may compensate, in part, for decreased ROMK activity. Finally, although collecting ducts from WNK4 D561A knockin animals exhibit normal potassium transport characteristics, more proximal segments, such as the CNT and DCT may still exhibit disordered ROMK function (140). The contribution of dysregulation of renal KCC in FHHt is unknown, though WNK4 FHHt mutants do not change the regulation of KCC by WNK4 activity in vitro (61).
C. Effects of WNK1 on potassium transport
Less information is available concerning effects of WNK1 on potassium transport in vivo. As noted above, in vitro data suggest that WNK1 inhibits ROMK activity, by stimulating its removal from the membrane via clathrin-dependent processes (77). In view of evidence that the intronic mutations in WNK1 may increase its abundance, one hypothesis suggests that increased WNK1 should increase the inhibition of ROMK activity. Mice heterozygous for WNK1 did not manifest abnormalities in serum K concentration, and the abundance of ROMK in these animals, and its distribution was not described (286). Mice transgenic for KS-WNK1 in kidney tubules exhibited increased ROMK abundance and hypokalemia (127), although mice with KS-WNK1 deleted did not show evidence of hyperkalemia (Hadchouel et al, ASN 2008). Thus, overall these results support the hypothesis that WNK1 does play a direct role in regulating renal K homeostasis and suggest that KS-WNK1 may act to inhibit WNK1 in this respect, but the data remain fragmentary and more work needs to be completed to solidify this picture.
E. WNK1 and cardiovascular system development
Mice lacking WNK1 were generated during a large-scale gene trap screen, and were found to die at around embryonic day 13.5 (E13.5) from unknown causes (286). Subsequently, transgenic mice expressing lacZ from a modified WNK1-containing BAC were generated (40). In this animal model, expression of KS-WNK1, which is also encoded by the WNK1 gene, was prevented by introducing a stop codon into exon 4a. The lacZ transgene was highly expressed in the heart during early development from E7.5 through E14.5. In the developing embryo lacZ expression was observed throughout the vasculature, including at the placenta and yolk sac (40), and was strongly expressed in the endothelium of both arteries and veins.
Recently, a conditional WNK1 knockout mouse was developed using the CRE-lox system, which has given more detailed insight into the precise role of WNK1 in cardiovascular development (264). Mice lacking WNK1 throughout the body died between E10.5 and E12.5, and displayed multiple cardiovascular abnormalities, resulting in pericardial edema and widespread hemorrhaging. The hearts of WNK1 knockout mice displayed smaller heart chamber size, and reduced myocardial trabeculation, and the dorsal aorta was smaller or collapsed. The yolk sacs of WNK1 knockout mice appeared thin and pale, and lacked larger vitelline vessels; a consequence of this defect was growth retardation. In the placenta, there was only superficial establishment of a circulatory network between the fetal and maternal blood vessels. Immunostaining of vascular endothelium using an antibody against platelet endothelial cell adhesion molecule 1 (PECAM1) revealed that disruption of WNK1 does not disrupt vasculogenesis, but rather leads to defects in sprouting angiogenesis and remodeling angiogenesis, as well as in cardiac development. Ectopic expression of the arterial marker neurophilin-1 (NP1) and the venous marker EphB4 was observed, but no differences in the expression levels of several genes involved in angiogenesis were found.
To determine whether endothelial or myocardial expression of WNK1 was necessary for embryonic viability, a mouse line in which a WNK1 cDNA was targeted to the ubiquitously active Rosa26 locus was generated (264). The presence of a transcriptional terminator prevents WNK1 expression, unless the terminator is excised by Cre recombinase. Tie2-Cre mice, which express Cre only in the endothelium, and the WNK1/Rosa26 line were then bred with WNK1 heterozygotes, and compound heterozygotes were then interbred to generate WNK1 knockouts in which WNK1 was expressed only in the endothelium. Endothelium-specific expression of WNK1 was sufficient to rescue the WNK1 knockout cardiovascular development phenotype. However, the majority of these mice died with the first day after birth, suggesting a critical role for WNK1 in postnatal life, as well as embryonic development. Using Sox2-Cre mice, which express Cre in the endothelium of the embryo, but not in extra-embryonic tissues (yolk sac and/or placenta), did not rescue WNK1 knockout mice, indicating that WNK1 expression in these tissues is required for embryogenesis. The generation of additional tissue-specific CRE-WNK1/Rosa26 rescue lines, as well as the production of tissue-specific WNK1 knockout mice, will help dissect the roles of WNK1 in postnatal physiology.
V WNK kinases and disease
A. Familial Hyperkalemic Hypertension
Familial Hyperkalemic Hypertension (or Gordon’s syndrome, or pseudohypoaldosteronism II) was first described in 1964 (172), and is inherited in an autosomal dominant manner (20, 56). Patients with FHHt all exhibit hyperkalemia, which appears to be the most consistent feature of the disease. Hypertension, while commonly present and sometimes severe, often appears later in life. Other characteristic features include mild metabolic acidosis, suppressed plasma renin activity, and aldosterone levels that are lower than would be expected, considering the hyperkalemia. Infusing NaCl does not increase urinary potassium excretion in patients with FHHt, as it does in normal individuals, whereas infusing non-chloride salts of Na+ does increase K+ excretion to normal levels (206, 227). Patients are often highly sensitive to thiazide diuretics, which can typically correct both the hyperkalemia and hypertension (135, 189, 282).
As described in [sections II.B.1. and II.C.4.], the genetic basis of FHHt was identified as mutations in WNK1 or WNK4. The roles of the WNK kinases in the regulation of renal ion transport are discussed in [section IV.]. Four models for mechanisms by which mutations in WNK4 lead to FHHt have been presented (Figure 12). These models may not be mutually exclusive. In view of the increased WNK1 message detected in the white cells of patients with FHHt, several groups have suggested that FHHt caused by WNK1 intronic mutations is likely to reflect an increase in full length WNK1, or a decrease in the ratio of KS-WNK1/WNK1 (Figure 13) (115, 127, 224, 249). In either case, increased WNK1 would stimulate NCC activity via effects on SPAK, and could also inhibit WNK4. The precise mechanisms by which defects in WNK kinase signaling lead to the FHHt phenotype, however, remain unresolved.
Figure 13. Model of mechanism of WNK1-induced FHHt.
In this figure, an arrow indicates stimulation, a large arrow indicates increased stimulation, and a T shape indicates inhibition. Under normal circumstances, the activity of NCC and ROMK is set by the balance between KS-WNK1 and WNK1. KS-WNK1 inhibits WNK1. WNK1 inhibits ROMK. WNK1 inhibits WNK4, which inhibits NCC. WNK1 activates SPAK, which stimulates NCC. When expression levels of WNK1 are increased, relative to KS-WNK1, WNK4 suppression is increased, SPAK activation is increased, and ROMK suppression is increased. See text for details.
B. Essential Hypertension
Essential hypertension, defined as chronically elevated blood pressure occurring in the absence of identifiable causes, affects 25% of the adult population in the developed world and is a major independent risk factor for stroke, myocardial infarction, and congestive heart failure. Although essential hypertension has no specific identifiable causes, many contributing factors, both genetic and environmental, have been described. Since the finding that mutations in WNKs 1 and 4 cause FHHt (258), and the observation that mice heterozygous for WNK1 have lower blood pressure (286), single nucleotide polymorphisms (SNPs) in the WNKs have become candidates for association with essential hypertension. One important facet of SNP analysis that is often overlooked is the role of polymorphisms that do not change coding, or are located in introns. Intronic SNPs in particular are often disregarded, but may play an important role in determining the expression level of the encoded gene. The FHHt-causing mutations in WNK1 involve large deletions of intron 1, which have been proposed to increase its expression level (258); it is therefore possible that more subtle nucleotide changes may also affect WNK1 expression levels.
Numerous SNPs have been identified in WNK1 (Table 5). In a study of Japanese patients, 36 SNPs were identified, with an intron 3 SNP and the T1056P SNP being highly frequent (109). However, none of these were associated with differences in blood pressure in this study. A more recent Japanese study showed the T1056P SNP has a dose-dependent association with increased blood pressure (169), as did M1808I and an intron 10 SNP. Analysis of 4 haplotypes (containing 2 additional SNPs), with population frequencies ranging from 8–48%, identified a haplotype associated with increased blood pressure, and one associated with reduced blood pressure. For 2 haplotypes, including that associated with lower blood pressure, the systolic and diastolic blood pressure levels in individuals with a high Na/K intake ratio were significantly higher than those with a low Na/K intake ratio. Blood pressures in carriers of the haplotype associated with increased blood pressure did not vary with the ration of Na/K intake (169). If these data are confirmed, they suggest that genetic differences in the WNK1 gene influence blood pressure response to dietary sodium and potassium intake. Given the effects of changes in dietary potassium and sodium on renal levels of WNK1 and KS-WNK1 [section II.B.1.], it is possible that the SNPs identified modulate the functional interactions between these isoforms depending on dietary electrolyte intake.
Several other studies have examined the association of WNK1 SNPs with blood pressure. Tobin and colleagues examined 9 SNPs spread across the entire WNK1 gene, and identified 4 associated with variation in mean 24 hour systolic blood pressure, and 5 with mean 24 hour diastolic blood pressure; SNPs in intron 10 and intron 23 were associated with variation of both (237). In children, 3 SNPs within the coding region of WNK1 (exons 4, 10 and 11) were linked with the rate of change of diastolic blood pressure with increasing age (238). Another group identified 3 SNPs associated with differences in ambulatory blood pressure responses to thiazide diuretics (240). A SNP in the 5′ regulatory region of WNK1 (approximately 3kb from the promoter) displayed a nominal association with hypertension severity (158). A recent meta-analysis found multiple SNPs and haplotypes significantly associated with blood pressure, essential hypertension and urinary potassium excretion (157). Interestingly, the SNP displaying the strongest association with these three parameters is located in intron 1, the intron mutated in FHHt. In addition, several rare haplotypes that displayed large blood-pressure lowering effects were identified. Another recent meta-analysis of 3200 Italian subjects identified association of WNK1 SNPs with systolic and diastolic blood pressure, but only when blood pressure was measured in the clinic (171). Finally, a WNK1 intron 10 SNP is associated with blood pressure variation on its own, but also tracked with a greater increase in blood pressure, thiazide sensitivity and reduced Na excretion when present in combination with SNPs for α-adducin and Nedd4-2, which alone displayed no associations (171). More SNPs in WNK1 are likely to be identified and associated with essential hypertension in the future.
Based on the location of human WNK4 on chromosome 17q, a region previously identified as a blood pressure quantitative trait locus (10, 93), Erhlich and colleagues searched for functional SNPs within the coding region of WNK4 (51) (Table 5). Eight SNPs that were identified in African American subjects, but only the single SNP (located within intron 10) identified in white subjects was associated with hypertension. However, subsequent studies of larger cohorts of white subjects have not detected an association between the intron 10 SNP and hypertension (14, 219). Several intronic SNPs were identified within a large Japanese cohort (1,818 subjects), but only one of these (C14717T, in intron 14) had a significant association with hypertension in males (109). WNK4 SNPs within exons have also been identified in another Japanese cohort (M546V, P556T and P1173T) (100), and a Chinese cohort from a desert region with a high salt intake and 35% prevalence of hypertension (A589S) (226). None of these mutations are within the acidic motif that is mutated in FHHt, but three of them lie close to it, and the other (P1173T) lies close to known sites of WNK4 phosphorylation. Overall, it appears that SNPs in WNK4 are relatively uncommon; although this makes the contribution of common WNK4 SNPs to hypertension less likely, rare polymorphisms with larger effects are increasingly recognized as contributing to human disease, and WNK4 remains a candidate gene.
C. Hereditary Sensory and Autonomic Neuropathy Type II (HSANII)
Hereditary sensory and autonomic neuropathies (HSANs) are a clinically diverse group of neuropathies that primarily affect sensory nerves, and can be transmitted as either autosomal dominant or autosomal recessive traits. HSANs are classified into types I-IV, depending on inheritance pattern, age of onset and clinical differences (131), and so far mutations in seven genes have been identified as causative. HSANII is a severe early-onset autosomal recessive disorder characterized by loss of heat, pain and touch perception, particularly in the lower limbs. Secondary complications include muscle atrophy, ulcerations and spontaneous amputation of digits.
Initially, linkage analysis of two HSANII pedigrees in Newfoundland identified a 1.06Mb region that included WNK1, but no mutations in WNK1 or other transcripts within the region were identified (111). Instead, a highly conserved ORF derived from a single exon, and encoding a 434 amino acid named HSN2, was identified within intron 8 of the WNK1 gene. Sequence analysis of affected individuals revealed deletion mutations that resulted in truncation, a finding subsequently described in other HSANII pedigrees (192, 193). More recently, an individual was identified with classic HSANII symptoms who surprisingly only carried a heterozygous mutation in HSN2 (211). Analysis of the WNK1 coding sequence identified a deletion mutation within exon 6 that results in truncation of WNK1. Detailed characterization of mRNA species containing HSN2 revealed that HSN2 is in fact a novel WNK1 exon, expressed exclusively in nervous system tissues. HSN2 is present in mRNA from brain, spinal cord, dorsal root ganglia and sciatic nerve, but in brain and spinal cord an additional exon, 8b, was identified (211). Interestingly Western blotting suggests that HSN2-containing WNK1 variants are generated through usage of the kidney-specific promoter, which results in a form of WNK1 lacking the kinase domain [section II.A.1.], since these variants are significantly smaller than if they were derived from the upstream promoter (230KDa versus 270KDa). However, the lack of antibodies specifically recognizing KS-WNK1 has prevented confirmation of this observation.
All of the mutations associated with HSANII so far identified are predicted to lead to early mRNA termination. Clarification of which promoter is used to generate the nervous system forms of WNK1 will be particularly helpful, since it will provide insight into the protein form of WNK1 involved in the disease. For example, early mRNA truncation often results in a completely non-functional protein, which would preclude a role for full-length WNK1, since disruption of WNK1 expression is embryonic lethal in mice (286). Targeted disruption of KS-WNK1, on the other hand, does not affect viability (Hadchouel et al. Abstract, ASN 2008). In both cases, any truncated protein expressed would lack the C-terminal coiled-coil motif, suggesting a possible role for the C-terminal domain.
One possible disease mechanism may involve dysregulation of TRP channels expressed in sensory neurons which are important for the transduction of chemical, thermal and mechanical signals (15). As discussed in [section III.E.8.], WNK kinases regulate the activity of several TRP channels. In particular, WNK1 inhibits in vitro activity of TRPV4 (57), which is expressed in sensory ganglia, skin and dorsal root ganglia, and plays a role in heat and mechanosensation (118). Interestingly, increased sensitivity to both warm and cold stimuli has been observed in heterozygous carriers of HSANII-causing mutations (129). Further support for a role for the WNKs in regulation of TRP channel function and peripheral sensory regulation in vivo is provided by the description of patients with a deletion in WNK3 displaying a high pain threshold (181) [section V.D.].
Peripheral neuropathy associated with agenesis of the corpus callosum (ACCPN) is an inherited progressive sensorimotor neuropathy. Individuals with ACCPN display mental retardation, dysmorphic features, complete or partial agenesis of the corpus callosum, and have an average life expectancy of 33 years of age (84). The underlying genetic defect in this disease results in production of a truncated form of KCC3 (84) that reaches the plasma membrane but is non-functional. Given the effects of the WNK kinases on KCC3 activity [section III.E.6.], and the role of WNK1 in HSANII, one could speculate a broader role for the WNK kinases in peripheral neuron function.
D. Autism
Autism spectrum disorders are characterized by impairments of communication, social interaction, and restricted and repetitive behaviors. While heritability has been observed, few genes have been identified as playing a contribution to their development, though the higher incidence in males suggests the involvement of X-linked genes. Recently, genetic analysis of affected male siblings identified a 470 kb X chromosome deletion (181). This deletion results in disruption of three genes, including WNK3, which is almost completely deleted; deletion occurs after exon 2, and any expressed WNK3 protein would be truncated at the beginning of the kinase domain.
These findings are informative, since they suggest that targeted disruption of WNK3 in mice may not affect viability, and could provide a model of autism. WNK3 modulates chloride transport, which plays a role in the maintenance of intracellular volume and the excitability of GABA-responsive neurons (97), and its expression co-localizes with components of the central GABA-ergic system. Multiple lines of evidence suggest an important role for GABA-ergic transmission in the etiology of autism (13). However, the fact that two other genes are also deleted, including PHF8, which has previously been reported to play a role in X-linked mental retardation (2, 114), indicate that deletion of WNK3 may be coincidental rather than causative.
E. Cancer
Consistent with their roles in regulating cell proliferation, and suggesting a possible role for the WNK kinases in tumorigenesis, several large scale screening studies have identified point mutations in members of the WNK family in a wide variety of cancers (Table 6). Several other lines of evidence lend support to the idea that the WNKs may be important players in the development of cancer.
Table 6.
Mutations in all WNK kinases isoforms are associated with cancer.
| WNK kinase | Type | Morphology | Nucleotide change | Amino acid change | Mutation type | Reference |
|---|---|---|---|---|---|---|
| 1 | Breast | Pleomorphic lobular carcinoma | G1255C | E419Q | Missense | (69, 222) |
| 1 | Breast | Invasive ductal carcinoma | C5395G | Q1799E | Missense | (69, 215, 222) |
| 1 | Breast | Pleomorphic lobular carcinoma | C6569G | S2190C | Missense | (69, 222) |
| 1 | Colorectal | A3596G | E1199G | Missense | (215) | |
| 1 | Lung | Adenocarcinoma | C7086A | F2362L | Missense | (36, 69) |
| 1 | Ovarian | Serous carcinoma | C2829T | Y943Y | Silent | (69) |
| 2 | Colorectal | Adenocarcinoma | 1922delC | P641.fs*2 | Frameshift deletion | (69) |
| 2 | Gastric | Adenocarcinoma | 4116delC | S1373fs*5 | Frameshift deletion | (69) |
| 2 | Lung | Adenocarcinoma | G5933T | S1978I | Missense | (36, 69) |
| 2 | Lung | Neuroendocrine carcinoma | G4856A | G1619E | Missense | (36, 69) |
| 2 | Ovarian | Serous carcinoma | G1486T | V396L | Missense | (69) |
| 2 | Ovarian | Mucinous carcinoma | 6642delC | T2215FS*31 | Frameshift deletion | (69) |
| 3 | Glioma | Glioblastoma multiforme | C2784T | H928H | Silent | (69) |
| 3 | Lung | Squamous cell carcinoma | C2561G | S854C | Missense | (69) |
| 3 | Lung | Large cell carcinoma | G4599T | L1533F | Missense | (69) |
| 3 | Renal | Clear cell carcinoma | C3809A | T1270N | Missense | (69) |
| 3 | Renal | Clear cell carcinoma | T4900C | S1634P | Missense | (69) |
| 4 | Gastric | Adenocarcinoma | 1786delG | V696fs*53 | T2215FS*31 | (69) |
| 4 | Melanoma | Metastatic | C1402T | L468L | Silent | (69) |
| 4 | Melanoma | C2974T | P992S | Missense | (69) | |
| 4 | Melanoma | C3154T | P1052S | Missense | (69) | |
| 4 | Ovarian | Mucinous carcinoma | C1302G | D434E | Missense | (69) |
WNK1 was identified in a degenerate PCR screen of malignant prostate tissues to assess the contribution of protein kinases to tumor development (148). A striking reduction in WNK1 expression was observed in F-11 tumor cells in which enzyme ganglioside 3-synthase was suppressed (287). Ganglioside 3 has previously been associated with brain tumors (16, 71, 72), suggesting that WNK1 may play a functional role in malignancy. As discussed in [section III.D.2], there is evidence of cross-talk between the SMAD and WNK signaling pathways (116), and this may be relevant in tumorigenesis (88). Finally, analysis of the human genome indicates that 3308 bp of the WNK1 gene run antisense to the spliced gene RAD52, raising the possibility of regulated alternate expression. RAD52 plays a key role in DNA double strand break repair (242) and along with WNK1, maps to chromosome locus 12p12.2-p13, a frequent site for allelic losses in breast and ovarian cancer (76, 212, 220). The tumor suppressors BRCA1 and BRCA2 regulate a RAD51-RAD52 complex, and mutations in BRCA1 and BRCA2 lead to dysregulation of this complex, and are well-known to play a role in the development of breast cancer (37).
An epigenetic approach, focusing on analysis of a recurrent site of aberrant methylation identified by restriction landmark genome scanning in astrocytomas and oligodendrogliomas, revealed that this site corresponds to a CpG island encompassing the putative transcription start site of WNK2 (82). Further analysis determined that in the majority of gliomas studied (31 total), the CpG island displayed increased methylation; in a few gliomas WNK2 was deleted. Compared with normal brain, WNK2 mRNA expression was reduced in the gliomas, and expression could be increased in vitro by a methylation inhibitor. Combined deletion of the chromosome arms 1p and 19q is frequently observed (in ~50%) in oligodendroglial tumors (151, 216, 250), and provides a strong predictor of positive response to chemotherapy and patient survival time (54, 138). Epigenetic silencing of WNK2 was found to be significantly associated with these deletions (82). Finally, expression of exogenous WNK2 in glioma cells lacking WNK2 inhibited colony-formation. A study of meningiomas, the most common form of primary tumor in the central nervous system in adults (161), resulted in similar findings, with altered WNK2 methylation detected in 83% of grade II and 71% of grade II meningiomas (94). Taken together, these data suggest that WNK2 acts as a tumor suppressor, and reduction of WNK2 expression by CpG methylation may play a role in tumorigenesis.
WNK2 was also cloned in a screen of a pancreatic cell line to identify activators of cytotoxic T lymphocytes (87). A WNK2 epitope could activate peripheral mononuclear blood cells from a patient with pancreatic cancer, and induce a cytotoxic response towards tumor cells.
Arsenic trioxide is extremely effective in the treatment of acute promyelocytic leukemia (205, 229), but the mechanisms underlying its effects are unclear. Exposure of a promyelocytic leukemia cell line to arsenic trioxide led to an inhibiton of cell proliferation, correlated with a 4-fold increase in WNK2 mRNA expression levels (145). These data provide further evidence for possible anti-proliferative effects of WNK2 that may be relevant in cancer.
F. Osteoporosis
Low bone mass density resulting from bone resorption exceeding bone formation is the primary characteristic of osteoporosis. The immune system plays an important role in mediating these processes (21, 119, 256). Microarray comparison of mRNA from B lymphocytes of postmenopausal women with either low or high bone mass density identified WNK1 as being downregulated in the low bone mass density group, a finding confirmed by real-time PCR (262). This study proposed that dysregulation of an estrogen receptor-MAP3K signaling network, of which WNK1 is a part, may play a role in the development of osteoporosis.
VI. WNKs as drug targets
Analysis of the physiological and pathophysiological roles of the WNK kinases is still in the embryonic stage, but an obvious therapeutic target for WNK-specific inhibitors (more specifically WNK1 and WNK3) is hypertension. At least 50% of hypertensive patients have a salt-sensitive component, and in approximately 30%, hypertension is predominantly due to abnormalities in renal sodium handling (255). Currently, only about 35% of patients undergoing treatment for hypertension have their blood pressure well-managed (34), suggesting the need for novel drugs. Cancer is also another disease whose treatment might benefit from inhibitors or stimulators of WNK kinases.
While a better understanding of WNK action in vivo is required, the development of specific inhibitors of the WNKs is already underway. Yagi and colleagues recently described an assay to screen WNK1 inhibitors and analyzed some of their biochemical properties (273). The kinetic mechanism for WNK1 interaction with its substrates ATP and a peptide (in this case OXSR1) was determined to be a random bi-bi sequential mechanism, and their noncompetitive inhibition patterns suggest they bind randomly to WNK1 to form a ternary complex (273). Therefore it should be possible to design inhibitors of both ATP binding and peptide-substrate binding. Screening of a panel of 86 commercially available kinase inhibitors revealed that only inhibitors of the Src kinases and EGFR, compete for ATP binding (273). Structural analysis suggested that specific WNK1 inhibitors could be designed by filling the space around Cysteine 250. However, given the high degree of homology between the kinase domains of members of the WNK family, it will be more challenging to design specific inhibitors that distinguish between WNKs.
As discussed in **[section III.D.1.)** WNK1 is able to bind and phosphorylate synaptotagmin 2 but WNK4 phosphorylates synaptotagmin 2 much less effectively (117, 269). Min and colleagues identified two residues (V318 and A448) in the substrate binding groove that might be involved in substrate recognition (143). Mutation of these residues prevented interactions between WNK1 and synaptotagmin 2, and reduced phosphorylation by 40–50%. In WNK4 these two residues are replaced by a glutamate and a lysine; in WNKs 2 and 3, the valine is conserved, but the alanine is not, raising the possibility that there may be substrate overlap between WNKs 1–3. Structural analysis of the kinase domains of all the WNK kinases and mutatgenesis studies will assist with development of isoform-specific inhibitors.
VII Conclusions and Perspectives
In just ten years, the WNK kinases have become a focus of intense interest for their pleiotropic actions and disease associations. A great deal has been learned about their targets, substrates, and mechanisms of action. Interest was initially focused on diseases of electrolyte transport by the kidney, but it is now recognized that this kinase family has much broader effects. Even simple physiological questions, however, such as the nature of effects of WNKs on renal Na and K potassium transport, remain remarkably controversial. Yet, it is hoped that continuing efforts during the next decades will illuminate that nature of the WNK kinase pathway and open the door to new drugs and treatments, just as the discovery of the renin/angiotensin system did a century ago.
Acknowledgments
This work was supported by grants from the NIH (K01 DK076617 to JM and DK51496 to DE). DE is also supported by a Merit Review from the Department of Veterans Affairs and a Grant-In-Aid from the Amerian Heart Association. Prior support from the American Society of Nephrology is also acknowledged.
References
- 1.Abdallah JG, Schrier RW, Edelstein C, Jennings SD, Wyse B, Ellison DH. Loop diuretic infusion increases thiazide-sensitive Na(+)/Cl(−)-cotransporter abundance: role of aldosterone. J Am Soc Nephrol. 2001;12:1335–1341. doi: 10.1681/ASN.V1271335. [DOI] [PubMed] [Google Scholar]
- 2.Abidi FE, Miano MG, Murray JC, Schwartz CE. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clinical genetics. 2007;72:19–22. doi: 10.1111/j.1399-0004.2007.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achard JM, Warnock DG, Disse-Nicodeme S, Fiquet-Kempf B, Corvol P, Fournier A, Jeunemaitre X. Familial hyperkalemic hypertension: phenotypic analysis in a large family with the WNK1 deletion mutation. AmJMed. 2003;114:495–498. doi: 10.1016/s0002-9343(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 4.Adragna NC, Chen Y, Delpire E, Lauf PK, Morris M. Hypertension in K-Cl cotransporter-3 knockout mice. Adv Exp Med Biol. 2004;559:379–385. doi: 10.1007/0-387-23752-6_35. [DOI] [PubMed] [Google Scholar]
- 5.Adragna NC, Di Fulvio M, Lauf PK. Regulation of K-Cl cotransport: from function to genes. J Membr Biol. 2004;201:109–137. doi: 10.1007/s00232-004-0695-6. [DOI] [PubMed] [Google Scholar]
- 6.Ahlstrom R, Yu AS. Characterization of the kinase activity of a WNK4 protein complex. Am J Physiol Renal Physiol. 2009;297:F685–F692. doi: 10.1152/ajprenal.00358.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amlal H, Paillard M, Bichara M. Cl--dependent NH4+ transport mechanisms in medullary thick ascending limb cells. Am J Physiol Cell Physiol. 1994;267:C1607–C1615. doi: 10.1152/ajpcell.1994.267.6.C1607. [DOI] [PubMed] [Google Scholar]
- 8.Anselmo AN, Earnest S, Chen W, Juang YC, Kim SC, Zhao Y, Cobb MH. WNK1 and OSR1 regulate the Na+, K+, 2Cl- cotransporter in HeLa cells. Proc Natl Acad Sci U S A. 2006;103:10883–10888. doi: 10.1073/pnas.0604607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachmann S, Velázquez H, Obermüller N, Reilly RF, Moser D, Ellison DH. Expression of the thiazide-sensitive Na-Cl cotransporter by rabbit distal convoluted tubule cells. J Clin Invest. 1995;96:2510–2514. doi: 10.1172/JCI118311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baima J, Nicolaou M, Schwartz F, DeStefano AL, Manolis A, Gavras I, Laffer C, Elijovich F, Farrer L, Baldwin CT, Gavras H. Evidence for linkage between essential hypertension and a putative locus on human chromosome 17. Hypertension. 1999;34:4–7. doi: 10.1161/01.hyp.34.1.4. [DOI] [PubMed] [Google Scholar]
- 11.Bandtlow C, Dechant G. From cell death to neuronal regeneration, effects of the p75 neurotrophin receptor depend on interactions with partner subunits. Sci STKE. 2004;2004:pe24. doi: 10.1126/stke.2352004pe24. [DOI] [PubMed] [Google Scholar]
- 12.Bartsch J, Truss M, Bode J, Beato M. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc Natl Acad Sci U S A. 1996;93:10741–10746. doi: 10.1073/pnas.93.20.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belmonte MK, Cook EH, Jr, Anderson GM, Rubenstein JL, Greenough WT, Beckel-Mitchener A, Courchesne E, Boulanger LM, Powell SB, Levitt PR, Perry EK, Jiang YH, DeLorey TM, Tierney E. Autism as a disorder of neural information processing: directions for research and targets for therapy. Molecular psychiatry. 2004;9:646–663. doi: 10.1038/sj.mp.4001499. [DOI] [PubMed] [Google Scholar]
- 14.Benjafield AV, Katyk K, Morris BJ. Association of EDNRA, but not WNK4 or FKBP1B, polymorphisms with essential hypertension. Clinical genetics. 2003;64:433–438. doi: 10.1034/j.1399-0004.2003.00148.x. [DOI] [PubMed] [Google Scholar]
- 15.Bevan S, Andersson DA. TRP channel antagonists for pain--opportunities beyond TRPV1. Curr Opin Investig Drugs. 2009;10:655–663. [PubMed] [Google Scholar]
- 16.Birkle S, Zeng G, Gao L, Yu RK, Aubry J. Role of tumor-associated gangliosides in cancer progression. Biochimie. 2003;85:455–463. doi: 10.1016/s0300-9084(03)00006-3. [DOI] [PubMed] [Google Scholar]
- 17.Boockvar JA, Kapitonov D, Kapoor G, Schouten J, Counelis GJ, Bogler O, Snyder EY, McIntosh TK, O’Rourke DM. Constitutive EGFR signaling confers a motile phenotype to neural stem cells. Molecular and cellular neurosciences. 2003;24:1116–1130. doi: 10.1016/j.mcn.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Bostanjoglo M, Reeves WB, Reilly RF, Velazquez H, Robertson N, Litwack G, Morsing P, Dorup J, Bachmann S, Ellison DH. 11Beta-hydroxysteroid dehydrogenase, mineralocorticoid receptor, and thiazide-sensitive Na-Cl cotransporter expression by distal tubules. J Am Soc Nephrol. 1998;9:1347–1358. doi: 10.1681/ASN.V981347. [DOI] [PubMed] [Google Scholar]
- 19.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 20.Brautbar N, Levi J, Rosler A, Leitesdorf E, Djaldeti M, Epstein M, Kleeman CR. Familial hyperkalemia, hypertension, and hyporeninemia with normal aldosterone levels. A tubular defect in potassium handling. Arch Intern Med. 1978;138:607–610. [PubMed] [Google Scholar]
- 21.Breuil V, Ticchioni M, Testa J, Roux CH, Ferrari P, Breittmayer JP, Albert-Sabonnadiere C, Durant J, De Perreti F, Bernard A, Euller-Ziegler L, Carle GF. Immune changes in post-menopausal osteoporosis: the Immunos study. Osteoporos Int. 21:805–814. doi: 10.1007/s00198-009-1018-7. [DOI] [PubMed] [Google Scholar]
- 22.Cahill MA, Ernst WH, Janknecht R, Nordheim A. Regulatory squelching. FEBS Lett. 1994;344:105–108. doi: 10.1016/0014-5793(94)00320-3. [DOI] [PubMed] [Google Scholar]
- 23.Cai H, Cebotaru V, Wang YH, Zhang XM, Cebotaru L, Guggino SE, Guggino WB. WNK4 kinase regulates surface expression of the human sodium chloride cotransporter in mammalian cells. Kidney Int. 2006;69:2162–2170. doi: 10.1038/sj.ki.5000333. [DOI] [PubMed] [Google Scholar]
- 24.Calo LA, Montisci R, Scognamiglio R, Davis PA, Pagnin E, Schiavo S, Mormino P, Semplicini A, Palatini P, D’Angelo A, Pessina AC. High angiotensin II state without cardiac remodeling (Bartter’s and Gitelman’s syndromes): are angiotensin II type 2 receptors involved? J Endocrinol Invest. 2009;32:832–836. doi: 10.1007/BF03345754. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Chen Y, Xu BE, Juang YC, Stippec S, Zhao Y, Cobb MH. Regulation of a third conserved phosphorylation site in SGK1. J Biol Chem. 2009;284:3453–3460. doi: 10.1074/jbc.M807502200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, Kadowaki T, Hay N. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes & development. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiga M, Rai T, Yang SS, Ohta A, Takizawa T, Sasaki S, Uchida S. Dietary salt regulates the phosphorylation of OSR1/SPAK kinases and the sodium chloride cotransporter through aldosterone. Kidney Int. 2008;74:1403–1409. doi: 10.1038/ki.2008.451. [DOI] [PubMed] [Google Scholar]
- 28.Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 29.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 30.Choate KA, Kahle KT, Wilson FH, Nelson-Williams C, Lifton RP. WNK1, a kinase mutated in inherited hypertension with hyperkalemia, localizes to diverse Cl- -transporting epithelia. Proc Natl Acad Sci U S A. 2003;100:663–668. doi: 10.1073/pnas.242728499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coles LC, Shaw PE. PAK1 primes MEK1 for phosphorylation by Raf-1 kinase during cross-cascade activation of the ERK pathway. Oncogene. 2002;21:2236–2244. doi: 10.1038/sj.onc.1205302. [DOI] [PubMed] [Google Scholar]
- 32.Cope G, Murthy M, Golbang AP, Hamad A, Liu CH, Cuthbert AW, O’Shaughnessy KM. WNK1 affects surface expression of the ROMK potassium channel independent of WNK4. J Am Soc Nephrol. 2006;17:1867–1874. doi: 10.1681/ASN.2005111224. [DOI] [PubMed] [Google Scholar]
- 33.Cortright DN, Szallasi A. TRP channels and pain. Current pharmaceutical design. 2009;15:1736–1749. doi: 10.2174/138161209788186308. [DOI] [PubMed] [Google Scholar]
- 34.Cutler JA, Sorlie PD, Wolz M, Thom T, Fields LE, Roccella EJ. Trends in hypertension prevalence, awareness, treatment, and control rates in United States adults between 1988–1994 and 1999–2004. Hypertension. 2008;52:818–827. doi: 10.1161/HYPERTENSIONAHA.108.113357. [DOI] [PubMed] [Google Scholar]
- 35.DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 36.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, Parker A, O’Meara S, Avis T, Barthorpe S, Brackenbury L, Buck G, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh BT, Yuen ST, Lakhani SR, Easton DF, Weber BL, Goldstraw P, Nicholson AG, Wooster R, Stratton MR, Futreal PA. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 37.De Greve J, Sermijn E, De Brakeleer S, Ren Z, Teugels E. Hereditary breast cancer: from bench to bedside. Current opinion in oncology. 2008;20:605–613. doi: 10.1097/CCO.0b013e3283139173. [DOI] [PubMed] [Google Scholar]
- 38.de Los Heros P, Kahle KT, Rinehart J, Bobadilla NA, Vazquez N, San Cristobal P, Mount DB, Lifton RP, Hebert SC, Gamba G. WNK3 bypasses the tonicity requirement for K-Cl cotransporter activation via a phosphatase-dependent pathway. Proc Natl Acad Sci U S A. 2006;103:1976–1981. doi: 10.1073/pnas.0510947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaloy C, Elvira-Matelot E, Clemessy M, Zhou XO, Imbert-Teboul M, Houot AM, Jeunemaitre X, Hadchouel J. Deletion of WNK1 first intron results in misregulation of both isoforms in renal and extrarenal tissues. Hypertension. 2008;52:1149–1154. doi: 10.1161/HYPERTENSIONAHA.108.120899. [DOI] [PubMed] [Google Scholar]
- 40.Delaloy C, Hadchouel J, Imbert-Teboul M, Clemessy M, Houot AM, Jeunemaitre X. Cardiovascular expression of the mouse WNK1 gene during development and adulthood revealed by a BAC reporter assay. Am J Pathol. 2006;169:105–118. doi: 10.2353/ajpath.2006.051290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaloy C, Lu J, Houot AM, Disse-Nicodeme S, Gasc JM, Corvol P, Jeunemaitre X. Multiple promoters in the WNK1 gene: one controls expression of a kidney-specific kinase-defective isoform. Mol Cell Biol. 2003;23:9208–9221. doi: 10.1128/MCB.23.24.9208-9221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delany NS, Hurle M, Facer P, Alnadaf T, Plumpton C, Kinghorn I, See CG, Costigan M, Anand P, Woolf CJ, Crowther D, Sanseau P, Tate SN. Identification and characterization of a novel human vanilloid receptor-like protein, VRL-2. Physiological genomics. 2001;4:165–174. doi: 10.1152/physiolgenomics.2001.4.3.165. [DOI] [PubMed] [Google Scholar]
- 43.Delpire E, Gagnon KB. SPAK and OSR1: STE20 kinases involved in the regulation of ion homoeostasis and volume control in mammalian cells. Biochem J. 2008;409:321–331. doi: 10.1042/BJ20071324. [DOI] [PubMed] [Google Scholar]
- 44.Denton J, Nehrke K, Yin X, Morrison R, Strange K. GCK-3, a newly identified Ste20 kinase, binds to and regulates the activity of a cell cycle-dependent ClC anion channel. J Gen Physiol. 2005;125:113–125. doi: 10.1085/jgp.200409215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorwart MR, Shcheynikov N, Wang Y, Stippec S, Muallem S. SLC26A9 is a Cl(−) channel regulated by the WNK kinases. J Physiol. 2007;584:333–345. doi: 10.1113/jphysiol.2007.135855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowd BF, Forbush B. PASK (proline-alanine-rich STE20-related kinase), a regulatory kinase of the Na-K-Cl cotransporter (NKCC1) J Biol Chem. 2003;278:27347–27353. doi: 10.1074/jbc.M301899200. [DOI] [PubMed] [Google Scholar]
- 47.Downward J. Mechanisms and consequences of activation of protein kinase B/Akt. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 48.Eblen ST, Slack JK, Weber MJ, Catling AD. Rac-PAK signaling stimulates extracellular signal-regulated kinase (ERK) activation by regulating formation of MEK1-ERK complexes. Mol Cell Biol. 2002;22:6023–6033. doi: 10.1128/MCB.22.17.6023-6033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellison DH, Velazquez H, Wright FS. Stimulation of distal potassium secretion by low lumen chloride in the presence of barium. Am J Physiol. 1985;248:F638–649. doi: 10.1152/ajprenal.1985.248.5.F638. [DOI] [PubMed] [Google Scholar]
- 50.Ellison DH, Velázquez H, Wright FS. Adaptation of the distal convoluted tubule of the rat: Structural and functional effects of dietary salt intake and chronic diuretic infusion. J Clin Invest. 1989;83:113–126. doi: 10.1172/JCI113847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erlich PM, Cui J, Chazaro I, Farrer LA, Baldwin CT, Gavras H, DeStefano AL. Genetic Variants of WNK4 in Whites and African Americans With Hypertension. Hypertension. 2003 Apr 28; doi: 10.1161/01.HYP.0000070025.30572.91. [DOI] [PubMed] [Google Scholar]
- 52.Everett LA, Morsli H, Wu DK, Green ED. Expression pattern of the mouse ortholog of the Pendred’s syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc Natl Acad Sci U S A. 1999;96:9727–9732. doi: 10.1073/pnas.96.17.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evers EE, Zondag GC, Malliri A, Price LS, ten Klooster JP, van der Kammen RA, Collard JG. Rho family proteins in cell adhesion and cell migration. Eur J Cancer. 2000;36:1269–1274. doi: 10.1016/s0959-8049(00)00091-5. [DOI] [PubMed] [Google Scholar]
- 54.Fallon KB, Palmer CA, Roth KA, Nabors LB, Wang W, Carpenter M, Banerjee R, Forsyth P, Rich K, Perry A. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J Neuropathol Exp Neurol. 2004;63:314–322. doi: 10.1093/jnen/63.4.314. [DOI] [PubMed] [Google Scholar]
- 55.Fang L, Garuti R, Kim BY, Wade JB, Welling PA. The ARH adaptor protein regulates endocytosis of the ROMK potassium secretory channel in mouse kidney. J Clin Invest. 2009;119:3278–3289. doi: 10.1172/JCI37950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farfel Z, Iaina A, Rosenthal T, Waks U, Shibolet S, Gafni J. Familial hyperpotassemia and hypertension accompanied by normal plasma aldosterone levels: possible hereditary cell membrane defect. Arch Intern Med. 1978;138:1828–1832. [PubMed] [Google Scholar]
- 57.Fu Y, Subramanya A, Rozansky D, Cohen DM. WNK kinases influence TRPV4 channel function and localization. Am J Physiol Renal Physiol. 2006;290:F1305–1314. doi: 10.1152/ajprenal.00391.2005. [DOI] [PubMed] [Google Scholar]
- 58.Gagnon KB, England R, Delpire E. Volume sensitivity of cation-Cl-cotransporters is modulated by the interaction of two kinases: Ste20-related proline-alanine-rich kinase and WNK4. Am J Physiol Cell Physiol. 2006;290:C134–142. doi: 10.1152/ajpcell.00037.2005. [DOI] [PubMed] [Google Scholar]
- 59.Gagnon KB, England R, Diehl L, Delpire E. Apoptosis-associated tyrosine kinase scaffolding of protein phosphatase 1 and SPAK reveals a novel pathway for Na-K-2C1 cotransporter regulation. Am J Physiol Cell Physiol. 2007;292:C1809–1815. doi: 10.1152/ajpcell.00580.2006. [DOI] [PubMed] [Google Scholar]
- 60.Gamba G. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev. 2005;85:423–493. doi: 10.1152/physrev.00011.2004. [DOI] [PubMed] [Google Scholar]
- 61.Garzon-Muvdi T, Pacheco-Alvarez D, Gagnon KB, Vazquez N, Ponce-Coria J, Moreno E, Delpire E, Gamba G. WNK4 kinase is a negative regulator of K+-Cl- cotransporters. Am J Physiol Renal Physiol. 2007;292:F1197–1207. doi: 10.1152/ajprenal.00335.2006. [DOI] [PubMed] [Google Scholar]
- 62.Gauthier BR, Wollheim CB. Synaptotagmins bind calcium to release insulin. American journal of physiology. 2008;295:E1279–1286. doi: 10.1152/ajpendo.90568.2008. [DOI] [PubMed] [Google Scholar]
- 63.Geppert M, Goda Y, Hammer RE, Li C, Rosahl TW, Stevens CF, Sudhof TC. Synaptotagmin I: a major Ca2+ sensor for transmitter release at a central synapse. Cell. 1994;79:717–727. doi: 10.1016/0092-8674(94)90556-8. [DOI] [PubMed] [Google Scholar]
- 64.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 65.Glover M, Zuber AM, O’Shaughnessy KM. Renal and brain isoforms of WNK3 have opposite effects on NCCT expression. J Am Soc Nephrol. 2009;20:1314–1322. doi: 10.1681/ASN.2008050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Golbang AP, Cope G, Hamad A, Murthy M, Liu CH, Cuthbert AW, O’Shaughnessy KM. Regulation of the Expression of the Na/Cl cotransporter (NCCT) by WNK4 and WNK1: evidence that accelerated dynamin-dependent endocytosis is not involved. Am J Physiol Renal Physiol. 2006;291:F1369–1376. doi: 10.1152/ajprenal.00468.2005. [DOI] [PubMed] [Google Scholar]
- 67.Golbang AP, Murthy M, Hamad A, Liu CH, Cope G, Van’t Hoff W, Cuthbert A, O’Shaughnessy KM. A new kindred with pseudohypoaldosteronism type II and a novel mutation (564D>H) in the acidic motif of the WNK4 gene. Hypertension. 2005;46:295–300. doi: 10.1161/01.HYP.0000174326.96918.d6. [DOI] [PubMed] [Google Scholar]
- 68.Gordon RD, Hodsman GP. The syndrome of hypertension and hyperkalaemia without renal failure: long term correction by thiazide diuretic. Scott Med J. 1986;31:43–44. doi: 10.1177/003693308603100114. [DOI] [PubMed] [Google Scholar]
- 69.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haila S, Hastbacka J, Bohling T, Karjalainen-Lindsberg ML, Kere J, Saarialho-Kere U. SLC26A2 (diastrophic dysplasia sulfate transporter) is expressed in developing and mature cartilage but also in other tissues and cell types. J Histochem Cytochem. 2001;49:973–982. doi: 10.1177/002215540104900805. [DOI] [PubMed] [Google Scholar]
- 71.Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci U S A. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hakomori S. Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer Res. 1996;56:5309–5318. [PubMed] [Google Scholar]
- 73.Halperin ML, Kamel KS. Dynamic interactions between integrative physiology and molecular medicine: the key to understand the mechanism of action of aldosterone in the kidney. Can J Physiol Pharmacol. 2000;78:587–594. [PubMed] [Google Scholar]
- 74.Harada Y, Sanada K, Fukada Y. Circadian activation of bullfrog retinal mitogen-activated protein kinase associates with oscillator function. J Biol Chem. 2000;275:37078–37085. doi: 10.1074/jbc.M004706200. [DOI] [PubMed] [Google Scholar]
- 75.Hastbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, Hamilton BA, Kusumi K, Trivedi B, Weaver A, et al. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 76.Hatta Y, Takeuchi S, Yokota J, Koeffler HP. Ovarian cancer has frequent loss of heterozygosity at chromosome 12p12.3–13.1 (region of TEL and Kip1 loci) and chromosome 12q23-ter: evidence for two new tumour-suppressor genes. Br J Cancer. 1997;75:1256–1262. doi: 10.1038/bjc.1997.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He G, Wang HR, Huang SK, Huang CL. Intersectin links WNK kinases to endocytosis of ROMK1. J Clin Invest. 2007;117:1078–1087. doi: 10.1172/JCI30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hisamoto N, Moriguchi T, Urushiyama S, Mitani S, Shibuya H, Matsumoto K. Caenorhabditis elegans WNK-STE20 pathway regulates tube formation by modulating ClC channel activity. EMBO reports. 2008;9:70–75. doi: 10.1038/sj.embor.7401128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoenderop JG, van Leeuwen JP, van der Eerden BC, Kersten FF, van der Kemp AW, Merillat AM, Waarsing JH, Rossier BC, Vallon V, Hummler E, Bindels RJ. Renal Ca2+ wasting, hyperabsorption, and reduced bone thickness in mice lacking TRPV5. J Clin Invest. 2003;112:1906–1914. doi: 10.1172/JCI19826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holden S, Cox J, Raymond FL. Cloning, genomic organization, alternative splicing and expression analysis of the human gene WNK3 (PRKWNK3) Gene. 2004;335:109–119. doi: 10.1016/j.gene.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 81.Hong-Hermesdorf A, Brux A, Gruber A, Gruber G, Schumacher K. A WNK kinase binds and phosphorylates V-ATPase subunit C. FEBS Lett. 2006;580:932–939. doi: 10.1016/j.febslet.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 82.Hong C, Moorefield KS, Jun P, Aldape KD, Kharbanda S, Phillips HS, Costello JF. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc Natl Acad Sci U S A. 2007;104:10974–10979. doi: 10.1073/pnas.0700683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoover LL, Kubalak SW. Holding their own: the noncanonical roles of Smad proteins. Science signaling. 2008;1:pe48. doi: 10.1126/scisignal.146pe48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Howard HC, Mount DB, Rochefort D, Byun N, Dupre N, Lu J, Fan X, Song L, Riviere JB, Prevost C, Horst J, Simonati A, Lemcke B, Welch R, England R, Zhan FQ, Mercado A, Siesser WB, George AL, Jr, McDonald MP, Bouchard JP, Mathieu J, Delpire E, Rouleau GA. The K-Cl cotransporter KCC3 is mutant in a severe peripheral neuropathy associated with agenesis of the corpus callosum. Nat Genet. 2002;32:384–392. doi: 10.1038/ng1002. [DOI] [PubMed] [Google Scholar]
- 85.Hu SH, Latham CF, Gee CL, James DE, Martin JL. Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc Natl Acad Sci U S A. 2007;104:8773–8778. doi: 10.1073/pnas.0701124104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Inoue R, Jian Z, Kawarabayashi Y. Mechanosensitive TRP channels in cardiovascular pathophysiology. Pharmacol Ther. 2009;123:371–385. doi: 10.1016/j.pharmthera.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 87.Ito M, Shichijo S, Tsuda N, Ochi M, Harashima N, Saito N, Itoh K. Molecular basis of T cell-mediated recognition of pancreatic cancer cells. Cancer Res. 2001;61:2038–2046. [PubMed] [Google Scholar]
- 88.Javelaud D, Mauviel A. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-beta: implications for carcinogenesis. Oncogene. 2005;24:5742–5750. doi: 10.1038/sj.onc.1208928. [DOI] [PubMed] [Google Scholar]
- 89.Jefferies KC, Cipriano DJ, Forgac M. Function, structure and regulation of the vacuolar (H+)-ATPases. Arch Biochem Biophys. 2008;476:33–42. doi: 10.1016/j.abb.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang Y, Cong P, Williams SR, Zhang W, Na T, Ma HP, Peng JB. WNK4 regulates the secretory pathway via which TRPV5 is targeted to the plasma membrane. Biochem Biophys Res Commun. 2008;375:225–229. doi: 10.1016/j.bbrc.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang Y, Ferguson WB, Peng JB. WNK4 enhances TRPV5-mediated calcium transport: potential role in hypercalciuria of familial hyperkalemic hypertension caused by gene mutation of WNK4. Am J Physiol Renal Physiol. 2007;292:F545–554. doi: 10.1152/ajprenal.00187.2006. [DOI] [PubMed] [Google Scholar]
- 92.Jiang ZY, Zhou QL, Holik J, Patel S, Leszyk J, Coleman K, Chouinard M, Czech MP. Identification of WNK1 as a substrate of Akt/protein kinase B and a negative regulator of insulin-stimulated mitogenesis in 3T3-L1 cells. J Biol Chem. 2005;280:21622–21628. doi: 10.1074/jbc.M414464200. [DOI] [PubMed] [Google Scholar]
- 93.Julier C, Delepine M, Keavney B, Terwilliger J, Davis S, Weeks DE, Bui T, Jeunemaitre X, Velho G, Froguel P, Ratcliffe P, Corvol P, Soubrier F, Lathrop GM. Genetic susceptibility for human familial essential hypertension in a region of homology with blood pressure linkage on rat chromosome 10. Hum Mol Genet. 1997;6:2077–2085. doi: 10.1093/hmg/6.12.2077. [DOI] [PubMed] [Google Scholar]
- 94.Jun P, Hong C, Lal A, Wong JM, McDermott MW, Bollen AW, Plass C, Held WA, Smiraglia DJ, Costello JF. Epigenetic silencing of the kinase tumor suppressor WNK2 is tumor-type and tumor-grade specific. Neuro Oncol. 2009;11:414–422. doi: 10.1215/15228517-2008-096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kahle KT, Gimenez I, Hassan H, Wilson FH, Wong RD, Forbush B, Aronson PS, Lifton RP. WNK4 regulates apical and basolateral Cl- flux in extrarenal epithelia. Proc Natl Acad Sci U S A. 2004;101:2064–2069. doi: 10.1073/pnas.0308434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kahle KT, Macgregor GG, Wilson FH, Van Hoek AN, Brown D, Ardito T, Kashgarian M, Giebisch G, Hebert SC, Boulpaep EL, Lifton RP. Paracellular Cl- permeability is regulated by WNK4 kinase: insight into normal physiology and hypertension. Proc Natl Acad Sci U S A. 2004;101:14877–14882. doi: 10.1073/pnas.0406172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kahle KT, Rinehart J, de Los Heros P, Louvi A, Meade P, Vazquez N, Hebert SC, Gamba G, Gimenez I, Lifton RP. WNK3 modulates transport of Cl- in and out of cells: implications for control of cell volume and neuronal excitability. Proc Natl Acad Sci U S A. 2005;102:16783–16788. doi: 10.1073/pnas.0508307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kahle KT, Wilson FH, Leng Q, Lalioti MD, O’Connell AD, Dong K, Rapson AK, MacGregor GG, Giebisch G, Hebert SC, Lifton RP. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet. 2003;35:372–376. doi: 10.1038/ng1271. [DOI] [PubMed] [Google Scholar]
- 99.Kaissling B, Bachmann S, Kriz W. Structural adaptation of the distal convoluted tubule to prolonged furosemide treatment. Am J Physiol. 1985;248:F374–F381. doi: 10.1152/ajprenal.1985.248.3.F374. [DOI] [PubMed] [Google Scholar]
- 100.Kamide K, Takiuchi S, Tanaka C, Miwa Y, Yoshii M, Horio T, Mannami T, Kokubo Y, Tomoike H, Kawano Y, Miyata T. Three novel missense mutations of WNK4, a kinase mutated in inherited hypertension, in Japanese hypertensives: implication of clinical phenotypes. Am J Hypertens. 2004;17:446–449. doi: 10.1016/j.amjhyper.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 101.Kaneko T, Li L, Li SS. The SH3 domain--a family of versatile peptide- and protein-recognition module. Front Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 102.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J. 2000;14:231–241. [PubMed] [Google Scholar]
- 103.Ke B, Oh E, Thurmond DC. Doc2beta is a novel Munc18c-interacting partner and positive effector of syntaxin 4-mediated exocytosis. J Biol Chem. 2007;282:21786–21797. doi: 10.1074/jbc.M701661200. [DOI] [PubMed] [Google Scholar]
- 104.Kim SH, Kim KX, Raveendran NN, Wu T, Pondugula SR, Marcus DC. Regulation of ENaC-mediated sodium transport by glucocorticoids in Reissner’s membrane epithelium. Am J Physiol Cell Physiol. 2009;296:C544–557. doi: 10.1152/ajpcell.00338.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Knauf F, Yang CL, Thomson RB, Mentone SA, Giebisch G, Aronson PS. Identification of a chloride-formate exchanger expressed on the brush border membrane of renal proximal tubule cells. Proc Natl Acad Sci U S A. 2001;98:9425–9430. doi: 10.1073/pnas.141241098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Knoers NV, Levtchenko EN. Gitelman syndrome. Orphanet J Rare Dis. 2008;3:22. doi: 10.1186/1750-1172-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ko GY, Ko ML, Dryer SE. Circadian regulation of cGMP-gated cationic channels of chick retinal cones. Erk MAP Kinase and Ca2+/calmodulin-dependent protein kinase II. Neuron. 2001;29:255–266. doi: 10.1016/s0896-6273(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 108.Kobayashi T, Deak M, Morrice N, Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J. 1999;344(Pt 1):189–197. [PMC free article] [PubMed] [Google Scholar]
- 109.Kokubo Y, Kamide K, Inamoto N, Tanaka C, Banno M, Takiuchi S, Kawano Y, Tomoike H, Miyata T. Identification of 108 SNPs in TSC, WNK1, and WNK4 and their association with hypertension in a Japanese general population. Journal of human genetics. 2004;49:507–515. doi: 10.1007/s10038-004-0181-0. [DOI] [PubMed] [Google Scholar]
- 110.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 111.Lafreniere RG, MacDonald ML, Dube MP, MacFarlane J, O’Driscoll M, Brais B, Meilleur S, Brinkman RR, Dadivas O, Pape T, Platon C, Radomski C, Risler J, Thompson J, Guerra-Escobio AM, Davar G, Breakefield XO, Pimstone SN, Green R, Pryse-Phillips W, Goldberg YP, Younghusband HB, Hayden MR, Sherrington R, Rouleau GA, Samuels ME. Identification of a novel gene (HSN2) causing hereditary sensory and autonomic neuropathy type II through the Study of Canadian Genetic Isolates. Am J Hum Genet. 2004;74:1064–1073. doi: 10.1086/420795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lalioti MD, Zhang J, Volkman HM, Kahle KT, Hoffmann KE, Toka HR, Nelson-Williams C, Ellison DH, Flavell R, Booth CJ, Lu Y, Geller DS, Lifton RP. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat Genet. 2006;38:1124–1132. doi: 10.1038/ng1877. [DOI] [PubMed] [Google Scholar]
- 113.Lambert JR, Nordeen SK. Steroid-selective initiation of chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter is controlled by the site of promoter integration. J Biol Chem. 1998;273:32708–32714. doi: 10.1074/jbc.273.49.32708. [DOI] [PubMed] [Google Scholar]
- 114.Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, Lespinasse J, Van Esch H, Lacombe D, Goizet C, Phan-Dinh Tuy F, van Bokhoven H, Fryns JP, Chelly J, Ropers HH, Moraine C, Hamel BC, Briault S. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. Journal of medical genetics. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lazrak A, Liu Z, Huang CL. Antagonistic regulation of ROMK by long and kidney-specific WNK1 isoforms. Proc Natl Acad Sci U S A. 2006;103:1615–1620. doi: 10.1073/pnas.0510609103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee BH, Chen W, Stippec S, Cobb MH. Biological cross-talk between WNK1 and the transforming growth factor beta-Smad signaling pathway. J Biol Chem. 2007;282:17985–17996. doi: 10.1074/jbc.M702664200. [DOI] [PubMed] [Google Scholar]
- 117.Lee BH, Min X, Heise CJ, Xu BE, Chen S, Shu H, Luby-Phelps K, Goldsmith EJ, Cobb MH. WNK1 phosphorylates synaptotagmin 2 and modulates its membrane binding. Mol Cell. 2004;15:741–751. doi: 10.1016/j.molcel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 118.Lee H, Caterina MJ. TRPV channels as thermosensory receptors in epithelial cells. Pflugers Arch. 2005;451:160–167. doi: 10.1007/s00424-005-1438-y. [DOI] [PubMed] [Google Scholar]
- 119.Leibbrandt A, Penninger JM. RANK/RANKL: regulators of immune responses and bone physiology. Ann N Y Acad Sci. 2008;1143:123–150. doi: 10.1196/annals.1443.016. [DOI] [PubMed] [Google Scholar]
- 120.Lenertz LY, Lee BH, Min X, Xu BE, Wedin K, Earnest S, Goldsmith EJ, Cobb MH. Properties of WNK1 and implications for other family members. J Biol Chem. 2005;280:26653–26658. doi: 10.1074/jbc.M502598200. [DOI] [PubMed] [Google Scholar]
- 121.Leng Q, Kahle KT, Rinehart J, MacGregor GG, Wilson FH, Canessa CM, Lifton RP, Hebert SC. WNK3, a kinase related to genes mutated in hereditary hypertension with hyperkalaemia, regulates the K+ channel ROMK1 (Kir1.1) J Physiol. 2006;571:275–286. doi: 10.1113/jphysiol.2005.102202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lewis MJ, Lewis EH, 3rd, Amos JA, Tsongalis GJ. Cystic fibrosis. Am J Clin Pathol. 2003;120 (Suppl):S3–13. doi: 10.1309/DUUVFR11W595V7FE. [DOI] [PubMed] [Google Scholar]
- 123.Li C, Li Y, Li Y, Liu H, Sun Z, Lu J, Zhao Y. Glucocorticoid repression of human with-no-lysine (K) kinase-4 gene expression is mediated by the negative response elements in the promoter. Journal of molecular endocrinology. 2008;40:3–12. doi: 10.1677/JME-07-0049. [DOI] [PubMed] [Google Scholar]
- 124.Li M, Zhao Y, Li Y, Li C, Chen F, Mao J, Zhang Y. Upregulation of human with-no-lysine kinase-4 gene expression by GATA-1 acetylation. The international journal of biochemistry & cell biology. 2009;41:872–878. doi: 10.1016/j.biocel.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 125.Liedtke W, Choe Y, Marti-Renom MA, Bell AM, Denis CS, Sali A, Hudspeth AJ, Friedman JM, Heller S. Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell. 2000;103:525–535. doi: 10.1016/s0092-8674(00)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liedtke W, Friedman JM. Abnormal osmotic regulation in trpv4−/− mice. Proc Natl Acad Sci U S A. 2003;100:13698–13703. doi: 10.1073/pnas.1735416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Z, Wang HR, Huang CL. Regulation of ROMK channel and K+ homeostasis by kidney-specific WNK1 kinase. J Biol Chem. 2009;284:12198–12206. doi: 10.1074/jbc.M806551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Loffing J, Loffing-Cueni D, Valderrabano V, Klausli L, Hebert SC, Rossier BC, Hoenderop JG, Bindels RJ, Kaissling B. Distribution of transcellular calcium and sodium transport pathways along mouse distal nephron. Am J Physiol Renal Physiol. 2001;281:F1021–1027. doi: 10.1152/ajprenal.0085.2001. [DOI] [PubMed] [Google Scholar]
- 129.Loggia ML, Bushnell MC, Tetreault M, Thiffault I, Bherer C, Mohammed NK, Kuchinad AA, Laferriere A, Dicaire MJ, Loisel L, Mogil JS, Brais B. Carriers of recessive WNK1/HSN2 mutations for hereditary sensory and autonomic neuropathy type 2 (HSAN2) are more sensitive to thermal stimuli. J Neurosci. 2009;29:2162–2166. doi: 10.1523/JNEUROSCI.4633-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lohi H, Kujala M, Makela S, Lehtonen E, Kestila M, Saarialho-Kere U, Markovich D, Kere J. Functional characterization of three novel tissue-specific anion exchangers SLC26A7, -A8, and -A9. J Biol Chem. 2002;277:14246–14254. doi: 10.1074/jbc.M111802200. [DOI] [PubMed] [Google Scholar]
- 131.Low PA. Autonomic neuropathies. Current opinion in neurology. 2002;15:605–609. doi: 10.1097/00019052-200210000-00011. [DOI] [PubMed] [Google Scholar]
- 132.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 133.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 134.Mayan H, Munter G, Shaharabany M, Mouallem M, Pauzner R, Holtzman EJ, Farfel Z. Hypercalciuria in familial hyperkalemia and hypertension accompanies hyperkalemia and precedes hypertension: description of a large family with the Q565E WNK4 mutation. J Clin Endocrinol Metab. 2004;89:4025–4030. doi: 10.1210/jc.2004-0037. [DOI] [PubMed] [Google Scholar]
- 135.Mayan H, Vered I, Mouallem M, Tzadok-Witkon M, Pauzner R, Farfel Z. Pseudohypoaldosteronism type II: marked sensitivity to thiazides, hypercalciuria, normomagnesemia, and low bone mineral density. J Clin Endocrinol Metab. 2002;87:3248–3254. doi: 10.1210/jcem.87.7.8449. [DOI] [PubMed] [Google Scholar]
- 136.McCormick JA, Bhalla V, Pao AC, Pearce D. SGK1: a rapid aldosterone-induced regulator of renal sodium reabsorption. Physiology (Bethesda) 2005;20:134–139. doi: 10.1152/physiol.00053.2004. [DOI] [PubMed] [Google Scholar]
- 137.McCormick JA, Feng Y, Dawson K, Behne MJ, Yu B, Wang J, Wyatt AW, Henke G, Grahammer F, Mauro TM, Lang F, Pearce D. Targeted disruption of the protein kinase SGK3/CISK impairs postnatal hair follicle development. Mol Biol Cell. 2004;15:4278–4288. doi: 10.1091/mbc.E04-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McDonald JM, See SJ, Tremont IW, Colman H, Gilbert MR, Groves M, Burger PC, Louis DN, Giannini C, Fuller G, Passe S, Blair H, Jenkins RB, Yang H, Ledoux A, Aaron J, Tipnis U, Zhang W, Hess K, Aldape K. The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer. 2005;104:1468–1477. doi: 10.1002/cncr.21338. [DOI] [PubMed] [Google Scholar]
- 139.Meneton P, Jeunemaitre X, de Wardener HE, MacGregor GA. Links between dietary salt intake, renal salt handling, blood pressure, and cardiovascular diseases. Physiol Rev. 2005;85:679–715. doi: 10.1152/physrev.00056.2003. [DOI] [PubMed] [Google Scholar]
- 140.Meneton P, Loffing J, Warnock DG. Sodium and potassium handling by the aldosterone-sensitive distal nephron: the pivotal role of the distal and connecting tubule. Am J Physiol Renal Physiol. 2004;287:F593–601. doi: 10.1152/ajprenal.00454.2003. [DOI] [PubMed] [Google Scholar]
- 141.Mennitt PA, Wade JB, Ecelbarger CA, Palmer LG, Frindt G. Localization of ROMK channels in the rat kidney. J Am Soc Nephrol. 1997;8:1823–1830. doi: 10.1681/ASN.V8121823. [DOI] [PubMed] [Google Scholar]
- 142.Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, Levesque M, Allaire N, Perrin S, Sands B, Crowell T, Cate RL, McCoy JM, Pepinsky RB. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nature neuroscience. 2004;7:221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 143.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure (Camb) 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 144.Mizuno A, Matsumoto N, Imai M, Suzuki M. Impaired osmotic sensation in mice lacking TRPV4. Am J Physiol Cell Physiol. 2003;285:C96–101. doi: 10.1152/ajpcell.00559.2002. [DOI] [PubMed] [Google Scholar]
- 145.Momeny M, Zakidizaji M, Ghasemi R, Dehpour AR, Rahimi Balaei M, Abdolazimi Y, Ghavamzadeh A, Alimoghaddam K, Ghaffari SH. Arsenic trioxide induces apoptosis in NB-4, an acute promyelocytic leukemia cell line, through up-regulation of p73 via suppression of nuclear factor kappa B-mediated inhibition of p73 transcription and prevention of NF-kappaB-mediated induction of XIAP, cIAP2, BCL-X(L) and survivin. Medical Oncology. 2009 doi: 10.1007/s12032-009-9294-9. [DOI] [PubMed] [Google Scholar]
- 146.Moniz S, Matos P, Jordan P. WNK2 modulates MEK1 activity through the Rho GTPase pathway. Cell Signal. 2008;20:1762–1768. doi: 10.1016/j.cellsig.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 147.Moniz S, Verissimo F, Matos P, Brazao R, Silva E, Kotelevets L, Chastre E, Gespach C, Jordan P. Protein kinase WNK2 inhibits cell proliferation by negatively modulating the activation of MEK1/ERK1/2. Oncogene. 2007;26:6071–6081. doi: 10.1038/sj.onc.1210706. [DOI] [PubMed] [Google Scholar]
- 148.Moore TM, Garg R, Johnson C, Coptcoat MJ, Ridley AJ, Morris JD. PSK, a novel STE20-like kinase derived from prostatic carcinoma that activates the c-Jun N-terminal kinase mitogen-activated protein kinase pathway and regulates actin cytoskeletal organization. J Biol Chem. 2000;275:4311–4322. doi: 10.1074/jbc.275.6.4311. [DOI] [PubMed] [Google Scholar]
- 149.Moriguchi T, Urushiyama S, Hisamoto N, Iemura SI, Uchida S, Natsume T, Matsumoto K, Shibuya H. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem. 2005;280:42685–42693. doi: 10.1074/jbc.M510042200. [DOI] [PubMed] [Google Scholar]
- 150.Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- 151.Mueller W, Hartmann C, Hoffmann A, Lanksch W, Kiwit J, Tonn J, Veelken J, Schramm J, Weller M, Wiestler OD, Louis DN, von Deimling A. Genetic signature of oligoastrocytomas correlates with tumor location and denotes distinct molecular subsets. Am J Pathol. 2002;161:313–319. doi: 10.1016/S0002-9440(10)64183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Murakami-Kojima M, Nakamichi N, Yamashino T, Mizuno T. The APRR3 component of the clock-associated APRR1/TOC1 quintet is phosphorylated by a novel protein kinase belonging to the WNK family, the gene for which is also transcribed rhythmically in Arabidopsis thaliana. Plant & cell physiology. 2002;43:675–683. doi: 10.1093/pcp/pcf084. [DOI] [PubMed] [Google Scholar]
- 153.Murthy M, Cope G, O’Shaughnessy KM. The acidic motif of WNK4 is crucial for its interaction with the K channel ROMK. Biochem Biophys Res Commun. 2008;375:651–654. doi: 10.1016/j.bbrc.2008.08.076. [DOI] [PubMed] [Google Scholar]
- 154.Naesens M, Steels P, Verberckmoes R, Vanrenterghem Y, Kuypers D. Bartter’s and Gitelman’s syndromes: from gene to clinic. Nephron Physiol. 2004;96:p65–78. doi: 10.1159/000076752. [DOI] [PubMed] [Google Scholar]
- 155.Nakamichi N, Murakami-Kojima M, Sato E, Kishi Y, Yamashino T, Mizuno T. Compilation and characterization of a novel WNK family of protein kinases in Arabiodpsis thaliana with reference to circadian rhythms. Biosci Biotechnol Biochem. 2002 Nov;66:2429–2436. doi: 10.1271/bbb.66.2429. [DOI] [PubMed] [Google Scholar]
- 156.Naray-Fejes-Toth A, Snyder PM, Fejes-Toth G. The kidney-specific WNK1 isoform is induced by aldosterone and stimulates epithelial sodium channel-mediated Na+ transport. Proc Natl Acad Sci U S A. 2004;101:17434–17439. doi: 10.1073/pnas.0408146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Newhouse S, Farrall M, Wallace C, Hoti M, Burke B, Howard P, Onipinla A, Lee K, Shaw-Hawkins S, Dobson R, Brown M, Samani NJ, Dominiczak AF, Connell JM, Lathrop GM, Kooner J, Chambers J, Elliott P, Clarke R, Collins R, Laan M, Org E, Juhanson P, Veldre G, Viigimaa M, Eyheramendy S, Cappuccio FP, Ji C, Iacone R, Strazzullo P, Kumari M, Marmot M, Brunner E, Caulfield M, Munroe PB. Polymorphisms in the WNK1 gene are associated with blood pressure variation and urinary potassium excretion. PLoS One. 2009;4:e5003. doi: 10.1371/journal.pone.0005003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Newhouse SJ, Wallace C, Dobson R, Mein C, Pembroke J, Farrall M, Clayton D, Brown M, Samani N, Dominiczak A, Connell JM, Webster J, Lathrop GM, Caulfield M, Munroe PB. Haplotypes of the WNK1 gene associate with blood pressure variation in a severely hypertensive population from the British Genetics of Hypertension study. Hum Mol Genet. 2005;14:1805–1814. doi: 10.1093/hmg/ddi187. [DOI] [PubMed] [Google Scholar]
- 159.Nguyen Dinh Cat A, Ouvrard-Pascaud A, Tronche F, Clemessy M, Gonzalez-Nunez D, Farman N, Jaisser F. Conditional transgenic mice for studying the role of the glucocorticoid receptor in the renal collecting duct. Endocrinology. 2009;150:2202–2210. doi: 10.1210/en.2008-1531. [DOI] [PubMed] [Google Scholar]
- 160.Niederost B, Oertle T, Fritsche J, McKinney RA, Bandtlow CE. Nogo-A and myelin-associated glycoprotein mediate neurite growth inhibition by antagonistic regulation of RhoA and Rac1. J Neurosci. 2002;22:10368–10376. doi: 10.1523/JNEUROSCI.22-23-10368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Norden AD, Drappatz J, Wen PY. Advances in meningioma therapy. Current neurology and neuroscience reports. 2009;9:231–240. doi: 10.1007/s11910-009-0034-5. [DOI] [PubMed] [Google Scholar]
- 162.O’Reilly M, Marshall E, Macgillivray T, Mittal M, Xue W, Kenyon CJ, Brown RW. Dietary Electrolyte-Driven Responses in the Renal WNK Kinase Pathway In Vivo. J Am Soc Nephrol. 2006;17:2402–2413. doi: 10.1681/ASN.2005111197. [DOI] [PubMed] [Google Scholar]
- 163.O’Reilly M, Marshall E, Speirs HJ, Brown RW. WNK1, a Gene within a Novel Blood Pressure Control Pathway, Tissue-Specifically Generates Radically Different Isoforms with and without a Kinase Domain. J Am Soc Nephrol. 2003;14:2447–2456. doi: 10.1097/01.asn.0000089830.97681.3b. [DOI] [PubMed] [Google Scholar]
- 164.Obermüller N, Bernstein PL, Velázquez H, Reilly R, Moser D, Ellison DH, Bachmann S. Expression of the thiazide-sensitive Na-Cl cotransporter in rat and human kidney. Am J Physiol. 1995;269:F900–F910. doi: 10.1152/ajprenal.1995.269.6.F900. [DOI] [PubMed] [Google Scholar]
- 165.Oh E, Heise CJ, English JM, Cobb MH, Thurmond DC. WNK1 is a novel regulator of Munc18c-syntaxin 4 complex formation in SNARE-mediated vesicle exocytosis. J Biol Chem. 2007;282:32613–32622. doi: 10.1074/jbc.M706591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Oh E, Spurlin BA, Pessin JE, Thurmond DC. Munc18c heterozygous knockout mice display increased susceptibility for severe glucose intolerance. Diabetes. 2005;54:638–647. doi: 10.2337/diabetes.54.3.638. [DOI] [PubMed] [Google Scholar]
- 167.Ohta A, Rai T, Yui N, Chiga M, Yang SS, Lin SH, Sohara E, Sasaki S, Uchida S. Targeted disruption of the Wnk4 gene decreases phosphorylation of Na-Cl cotransporter, increases Na excretion, and lowers blood pressure. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp344. [DOI] [PubMed] [Google Scholar]
- 168.Ohta A, Yang SS, Rai T, Chiga M, Sasaki S, Uchida S. Overexpression of human WNK1 increases paracellular chloride permeability and phosphorylation of claudin-4 in MDCKII cells. Biochem Biophys Res Commun. 2006;349:804–808. doi: 10.1016/j.bbrc.2006.08.101. [DOI] [PubMed] [Google Scholar]
- 169.Osada Y, Miyauchi R, Goda T, Kasezawa N, Horiike H, Iida M, Sasaki S, Yamakawa-Kobayashi K. Variations in the WNK1 gene modulates the effect of dietary intake of sodium and potassium on blood pressure determination. Journal of human genetics. 2009;54:474–478. doi: 10.1038/jhg.2009.64. [DOI] [PubMed] [Google Scholar]
- 170.Pacheco-Alvarez D, Cristobal PS, Meade P, Moreno E, Vazquez N, Munoz E, Diaz A, Juarez ME, Gimenez I, Gamba G. The Na+:Cl− cotransporter is activated and phosphorylated at the amino-terminal domain upon intracellular chloride depletion. J Biol Chem. 2006;281:28755–28763. doi: 10.1074/jbc.M603773200. [DOI] [PubMed] [Google Scholar]
- 171.Padmanabhan S, Menni C, Lee WK, Laing S, Brambilla P, Sega R, Perego R, Grassi G, Cesana G, Delles C, Mancia G, Dominiczak AF. The effects of sex and method of blood pressure measurement on genetic associations with blood pressure in the PAMELA study. J Hypertens. 28:465–477. doi: 10.1097/HJH.0b013e32833594d7. [DOI] [PubMed] [Google Scholar]
- 172.Paver WK, Pauline GJ. Hypertension and hyperpotassemia without renal disease in a young male. Med J Aust. 1964;2:305–306. doi: 10.5694/j.1326-5377.1964.tb115766.x. [DOI] [PubMed] [Google Scholar]
- 173.Perez-Sayans M, Garcia-Garcia A, Reboiras-Lopez MD, Gandara-Vila P. Role of V-ATPases in solid tumors: importance of the subunit C (Review) International journal of oncology. 2009;34:1513–1520. doi: 10.3892/ijo_00000280. [DOI] [PubMed] [Google Scholar]
- 174.Piechotta K, Garbarini N, England R, Delpire E. Characterization of the interaction of the stress kinase SPAK with the Na+-K+-2Cl- cotransporter in the nervous system: evidence for a scaffolding role of the kinase. Journal of Biological Chemistry. 2003;278:52848–52856. doi: 10.1074/jbc.M309436200. [DOI] [PubMed] [Google Scholar]
- 175.Placzek M, Briscoe J. The floor plate: multiple cells, multiple signals. Nat Rev Neurosci. 2005;6:230–240. doi: 10.1038/nrn1628. [DOI] [PubMed] [Google Scholar]
- 176.Plotkin MD, Kaplan MR, Peterson LN, Gullans SR, Hebert SC, Delpire E. Expression of the Na+-K+-2Cl- cotransporter BSC2 in the nervous system. Am J Physiol Cell Physiol. 1997;272:C173–C183. doi: 10.1152/ajpcell.1997.272.1.C173. [DOI] [PubMed] [Google Scholar]
- 177.Plotkin MD, Kaplan MR, Verlander JW, Lee W-S, Brown D, Poch E, Gullans SR, Hebert SC. Localization of the thiazide sensitive Na-Cl cotransporter, rTSC1, in the rat kidney. Kidney Int. 1996;50:174–183. doi: 10.1038/ki.1996.300. [DOI] [PubMed] [Google Scholar]
- 178.Ponce-Coria J, San-Cristobal P, Kahle KT, Vazquez N, Pacheco-Alvarez D, de Los Heros P, Juarez P, Munoz E, Michel G, Bobadilla NA, Gimenez I, Lifton RP, Hebert SC, Gamba G. Regulation of NKCC2 by a chloride-sensing mechanism involving the WNK3 and SPAK kinases. Proc Natl Acad Sci U S A. 2008;105:8458–8463. doi: 10.1073/pnas.0802966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Proctor G, Linas S. Type 2 pseudohypoaldosteronism: new insights into renal potassium, sodium, and chloride handling. Am J Kidney Dis. 2006;48:674–693. doi: 10.1053/j.ajkd.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 180.Pullikuth AK, Catling AD. Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cell Signal. 2007;19:1621–1632. doi: 10.1016/j.cellsig.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Qiao Y, Liu X, Harvard C, Hildebrand MJ, Rajcan-Separovic E, Holden JJ, Lewis ME. Autism-associated familial microdeletion of Xp11.22. Clinical genetics. 2008;74:134–144. doi: 10.1111/j.1399-0004.2008.01028.x. [DOI] [PubMed] [Google Scholar]
- 182.Radhakrishnan A, Stein A, Jahn R, Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2009;284:25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- 184.Reilly RF, Ellison DH. Mammalian distal tubule: physiology, pathophysiology, and molecular anatomy. Physiol Rev. 2000;80:277–313. doi: 10.1152/physrev.2000.80.1.277. [DOI] [PubMed] [Google Scholar]
- 185.Richardson C, Rafiqi FH, Karlsson HK, Moleleki N, Vandewalle A, Campbell DG, Morrice NA, Alessi DR. Activation of the thiazide-sensitive Na+-Cl-cotransporter by the WNK-regulated kinases SPAK and OSR1. J Cell Sci. 2008;121:675–684. doi: 10.1242/jcs.025312. [DOI] [PubMed] [Google Scholar]
- 186.Rickman C, Medine CN, Bergmann A, Duncan RR. Functionally and spatially distinct modes of munc18-syntaxin 1 interaction. J Biol Chem. 2007;282:12097–12103. doi: 10.1074/jbc.M700227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rinehart J, Kahle KT, de Los Heros P, Vazquez N, Meade P, Wilson FH, Hebert SC, Gimenez I, Gamba G, Lifton RP. WNK3 kinase is a positive regulator of NKCC2 and NCC, renal cation-Cl- cotransporters required for normal blood pressure homeostasis. Proc Natl Acad Sci U S A. 2005;102:16777–16782. doi: 10.1073/pnas.0508303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rinehart J, Maksimova YD, Tanis JE, Stone KL, Hodson CA, Zhang J, Risinger M, Pan W, Wu D, Colangelo CM, Forbush B, Joiner CH, Gulcicek EE, Gallagher PG, Lifton RP. Sites of regulated phosphorylation that control K-Cl cotransporter activity. Cell. 2009;138:525–536. doi: 10.1016/j.cell.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ring AM, Cheng SX, Leng Q, Kahle KT, Rinehart J, Lalioti MD, Volkman HM, Wilson FH, Hebert SC, Lifton RP. WNK4 regulates activity of the epithelial Na+ channel in vitro and in vivo. Proc Natl Acad Sci U S A. 2007;104:4020–4024. doi: 10.1073/pnas.0611727104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ring AM, Leng Q, Rinehart J, Wilson FH, Kahle KT, Hebert SC, Lifton RP. An SGK1 site in WNK4 regulates Na+ channel and K+ channel activity and has implications for aldosterone signaling and K+ homeostasis. Proc Natl Acad Sci U S A. 2007;104:4025–4029. doi: 10.1073/pnas.0611728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ringold GM. Glucocorticoid regulation of mouse mammary tumor virus gene expression. Biochim Biophys Acta. 1979;560:487–508. doi: 10.1016/0304-419x(79)90014-3. [DOI] [PubMed] [Google Scholar]
- 192.Riviere JB, Verlaan DJ, Shekarabi M, Lafreniere RG, Benard M, Der Kaloustian VM, Shbaklo Z, Rouleau GA. A mutation in the HSN2 gene causes sensory neuropathy type II in a Lebanese family. Ann Neurol. 2004;56:572–575. doi: 10.1002/ana.20237. [DOI] [PubMed] [Google Scholar]
- 193.Rotthier A, Baets J, De Vriendt E, Jacobs A, Auer-Grumbach M, Levy N, Bonello-Palot N, Kilic SS, Weis J, Nascimento A, Swinkels M, Kruyt MC, Jordanova A, De Jonghe P, Timmerman V. Genes for hereditary sensory and autonomic neuropathies: a genotype-phenotype correlation. Brain. 2009;132:2699–2711. doi: 10.1093/brain/awp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rozansky DJ, Cornwall T, Subramanya AR, Rogers S, Yang YF, David LL, Zhu X, Yang CL, Ellison DH. Aldosterone mediates activation of the thiazide-sensitive Na-Cl cotransporter through an SGK1 and WNK4 signaling pathway. J Clin Invest. 2009;119:2601–2612. doi: 10.1172/JCI38323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Russell JM. Sodium-potassium-chloride cotransport. Physiol Rev. 2000;80:211–276. doi: 10.1152/physrev.2000.80.1.211. [DOI] [PubMed] [Google Scholar]
- 196.Sagi-Eisenberg R. The mast cell: where endocytosis and regulated exocytosis meet. Immunological reviews. 2007;217:292–303. doi: 10.1111/j.1600-065X.2007.00516.x. [DOI] [PubMed] [Google Scholar]
- 197.Sale EM, Hodgkinson CP, Jones NP, Sale GJ. A new strategy for studying protein kinase B and its three isoforms. Role of protein kinase B in phosphorylating glycogen synthase kinase-3, tuberin, WNK1, and ATP citrate lyase. Biochemistry. 2006;45:213–223. doi: 10.1021/bi050287i. [DOI] [PubMed] [Google Scholar]
- 198.San-Cristobal P, Pacheco-Alvarez D, Richardson C, Ring AM, Vazquez N, Rafiqi FH, Chari D, Kahle KT, Leng Q, Bobadilla NA, Hebert SC, Alessi DR, Lifton RP, Gamba G. Angiotensin II signaling increases activity of the renal Na-Cl cotransporter through a WNK4-SPAK-dependent pathway. Proc Natl Acad Sci U S A. 2009;106:4384–4389. doi: 10.1073/pnas.0813238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.San-Cristobal P, Ponce-Coria J, Vazquez N, Bobadilla NA, Gamba G. WNK3 and WNK4 amino-terminal domain defines their effect on the renal Na+-Cl-cotransporter. Am J Physiol Renal Physiol. 2008;295:F1199–1206. doi: 10.1152/ajprenal.90396.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sanada K, Hayashi Y, Harada Y, Okano T, Fukada Y. Role of circadian activation of mitogen-activated protein kinase in chick pineal clock oscillation. J Neurosci. 2000;20:986–991. doi: 10.1523/JNEUROSCI.20-03-00986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sandberg MB, Maunsbach AB, McDonough AA. Redistribution of distal tubule Na+-Cl- cotransporter (NCC) in response to a high-salt diet. Am J Physiol Renal Physiol. 2006;291:F503–508. doi: 10.1152/ajprenal.00482.2005. [DOI] [PubMed] [Google Scholar]
- 202.Sandberg MB, Riquier AD, Pihakaski-Maunsbach K, McDonough AA, Maunsbach AB. ANG II provokes acute trafficking of distal tubule Na+-Cl cotransporter to apical membrane. Am J Physiol Renal Physiol. 2007;293:F662–669. doi: 10.1152/ajprenal.00064.2007. [DOI] [PubMed] [Google Scholar]
- 203.Sandow J. Growth effects of insulin and insulin analogues. Archives of physiology and biochemistry. 2009;115:72–85. doi: 10.1080/13813450902835690. [DOI] [PubMed] [Google Scholar]
- 204.Sansom SC. Reemergence of the maxi K+ as a K+ secretory channel. Kidney Int. 2007;71:1322–1324. doi: 10.1038/sj.ki.5002234. author reply 1324–1325. [DOI] [PubMed] [Google Scholar]
- 205.Sanz MA, Grimwade D, Tallman MS, Lowenberg B, Fenaux P, Estey EH, Naoe T, Lengfelder E, Buchner T, Dohner H, Burnett AK, Lo-Coco F. Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113:1875–1891. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- 206.Schambelan M, Sebastian A, Rector FC., Jr Mineralocorticoid-resistant renal hyperkalemia without salt wasting (type II pseudohypoaldosteronism): role of increased renal chloride reabsorption. Kidney Int. 1981;19:716–727. doi: 10.1038/ki.1981.72. [DOI] [PubMed] [Google Scholar]
- 207.Schiavo G, Gu QM, Prestwich GD, Sollner TH, Rothman JE. Calcium-dependent switching of the specificity of phosphoinositide binding to synaptotagmin. Proc Natl Acad Sci U S A. 1996;93:13327–13332. doi: 10.1073/pnas.93.23.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton RP, Rossier BC. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. Embo J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- 209.Schmitt R, Ellison DH, Farman N, Rossier B, Reilly RF, Kunchaparty S, Reeves WB, Oberbäumer I, Tapp R, Bachmann S. Developmental expression of sodium entry pathways in rat distal nephron. Am J Physiol. 1999;276:F367–F381. doi: 10.1152/ajprenal.1999.276.3.F367. [DOI] [PubMed] [Google Scholar]
- 210.Shaharabany M, Holtzman EJ, Mayan H, Hirschberg K, Seger R, Farfel Z. Distinct pathways for the involvement of WNK4 in the signaling of hypertonicity and EGF. The FEBS journal. 2008;275:1631–1642. doi: 10.1111/j.1742-4658.2008.06318.x. [DOI] [PubMed] [Google Scholar]
- 211.Shekarabi M, Girard N, Riviere JB, Dion P, Houle M, Toulouse A, Lafreniere RG, Vercauteren F, Hince P, Laganiere J, Rochefort D, Faivre L, Samuels M, Rouleau GA. Mutations in the nervous system-specific HSN2 exon of WNK1 cause hereditary sensory neuropathy type II. J Clin Invest. 2008 doi: 10.1172/JCI34088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Shen Z, Denison K, Lobb R, Gatewood JM, Chen DJ. The human and mouse homologs of the yeast RAD52 gene: cDNA cloning, sequence analysis, assignment to human chromosome 12p12.2-p13, and mRNA expression in mouse tissues. Genomics. 1995;25:199–206. doi: 10.1016/0888-7543(95)80126-7. [DOI] [PubMed] [Google Scholar]
- 213.Shimizu T, Yoshitomi K, Nakamura M, Imai M. Site and mechanism of action of trichlormethiazide in rabbit distal nephron segments perfused in vitro. J Clin Invest. 1988;82:721–730. doi: 10.1172/JCI113653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sindic A, Chang MH, Mount DB, Romero MF. Renal physiology of SLC26 anion exchangers. Curr Opin Nephrol Hypertens. 2007;16:484–490. doi: 10.1097/MNH.0b013e3282e7d7d0. [DOI] [PubMed] [Google Scholar]
- 215.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 216.Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 217.Snyder PM, Olson DR, Thomas BC. SGK modulates Nedd4–2-mediated inhibition of ENaC. J Biol Chem. 2001 Nov 5; doi: 10.1074/jbc.C100623200. [DOI] [PubMed] [Google Scholar]
- 218.Song J, Hu X, Riazi S, Tiwari S, Wade JB, Ecelbarger CA. Regulation of blood pressure, the epithelial sodium channel (ENaC), and other key renal sodium transporters by chronic insulin infusion in rats. Am J Physiol Renal Physiol. 2006;290:F1055–1064. doi: 10.1152/ajprenal.00108.2005. [DOI] [PubMed] [Google Scholar]
- 219.Speirs HJ, Morris BJ. WNK4 intron 10 polymorphism is not associated with hypertension. Hypertension. 2004;43:766–768. doi: 10.1161/01.HYP.0000120121.43524.cd. [DOI] [PubMed] [Google Scholar]
- 220.Spirin KS, Simpson JF, Takeuchi S, Kawamata N, Miller CW, Koeffler HP. p27/Kip1 mutation found in breast cancer. Cancer Res. 1996;56:2400–2404. [PubMed] [Google Scholar]
- 221.Staub O, Abriel H, Plant P, Ishikawa T, Kanelis V, Saleki R, Horisberger JD, Schild L, Rotin D. Regulation of the epithelial Na+ channel by Nedd4 and ubiquitination. Kidney Int. 2000;57:809–815. doi: 10.1046/j.1523-1755.2000.00919.x. [DOI] [PubMed] [Google Scholar]
- 222.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, O’Meara S, Parker A, Tarpey P, Avis T, Barthorpe A, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh B, Yuen ST, Nicholson AG, Lakhani S, Easton DF, Weber BL, Stratton MR, Futreal PA, Wooster R. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 223.Subramanya AR, Liu J, Ellison DH, Wade JB, Welling PA. WNK4 Diverts the Thiazide-sensitive NaCl Cotransporter to the Lysosome and Stimulates AP-3 Interaction. J Biol Chem. 2009;284:18471–18480. doi: 10.1074/jbc.M109.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Subramanya AR, Yang CL, Zhu X, Ellison DH. Dominant-negative regulation of WNK1 by its kidney-specific kinase-defective isoform. Am J Physiol Renal Physiol. 2006;290:F619–624. doi: 10.1152/ajprenal.00280.2005. [DOI] [PubMed] [Google Scholar]
- 225.Sun X, Gao L, Yu RK, Zeng G. Down-regulation of WNK1 protein kinase in neural progenitor cells suppresses cell proliferation and migration. J Neurochem. 2006;99:1114–1121. doi: 10.1111/j.1471-4159.2006.04159.x. [DOI] [PubMed] [Google Scholar]
- 226.Sun ZJ, Li Y, Lu JY, Ding Q, Liang Y, Shi JP, Li-Ling J, Zhao YY. Association of Ala589Ser polymorphism of WNK4 gene with essential hypertension in a high-risk Chinese population. J Physiol Sci. 2009;59:81–86. doi: 10.1007/s12576-008-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Take C, Ikeda K, Kurasawa T, Kurokawa K. Increased chloride reabsorption as an inherited renal tubular defect in familial type II pseudohypoaldosteronism. N Engl J Med. 1991;324:472–476. doi: 10.1056/NEJM199102143240707. [DOI] [PubMed] [Google Scholar]
- 228.Talati G, Ohta A, Rai T, Sohara E, Naito S, Vandewalle A, Sasaki S, Uchida S. Effect of angiotensin II on the WNK-OSR1/SPAK-NCC phosphorylation cascade in cultured mpkDCT cells and in vivo mouse kidney. Biochem Biophys Res Commun. 393:844–848. doi: 10.1016/j.bbrc.2010.02.096. [DOI] [PubMed] [Google Scholar]
- 229.Tallman MS. What is the role of arsenic in newly diagnosed APL? Best practice & research. 2008;21:659–666. doi: 10.1016/j.beha.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 230.Tanaka M, Nishi M, Morimoto M, Sugimoto T, Kawata M. Imaging analysis of mineralocorticoid receptor and importins in single living cells by using GFP color variants. Cell and tissue research. 2005;320:447–453. doi: 10.1007/s00441-004-0984-5. [DOI] [PubMed] [Google Scholar]
- 231.Tatum R, Zhang Y, Lu Q, Kim K, Jeansonne BG, Chen YH. WNK4 phosphorylates ser(206) of claudin-7 and promotes paracellular Cl(−) permeability. FEBS Lett. 2007;581:3887–3891. doi: 10.1016/j.febslet.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 232.Thomas W, McEneaney V, Harvey BJ. Aldosterone-induced signalling and cation transport in the distal nephron. Steroids. 2008;73:979–984. doi: 10.1016/j.steroids.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 233.Thurmond DC, Kanzaki M, Khan AH, Pessin JE. Munc18c function is required for insulin-stimulated plasma membrane fusion of GLUT4 and insulin-responsive amino peptidase storage vesicles. Mol Cell Biol. 2000;20:379–388. doi: 10.1128/mcb.20.1.379-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Thurmond DC, Pessin JE. Discrimination of GLUT4 vesicle trafficking from fusion using a temperature-sensitive Munc18c mutant. Embo J. 2000;19:3565–3575. doi: 10.1093/emboj/19.14.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tian W, Fu Y, Garcia-Elias A, Fernandez-Fernandez JM, Vicente R, Kramer PL, Klein RF, Hitzemann R, Orwoll ES, Wilmot B, McWeeney S, Valverde MA, Cohen DM. A loss-of-function nonsynonymous polymorphism in the osmoregulatory TRPV4 gene is associated with human hyponatremia. Proc Natl Acad Sci U S A. 2009;106:14034–14039. doi: 10.1073/pnas.0904084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tian W, Salanova M, Xu H, Lindsley JN, Oyama TT, Anderson S, Bachmann S, Cohen DM. Renal expression of osmotically responsive cation channel TRPV4 is restricted to water-impermeant nephron segments. Am J Physiol Renal Physiol. 2004;287:F17–24. doi: 10.1152/ajprenal.00397.2003. [DOI] [PubMed] [Google Scholar]
- 237.Tobin MD, Raleigh SM, Newhouse S, Braund P, Bodycote C, Ogleby J, Cross D, Gracey J, Hayes S, Smith T, Ridge C, Caulfield M, Sheehan NA, Munroe PB, Burton PR, Samani NJ. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation. 2005;112:3423–3429. doi: 10.1161/CIRCULATIONAHA.105.555474. [DOI] [PubMed] [Google Scholar]
- 238.Tobin MD, Timpson NJ, Wain LV, Ring S, Jones LR, Emmett PM, Palmer TM, Ness AR, Samani NJ, Smith GD, Burton PR. Common variation in the WNK1 gene and blood pressure in childhood: the Avon Longitudinal Study of Parents and Children. Hypertension. 2008;52:974–979. doi: 10.1161/HYPERTENSIONAHA.108.118414. [DOI] [PubMed] [Google Scholar]
- 239.Tucker WC, Chapman ER. Role of synaptotagmin in Ca2+-triggered exocytosis. Biochem J. 2002;366:1–13. doi: 10.1042/BJ20020776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. WNK1 kinase polymorphism and blood pressure response to a thiazide diuretic. Hypertension. 2005;46:758–765. doi: 10.1161/01.HYP.0000186240.81996.57. [DOI] [PubMed] [Google Scholar]
- 241.Vallon V, Schroth J, Lang F, Kuhl D, Uchida S. Expression and phosphorylation of the Na+-Cl- cotransporter NCC in vivo is regulated by dietary salt, potassium, and SGK1. Am J Physiol Renal Physiol. 2009;297:F704–712. doi: 10.1152/ajprenal.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human Rad52 protein. Nature. 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 243.Velázquez H, Bartiss A, Bernstein PL, Ellison DH. Adrenal steroids stimulate thiazide-sensitive NaCl transport by the rat renal distal tubule. Am J Physiol. 1996;270:F211–F219. doi: 10.1152/ajprenal.1996.270.1.F211. [DOI] [PubMed] [Google Scholar]
- 244.Velázquez H, Náray-Fejes-Tóth A, Silva T, Andújar E, Reilly RF, Desir GV, Ellison DH. The distal convoluted tubule of the rabbit coexpresses NaCl cotransporter and 11b-hydroxysteroid dehydrogenase. Kidney Int. 1998;54:464–472. doi: 10.1046/j.1523-1755.1998.00036.x. [DOI] [PubMed] [Google Scholar]
- 245.Verissimo F, Jordan P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene. 2001;20:5562–5569. doi: 10.1038/sj.onc.1204726. [DOI] [PubMed] [Google Scholar]
- 246.Verissimo F, Silva E, Morris JD, Pepperkok R, Jordan P. Protein kinase WNK3 increases cell survival in a caspase-3-dependent pathway. Oncogene. 2006;25:4172–4182. doi: 10.1038/sj.onc.1209449. [DOI] [PubMed] [Google Scholar]
- 247.Vitari AC, Deak M, Collins BJ, Morrice N, Prescott AR, Phelan A, Humphreys S, Alessi DR. WNK1, the kinase mutated in an inherited high-blood-pressure syndrome, is a novel PKB (protein kinase B)/Akt substrate. Biochem J. 2004;378:257–268. doi: 10.1042/BJ20031692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon’s hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wade JB, Fang L, Liu J, Li D, Yang CL, Subramanya AR, Maouyo D, Mason A, Ellison DH, Welling PA. WNK1 kinase isoform switch regulates renal potassium excretion. Proc Natl Acad Sci U S A. 2006;103:8558–8563. doi: 10.1073/pnas.0603109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Walker C, du Plessis DG, Joyce KA, Fildes D, Gee A, Haylock B, Husband D, Smith T, Broome J, Warnke PC. Molecular pathology and clinical characteristics of oligodendroglial neoplasms. Ann Neurol. 2005;57:855–865. doi: 10.1002/ana.20496. [DOI] [PubMed] [Google Scholar]
- 251.Wang HR, Liu Z, Huang CL. Domains of WNK1 kinase in the regulation of ROMK1. Am J Physiol Renal Physiol. 2008 doi: 10.1152/ajprenal.90287.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wang X, Tournier C. Regulation of cellular functions by the ERK5 signalling pathway. Cell Signal. 2006;18:753–760. doi: 10.1016/j.cellsig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 253.Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X. The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant biology (Stuttgart, Germany) 2008;10:548–562. doi: 10.1111/j.1438-8677.2008.00072.x. [DOI] [PubMed] [Google Scholar]
- 254.Wang Z, Yang CL, Ellison DH. Comparison of WNK4 and WNK1 kinase and inhibiting activities. Biochem Biophys Res Commun. 2004;317:939–944. doi: 10.1016/j.bbrc.2004.03.132. [DOI] [PubMed] [Google Scholar]
- 255.Weinberger MH, Luft FC, Bloch R, Henry DP, Pratt JH, Weyman AE, Rankin LI, Murray RH, Willis LR, Grim CE. The blood pressure-raising effects of high dietary sodium intake: racial differences and the role of potassium. J Am Coll Nutr. 1982;1:139–148. doi: 10.1080/07315724.1982.10718981. [DOI] [PubMed] [Google Scholar]
- 256.Weitzmann MN, Pacifici R. T cells: unexpected players in the bone loss induced by estrogen deficiency and in basal bone homeostasis. Ann N Y Acad Sci. 2007;1116:360–375. doi: 10.1196/annals.1402.068. [DOI] [PubMed] [Google Scholar]
- 257.Wicks SJ, Lui S, Abdel-Wahab N, Mason RM, Chantry A. Inactivation of smad-transforming growth factor beta signaling by Ca(2+)-calmodulin-dependent protein kinase II. Mol Cell Biol. 2000;20:8103–8111. doi: 10.1128/mcb.20.21.8103-8111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 259.Wilson FH, Kahle KT, Sabath E, Lalioti MD, Rapson AK, Hoover RS, Hebert SC, Gamba G, Lifton RP. Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci U S A. 2003;100:680–684. doi: 10.1073/pnas.242735399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Woudenberg-Vrenken TE, Bindels RJ, Hoenderop JG. The role of transient receptor potential channels in kidney disease. Nature reviews. 2009;5:441–449. doi: 10.1038/nrneph.2009.100. [DOI] [PubMed] [Google Scholar]
- 261.Wu C, Lee SF, Furmaniak-Kazmierczak E, Cote GP, Thomas DY, Leberer E. Activation of myosin-I by members of the Ste20p protein kinase family. J Biol Chem. 1996;271:31787–31790. doi: 10.1074/jbc.271.50.31787. [DOI] [PubMed] [Google Scholar]
- 262.Xiao P, Chen Y, Jiang H, Liu YZ, Pan F, Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR, Deng HW. In vivo genome-wide expression study on human circulating B cells suggests a novel ESR1 and MAPK3 network for postmenopausal osteoporosis. J Bone Miner Res. 2008;23:644–654. doi: 10.1359/JBMR.080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xie J, Craig L, Cobb MH, Huang CL. Role of with-no-lysine [K] kinases in the pathogenesis of Gordon’s syndrome. PediatrNephrol. 2006;21:1231–1236. doi: 10.1007/s00467-006-0106-6. [DOI] [PubMed] [Google Scholar]
- 264.Xie J, Wu T, Xu K, Huang IK, Cleaver O, Huang CL. Endothelial-Specific Expression of WNK1 Kinase Is Essential for Angiogenesis and Heart Development in Mice. Am J Pathol. 2009 doi: 10.2353/ajpath.2009.090094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Xie Q, Welch R, Mercado A, Romero MF, Mount DB. Molecular characterization of the murine Slc26a6 anion exchanger: functional comparison with Slc26a1. Am J Physiol Renal Physiol. 2002;283:F826–838. doi: 10.1152/ajprenal.00079.2002. [DOI] [PubMed] [Google Scholar]
- 266.Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 267.Xu BE, Min X, Stippec S, Lee BH, Goldsmith EJ, Cobb MH. Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem. 2002;277:48456–48462. doi: 10.1074/jbc.M207917200. [DOI] [PubMed] [Google Scholar]
- 268.Xu BE, Stippec S, Chu PY, Lazrak A, Li XJ, Lee BH, English JM, Ortega B, Huang CL, Cobb MH. WNK1 activates SGK1 to regulate the epithelial sodium channel. Proc Natl Acad Sci U S A. 2005;102:10315–10320. doi: 10.1073/pnas.0504422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Xu BE, Stippec S, Lazrak A, Huang CL, Cobb MH. WNK1 activates SGK1 by a PI-3 kinase-dependent and non-catalytic mechanism. J Biol Chem. 2005;280:34218–34223. doi: 10.1074/jbc.M505735200. [DOI] [PubMed] [Google Scholar]
- 270.Xu BE, Stippec S, Lenertz L, Lee BH, Zhang W, Lee YK, Cobb MH. WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J Biol Chem. 2004;279:7826–7831. doi: 10.1074/jbc.M313465200. [DOI] [PubMed] [Google Scholar]
- 271.Xu J, Henriksnas J, Barone S, Witte D, Shull GE, Forte JG, Holm L, Soleimani M. SLC26A9 is expressed in gastric surface epithelial cells, mediates Cl-/HCO3- exchange, and is inhibited by NH4+ Am J Physiol Cell Physiol. 2005;289:C493–505. doi: 10.1152/ajpcell.00030.2005. [DOI] [PubMed] [Google Scholar]
- 272.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Yagi YI, Abe K, Ikebukuro K, Sode K. Kinetic mechanism and inhibitor characterization of WNK1 kinase. Biochemistry. 2009;48:10255–10266. doi: 10.1021/bi900666n. [DOI] [PubMed] [Google Scholar]
- 274.Yakymovych I, Ten Dijke P, Heldin CH, Souchelnytskyi S. Regulation of Smad signaling by protein kinase C. Faseb J. 2001;15:553–555. doi: 10.1096/fj.00-0474fje. [DOI] [PubMed] [Google Scholar]
- 275.Yamashita T, Fujitani M, Yamagishi S, Hata K, Mimura F. Multiple signals regulate axon regeneration through the Nogo receptor complex. Molecular neurobiology. 2005;32:105–111. doi: 10.1385/MN:32:2:105. [DOI] [PubMed] [Google Scholar]
- 276.Yamauchi K, Rai T, Kobayashi K, Sohara E, Suzuki T, Itoh T, Suda S, Hayama A, Sasaki S, Uchida S. Disease-causing mutant WNK4 increases paracellular chloride permeability and phosphorylates claudins. Proc Natl Acad Sci U S A. 2004;101:4690–4694. doi: 10.1073/pnas.0306924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Yan GX, Chen J, Yamada KA, Kleber AG, Corr PB. Contribution of shrinkage of extracellular space to extracellular K+ accumulation in myocardial ischaemia of the rabbit. J Physiol. 1996;490 ( Pt 1):215–228. doi: 10.1113/jphysiol.1996.sp021137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Yang C-L, Zhu X, Ellison DH. The thiazide-sensitive Na-Cl cotransporter is regulated by a WNK kinase signaling complex. J Clin Invest. 2007;117:3403–3411. doi: 10.1172/JCI32033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yang CL, Angell J, Mitchell R, Ellison DH. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest. 2003;111:1039–1045. doi: 10.1172/JCI17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yang CL, Liu X, Paliege A, Zhu X, Bachmann S, Dawson DC, Ellison DH. WNK1 and WNK4 modulate CFTR activity. Biochem Biophys Res Commun. 2007;353:535–540. doi: 10.1016/j.bbrc.2006.11.151. [DOI] [PubMed] [Google Scholar]
- 281.Yang CL, Zhu X, Wang Z, Subramanya AR, Ellison DH. Mechanisms of WNK1 and WNK4 interaction in the regulation of thiazide-sensitive NaCl cotransport. J Clin Invest. 2005;115:1379–1387. doi: 10.1172/JCI22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yang SS, Morimoto T, Rai T, Chiga M, Sohara E, Ohno M, Uchida K, Lin SH, Moriguchi T, Shibuya H, Kondo Y, Sasaki S, Uchida S. Molecular Pathogenesis of Pseudohypoaldosteronism Type II: Generation and Analysis of a Wnk4(D561A/+) Knockin Mouse Model. Cell Metab. 2007;5:331–344. doi: 10.1016/j.cmet.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 283.Yang SS, Yamauchi K, Rai T, Hiyama A, Sohara E, Suzuki T, Itoh T, Suda S, Sasaki S, Uchida S. Regulation of apical localization of the thiazide-sensitive NaCl cotransporter by WNK4 in polarized epithelial cells. Biochem Biophys Res Commun. 2005;330:410–414. doi: 10.1016/j.bbrc.2005.02.172. [DOI] [PubMed] [Google Scholar]
- 284.Yue P, Lin DH, Pan CY, Leng Q, Giebisch G, Lifton RP, Wang WH. Src family protein tyrosine kinase (PTK) modulates the effect of SGK1 and WNK4 on ROMK channels. Proc Natl Acad Sci U S A. 2009;106:15061–15066. doi: 10.1073/pnas.0907855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Zagorska A, Pozo-Guisado E, Boudeau J, Vitari AC, Rafiqi FH, Thastrup J, Deak M, Campbell DG, Morrice NA, Prescott AR, Alessi DR. Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol. 2007;176:89–100. doi: 10.1083/jcb.200605093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, BeltrandelRio H, Buxton EC, Edwards J, Finch RA, Friddle CJ, Gupta A, Hansen G, Hu Y, Huang W, Jaing C, Key BW, Jr, Kipp P, Kohlhauff B, Ma ZQ, Markesich D, Payne R, Potter DG, Qian N, Shaw J, Schrick J, Shi ZZ, Sparks MJ, Van Sligtenhorst I, Vogel P, Walke W, Xu N, Zhu Q, Person C, Sands AT. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zeng G, Gao L, Xia T, Gu Y, Yu RK. Expression of the mouse WNK1 gene in correlation with ganglioside GD3 and functional analysis of the mouse WNK1 promoter. Gene. 2005;344:233–239. doi: 10.1016/j.gene.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 288.Zhang W, Na T, Peng JB. WNK3 positively regulates epithelial calcium channels TRPV5 and TRPV6 via a kinase-dependent pathway. Am J Physiol Renal Physiol. 2008;295:F1472–1484. doi: 10.1152/ajprenal.90229.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhang Z, Xu X, Zhang Y, Zhou J, Yu Z, He C. LINGO-1 interacts with WNK1 to regulate nogo-induced inhibition of neurite extension. J Biol Chem. 2009;284:15717–15728. doi: 10.1074/jbc.M808751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Zhou B, Zhuang J, Gu D, Wang H, Cebotaru L, Guggino WB, Cai H. WNK4 enhances the degradation of NCC through a sortilin-mediated lysosomal pathway. J Am Soc Nephrol. 21:82–92. doi: 10.1681/ASN.2008121275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zhou QL, Park JG, Jiang ZY, Holik JJ, Mitra P, Semiz S, Guilherme A, Powelka AM, Tang X, Virbasius J, Czech MP. Analysis of insulin signalling by RNAi-based gene silencing. Biochem Soc Trans. 2004;32:817–821. doi: 10.1042/BST0320817. [DOI] [PubMed] [Google Scholar]