Digital MDA for enumeration of total nucleic acid contamination (original) (raw)
Abstract
Multiple displacement amplification (MDA) is an isothermal, sequence-independent method for the amplification of high molecular weight DNA that is driven by ϕ29 DNA polymerase (DNAP). Here we report digital MDA (dMDA), an ultrasensitive method for quantifying nucleic acid fragments of unknown sequence. We use the new assay to show that our custom ϕ29 DNAP preparation is free of contamination at the limit of detection of the dMDA assay (1 contaminating molecule per assay microliter). Contamination in commercially available preparations is also investigated. The results of the dMDA assay provide strong evidence that the so-called ‘template-independent’ MDA background can be attributed to high-molecular weight contaminants and is not primer-derived in the commercial kits tested. dMDA is orders of magnitude more sensitive than PCR-based techniques for detection of microbial genomic DNA fragments and opens up new possibilities for the ultrasensitive quantification of DNA fragments in a wide variety of application areas using MDA chemistry and off-the-shelf hardware developed for digital PCR.
INTRODUCTION
Multiple displacement amplification (MDA) is an isothermal, sequence-independent method for exponential amplification of high molecular weight DNA (1). MDA, which relies on the strong strand-displacement synthesis activity of ϕ29 DNAP, has revolutionized analyses of small-quantity DNA samples by providing a means to carry out high fidelity whole genome amplification while maintaining more uniform locus representation than competing techniques, especially those based on PCR (2–6). However, background amplification limits the application of MDA in the most demanding applications, i.e_._ for single-cell analysis, forensics and analysis of ancient samples (7–13).
There has been a debate in the literature whether this background amplification arises from high molecular weight DNA contaminants and/or from side products of the MDA reaction derived from the random primers used (4,5,11,14,15). In the best case, background amplification merely reduces product yield. In other cases, downstream analyses based on product quantification, genotyping or sequencing may be compromised.
In previous work using MDA in microfluidic devices, where amplification reactions are carried out in small volumes, typically 60 nl, we have observed contaminating sequences in <10% of nanoliter single-cell amplifications, suggesting that the concentration of contaminating molecules in the MDA reaction mix was lower than one per 60 nl. However, over a period of about a year, we noticed an increase in levels of bacterial DNA contaminating the commercial MDA reagents. After changing suppliers several times and continuing to experience high levels of reagent contamination, we set out to create our own ‘high-purity’ MDA reagent set and to develop an assay to measure contaminant levels directly. We found standard PCR methods (including digital PCR) relying on the 16S small subunit ribosomal RNA (ssu rRNA) gene locus to be inadequate, and developed a new method, digital MDA (or dMDA), for direct, quantitative, measurement of contaminating DNA fragments with extremely high sensitivity. The new method may have utility in other application areas as discussed below.
MATERIALS AND METHODS
Containers and buffers
All glassware and plasticware used for cell growth, protein extraction, protein purification, digital PCR (dPCR) and dMDA were factory new or cleaned with detergent in hot water, rinsed in hot water, treated with a 0.6% hypochlorite solution and rinsed with sterile, UV-treated water. All solutions not containing nucleotides, primers, DNA templates or enzymes were obtained in the most pure form available, then filtered at 0.2 µm and UV-treated (Stratalinker model 1800, 30 min, 1″ from source on ice) after mixing in any additives. Chemical additions were made as concentrated aqueous solutions, which were similarly filtered and UV-treated.
Commercial MDA reagents
The commercial MDA reagents and enzymes used in this study were all purchased in early 2010. The products used were: Repli-G Midi (Qiagen), GenomiPhi v2 (GE) and RepliPHI (Epicentre).
Gene synthesis
A gene encoding wild-type ϕ29 DNAP with cleavable hexahistidine and glutathione S-transferase affinity purification tags fused to the N-terminus was chemically synthesized with codons optimized for over-expression in Escherichia coli (GenScript).
Protein expression
Escherichia coli BL21 DE3 cells (Invitrogen) were transformed with the pGS-21a plasmid containing the ϕ29 DNAP construct by heat shock and plated on LB-agar, selecting for ampicillin resistance. The next day, a fresh colony was used to inoculate 2 l of autoclaved Terriffic Broth containing 100 mg/ml ampicillin and 100 U DNAse I (New England Biolabs). The cells were grown to an OD600 of 0.1 at 37°C in an air incubator, when the culture was induced with 200 μM IPTG and left to grow for 6 h at 30°C. Denaturing poly(acrylamide) gel electrophoresis (PAGE, Invitrogen NuPAGE 4–12% Bis–Tris gel, 1× MES running buffer, stained with SYPRO Ruby) confirmed over-expression of a protein approximating 90 kDa in the soluble fraction of the cell lysate.
Protein extraction
Cells were harvested by centrifugation at 800 g. The cells were resuspended in 10 ml lysis buffer (0.25 × PBS, 0.2% Tween-20, 5% glycerol, 1 mM EDTA, 0.2 mg/ml PEFAbloc, 5 mM DTT). Half of the suspension was frozen and 5 ml were diluted to a volume of 40 ml in lysis buffer. The cells were broken by sonication in a glass beaker set in ice at power setting 9 using a programmable probe-type sonicator (Misonix sonicator XL). sonication treatment (150 s) was applied in 10 s bursts with 30 s between bursts. After sonication, Magnesium Chloride was added to 2.5 mM, calcium chloride to 0.5 mM, lysozyme (Hampton Research) to 2 mg/ml, imidazole to 20 mM, along with 100 U DNAse I. The lysate was incubated with slow mixing at room temperature for 10 min, after which the sample was cooled on ice and sodium chloride added to 0.25 M. The lysate was clarified by centrifugation at 20 000 g and the supernatant filtered at 0.2 µm.
Protein purification
All subsequent steps were carried out inside a PCR workstation (Airclean Systems) inside a 4°C cold room. The lysate was loaded at 1 ml/min on a new 1 ml HisTrap HP column (GE Healthcare) equilibrated with His wash buffer (0.25× PBS, 0.2% Tween-20, 5% glycerol, 2.5 mM MgCl2, 0.5 mM CaCl2, 20 mM imidazole, 0.4 M NaCl and 5 mM DTT) using a syringe pump (Harvard Apparatus PHD 2000). All chromatographic steps were carried out using disposable plastic syringes and the syringe pump. The column was washed with 10 ml His wash buffer and the product eluted with 4 ml His Elution buffer (0.25× PBS, 0.2% Tween-20, 5% glycerol, 2.5 mM MgCl2, 0.5 mM CaCl2, 300 mM imidazole, 0.25 M NaCl and 5 mM DTT) at 1 ml/min. The sample was filtered at 0.2 µm, diluted to 20 ml with 0.01 M Tris pH 8 containing 0.2% Tween-20, 2.5 mM MgCl2, 0.5 mM CaCl2 and 5% glycerol. 100 U DNAse I (New England Biolabs) and 50 U RNAse A (Sigma Aldrich) were added and the sample incubated for 10 min at room temperature. The sample was then loaded at 0.4 ml/min on a 1 ml GSTrap column (GE Healthcare) equilibrated with His wash buffer. The column was washed with 10 column volumes His wash buffer and eluted at 1 ml/min in 4 ml GST elution buffer (50 mM Tris–HCl pH 8.0, 0.1 M NaCl, 0.2% Tween-20, 5% glycerol, 2.5 mM MgCl2, 20 mM reduced glutathione). Imidazole was added to the sample to 30 mM which was then applied at 1 ml/min to a fresh 1 ml HisTrap HP column equilibrated with His wash buffer. The column was washed with 10 ml His wash buffer and the product eluted in four fractions at 1 mg/ml with 4 ml His Elution buffer.
The fractions were filtered at 0.2 µm and stored in the cold room while a PAGE gel (Invitrogen NuPAGE 4–12% Bis–Tris gel, 1× MES running buffer, stained with SYPRO Ruby) following the expression and purification was run. The gel confirmed a high concentration of the target protein in the second fraction from the final HisTrap elution. Using the calculated extinction coefficient for the product at 280 nm, 163 230/M/cm, the concentration of the 0.8 ml fraction was determined at 4.3 mg/ml, indicating a net yield of ∼3.5 mg. The total expression was estimated in excess of 30 mg/l of culture; the purification procedures were intentionally biased towards product purity at the expense of yield. No effort was made to remove the three minor low molecular weight bands, which are most likely C-terminal fragments of the desired product that co-purify on the affinity resins. The product was diluted with His wash buffer and glycerol to 1.4 mg/ml (50% glycerol, 1.0 mg/ml WT equivalent weight). An additional 0.3% Tween-20 was supplemented before filtering at 0.2 µm. Aliquots (50 μl) of the enzyme were frozen at −60°C for storage.
Activity assays
The commercial and high-purity ϕ29 DNAP reagents were tested for activity in 50 μl MDA reactions. The template was 10 000 genomic equivalents E. coli K12 genomic DNA prepared by the DNeasy method for Gram-negative cells (Qiagen), denatured by alkaline treatment and neutralized according to the GenomiPhi v2 reaction protocol for chemical denaturation. Reaction master mixes were made up according to the manufacturers’ instructions, omitting any chemical or thermal template denaturation steps and supplementing 0.3% Tween-20 and 1× SYBR GREEN I (Invitrogen), used to follow the reactions in real time during a 16 h, 30°C incubation on the MX3005-P thermocycler (Stratagene). In all cases, a single master mix was subdivided immediately prior to enzyme addition. The crude products were also analyzed by agarose gel electrophoresis to assess the presence of high molecular weight amplified DNA. Endonuclease activity was tested by incubation of enzyme samples with 1 μg ϕ × 174 virion (ss, closed circular) DNA (New England Biolabs) at 37°C for 4 h in DNAse I reaction buffer (New England Biolabs).
DNA shearing
Twenty nanograms E. coli K12 genomic DNA [again prepared by the DNeasy method (Qiagen) for Gram-negative cells] was diluted in 200 μl 20 mM Tris, pH 7.5, containing 0.05% Tween-20. The DNA was sheared using a Hydroshear device (Digilab) with the standard shearing assembly (speed code 12, 20 cycles). Shearing to a size range between 3 and 5 kb was confirmed by agarose gel electrophoresis (data not shown).
Digital PCR
Digital PCR targeting microbial ssu rRNA gene sequences was carried out in 12.765 digital arrays (Fluidigm) according to the manufacturer's protocol on a Biomark thermocycler. Reaction mixes were set up in a PCR workstation (Airclean Systems), while templates were mixed in and the chip loaded on a clean benchtop in the open lab. In all cases, a single master mix was subdivided immediately prior to enzyme addition, thorough mixing and application to the digital array sample wells. For signal generation, we used the universal Taqman scheme (16,17) and the locked nucleic acid FAM probe #149 (Roche). The broad-specificity ssu rRNA primers (18), with sequences 515F GTGCCAGCMGCCGCGGTAA, 515F-UPL GGCGGCGAGTGCCAGCMGCCGCGGTAA and 1391R GACGGGCGGTGWGTRCA, were obtained from Integrated DNA Technologies (IDT). A standard UNG (2 min, 50°C), hot-start (5 min, 95°C), two-step (95°C, 30 s; 65°C, 60 s; 45–50 cycles) thermocycling program was used. The dPCR fluorescence image presented represents the reaction endpoint and is background-subtracted, with the contrast (linearly) scaled to best depict the positive reaction chambers.
Digital MDA
Digital MDA was carried out in 12.765 digital arrays on a Biomark thermocycler (Fluidigm). Reaction master mixes were made up according to the manufacturers’ instructions in the PCR workstation, omitting template denaturation steps and supplementing 0.3% Tween-20 and 1× SYBR GREEN I (Invitrogen). No template control reactions were loaded on the chip in the PCR workstation, while template-containing solutions were loaded in the open on a clean benchtop. In all cases, a single master mix was subdivided immediately prior to enzyme addition, thorough mixing and application to the digital array sample wells. The thermal program consisted of a 16 h incubation at 30°C, with an image recorded every 15 min. Escherichia coli genomic DNA was not denatured prior to amplification. Intact λ DNA was denatured by alkaline treatment (0.4 M KOH, 10 mM EDTA) and neutralized prior to mixing with amplification reagents. Average fluorescence intensities were extracted by averaging the pixel intensity values from the relevant image regions. In all cases, the dMDA fluorescence images presented represent the reaction endpoint and are background-subtracted, with the contrast (linearly) scaled to best depict the positive reaction chambers.
RESULTS
To produce a ϕ29 DNAP sample free of nucleic acid contamination, we followed an affinity chromatography strategy, taking advantage of nuclease treatment at several steps to digest contaminating DNA and RNA. Furthermore, all glassware and plasticware were factory new or washed with detergent and hypochlorite as described in the ‘Materials and Methods’ section. All solutions were prepared from the cleanest available components, then filtered at 0.2 µm and UV-treated to eliminate contaminants and degrade remaining nucleic acids. All open-tube steps following clarification of the lysate were carried out in a PCR workstation located inside a coldroom.
We designed a synthetic gene encoding wild-type ϕ29 DNAP with dual, cleavable, N-terminal affinity tags (hexahistidine and glutathione S-transferase). The expression plasmid was transformed into E. coli, which was grown on the liter scale in DNAse-treated media and induced to over-produce the target enzyme by standard methods. The cells were lysed by sonication and lysozyme treatment and the lysate was spiked with DNAse I. The protein was purified by serial affinity chromatography steps on metal-affinity and glutathione resin. After an initial metal-affinity step with limiting resin, the preparation was treated with DNAse I and RNAse A before subsequent affinity purifications on the glutathione resin and fresh metal-affinity resin to remove the DNAse and RNAse endonuclease activities. Figure 1A shows a denaturing and reducing acrylamide gel following the steps of the purification. The major purification product occurs just below the 98 kDa marker, consistent with the expected 93 kDa molecular weight of the target construct. The bands in the purified product <63 kDa are most likely C-terminal deletions of the designed product: no effort was made to remove these. We note that affixing one tag to each end of the protein would enable selection of the full-length product using the same purification procedure. The purification netted 3.5 mg of the tagged product based on the absorbance of the sample at 280 nm.
Figure 1.
Expression and activity of the high-purity φ29 DNAP. (A) Denaturing acrylamide gel following expression and purification of affinity-tagged φ29 DNAP. Lanes show content of crude lysate, column flow-throughs (FT) and eluates. The arrow indicates the expression target. Bands in product below 63 kDa marker in the final product (second Ni affinity elution) are most likely C-terminal deletions of the designed product: no effort was made to remove these. (B) Agarose gel showing MDA products from μl-scale reactions using 10 000 GE E. coli genomic DNA as template. The variously sourced enzymes all produced high molecular weight DNA products.
To test the activity of the purified enzyme, we ran 50 μl MDA reactions using E. coli genomic DNA as a template with 1× SYBR GREEN I. All four reactions gave a strong fluorescence signal a few hours into the reaction (data not shown). Figure 1B shows the presence of high molecular weight DNA products on an agarose gel in reactions driven by the high-purity enzyme and three commercial ϕ29 DNAP preparations. This indicates that the affinity tags on our ϕ29 DNAP preparation do not eliminate the polymerase activity of the enzyme in MDA. Observing this, we chose not to cleave the tags, since the biochemical steps necessary to do so have the potential to reintroduce contamination. We also tested for residual nuclease activity by incubating 1 μg of single-stranded circular template with the enzyme for 4 h. The high-purity enzyme and the commercial preps tested negative for endonuclease activity (data not shown).
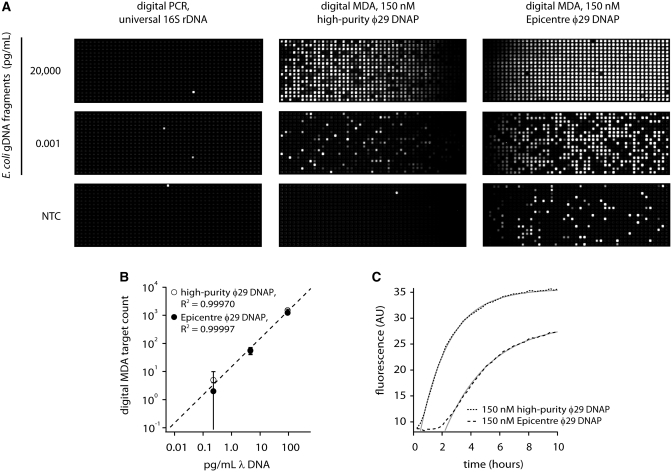
To compare the sensitivity of dMDA with dPCR for highly fragmented DNA, we prepared 3–5 kb fragments of E. coli genomic DNA and used the same fragment preparation in dPCR and dMDA assays in 12.765 digital arrays. The dPCR assay (Figure 2A), using broad-specificity ssu rRNA primers (18) shows poor sensitivity for the genomic fragments at concentrations up to nanograms per millileter, although a ssu rRNA amplicon was efficiently detected at 4000 copies per milliliter under the same conditions in the same chip (data not shown). In contrast, dMDA (Figure 2A) easily detects the fragments at reaction concentrations down to 1 fg/ml, demonstrating much improved sensitivity compared with the PCR-based assay in enumeration of low-quality DNA fragments. The scoring of positive spots is straightforward in digital MDA due to the black and white, or ‘digital’, nature of the amplification and faithful segregation of the reaction products within their chambers of origin.
Figure 2.
Digital MDA versus dPCR quantification of 3–5 kb E. coli genomic DNA fragments. (A) Digital assay panels, 765 reactions per panel. Digital PCR does not effectively detect the E. coli genome fragments (positive control panel with amplicon template not shown). All three digital PCR panels and the positive control reaction were prepared from the same reaction mix and run in a single chip. Digital MDA easily detects E. coli genome fragments down to femptograms per microliter. The Epicentre enzyme shows higher background in the no template control than the high-purity enzyme prepared in-house. All six digital MDA panels shown were prepared using the same reaction mix (except for the enzyme) and run in a single chip. (B) Calibration curve showing quantification of denatured λ DNA. Panels were scored at 1.5 h for high-purity φ29 DNAP reactions and at 4 h for Epicentre φ29 DNAP reactions. Each datapoint corresponds to a single digital assay panel with error bars that represent 95% confidence intervals based on event counting statistics. The coefficient of determination, R2, exceeded 0.999 in both cases. (C) Time-dependent appearance of digital MDA signal in digital amplification of denatured λ DNA. Both the high-purity and Epicenter φ29 DNAP preparations show strong MDA activity. All the data presented in parts (B) and (C) were obtained from a single chip run.
Figure 2A shows replicate dMDA experiments driven by the ϕ29 DNAP we prepared and a commercial preparation (Epicentre). Interestingly, the no template control (NTC) reactions give different results. The high-purity enzyme panel shows only one spot, indicating that the reagents used to make up the MDA reaction buffer (which included the ‘reaction buffer’ and ‘sample buffer’ components an exceptionally clean GE GenomiPhi v2 lot) and by extension, the high-purity enzyme, appear largely free of contamination from amplifiable DNA fragments. In contrast, the NTC reaction with Epicentre enzyme (dissolved in the same MDA reaction buffer) shows many spots, indicating a detectable level of contamination with MDA-amplifiable DNA fragments in the commercial enzyme preparation. The NTC reactions each contained 150 nM enzyme, indicating that several hundred MDA-active contaminating fragments are present per microliter (100 U or 0.1 μg) of the Epicentre enzyme. The number of contaminant molecules detected here represents a lower limit. Tiny or heavily damaged DNA fragments may not be detected and since no denaturation of the enzyme solutions was carried out (in order to avoid compromising the enzyme activity), neither would fully duplexed molecules be detected.
To assess digital MDA as a quantitative method for nucleic acid determination, we carried out assays on a monodisperse template, λ DNA. In order to maximize the sensitivity of dMDA by rendering all the template molecules susceptible to MDA, we denatured the input material by alkaline treatment prior to dilution. The results are shown in Figure 2B, which indicate a faithful linear response of digital MDA counts to λ DNA concentration using both enzyme samples.
The intensity time course from the calibration experiment provides another basis for assessment of high-purity enzyme's specific activity (Figure 2C). The measurement of ϕ29 DNAP activity is non-trivial due to the presence of two catalytic sites with opposing enzymatic activities whose relative balance depends on substrate concentrations. For strand-displacement synthesis, a third (helicase) activity enters the equation, with its own dependencies on the nature of the substrates and reaction conditions. Nonetheless, early rates of average fluorescence increase give an indication of the polymerase activity in MDA under a specific condition. The timecourse in Figure 2C shows the appearance of fluorescence in amplification of the denatured λ DNA by equimolar amounts of the high-purity ϕ29 DNAP and the Epicentre ϕ29 DNAP. Both preparations give strong amplification within the first few hours, with the signal rising faster and higher in the reactions with the high-purity ϕ29 DNAP in this instance. In other head-to-head experiments, we have observed similar timing and rates of signal increase, leading us to believe that the high-purity ϕ29 DNAP is comparable in specific activity to that which we obtained from Epicentre. Thus we assign the activity of our 15 µM stock at 1000 U/µl to match the specific molar activity of the Epicentre ϕ29 DNAP.
To directly compare contaminant levels in different preparations of ϕ29 DNAP, we ran NTC dMDA assays using enzyme from four sources and the same reaction buffer employed in Figure 2. The results were quantified by counting spots and are plotted in Figure 3B, which reveals high contamination levels in the enzyme samples obtained from all three manufacturers and very low levels in our high-purity sample. We also made a more stringent test for contamination in the high-purity ϕ29 DNAP sample by over-loading a digital MDA assay with a 10-fold higher concentration of the high-purity enzyme. In order to maintain parity in enzyme activity, the excess ϕ29 DNAP was heat-killed (65°C for 10 min). No positive spots were detected, indicating that the high-purity ϕ29 DNAP has fewer than one MDA-active contaminating fragment per 500 U. This also implies that nonzero contaminant counts observed in NTC assays with the high-purity ϕ29 DNAP preparation (visible in Figure 2, ‘high-purity’ NTC and Figure 3B, ‘high-purity’) most likely arise from a source other than the high-purity enzyme.
Figure 3.
Digital MDA on Fluidigm 12.765 digital array reveals varying levels of contamination in φ29 DNAP from three commercial providers. φ29 DNAP prepared in-house shows little contamination. A single reaction mix including exceptionally clean lots of the GE MDA ‘reaction’ and ‘sample’ buffers were used in all cases. (A) Example image of dMDA assay endpoint (all four panels are from a single chip run). Readout is digital, indicating contaminants, but not primer-derived products, underlie background amplification. (B) Quantification of dMDA assay results, indicating contaminant levels in the various enzyme preparations. The boxplots show data quartile ranges, median (black line) and outlier values (crosses). Results from several enzyme lots are pooled for each manufacturer, with 9–13 total dMDA assays per enzyme source.
The level and nature of contamination varies widely across enzyme and reaction buffer lots from all the manufacturers. Some of the reaction buffer lots we tested revealed contamination in the range of thousands of fragments per reaction microliter. The sequence-independence and high sensitivity of the dMDA assay can be used to test individual lots of enzymes and buffers for critical applications and determine the number of contaminating fragments per unit enzyme activity and per microliter reaction volume.
DISCUSSION
Digital assays for nucleic acid detection work by direct or indirect counting of analyte molecules and require single-molecule detection sensitivity. Digital assays requiring enzymatic amplification to generate higher signal levels for counting rely on the physical segregation of analyte molecules by limiting dilution such that individual analyte molecules can be separately amplified and counted (19). This is the case for both dPCR (20,21) and dMDA. We have developed a quantitative and sensitive method for assaying nucleic acid contamination based on carrying out a large number of single-molecule MDA reactions in a microfluidic device. Termed digital MDA in analog to digital PCR, the technique is useful to enumerate the number of high molecular weight MDA-active template molecules of unknown sequence in a sample.
Until development of the dMDA assay, we found the quantification of contaminating bacterial DNA in MDA reagents to be surprisingly difficult. In principle, real-time MDA could be used to quantify contaminates, although calibrating such an assay presents a problem when the template characteristics are unknown. We could achieve good sensitivity by running universal 16S QPCR on the products of NTC MDA reactions. However, because the degree of pre-amplification by MDA was unknown (both in the gross sense and in a locus-specific sense as a consequence of MDA amplification bias), the result was not quantitative and could not be effectively used to compare different reagent sets or the effects of different reagent treatments. On the other hand, if we strove for a fully quantitative readout by direct QPCR, we found no consistent signal above the baseline amplification of microbial DNA from the PCR reagents themselves. Microfluidic dPCR addresses the problem of interference by PCR reagent contamination by reducing the assay volume, but with an unacceptable tradeoff in detection sensitivity. For example, even under ideal conditions, 1000 GE of E. coli DNA per milliliter in 10 kb fragments will give only two spots per panel on the 12.765 digital array (in PCR mode) and would be undetectable on the 48.770 digital array (in PCR mode).
Because the ssu rRNA gene makes up <1% of many bacterial genomes, there is a significant sensitivity gain to be had in using the MDA reaction rather than PCR for fragment detection, since any fragment, rather than just ssu rRNA gene-containing fragments, can be detected by MDA. For determination of MDA reagent contamination, there are the further advantages that no interference from additional reagents need be introduced and that the presence of MDA-inactive substrates is of no consequence since they cannot compete with amplification of desired templates, although MDA-inactive substrates would formally contribute to false-negative counts in quantification applications. The performance advantage of dMDA over dPCR is sizable for assays of highly fragmented or damaged DNA, as demonstrated in Figure 2 and expected in specific application areas such as forensics, studies of ancient DNA and astrobiology.
The high-purity ϕ29 DNAP reagent is useful not only to support the dMDA quantification method, but also for our original goal of carrying out single-cell MDA reactions free of contamination. Table 1 indicates the expected number of contaminating DNA fragments in MDA reactions at three scales: 6, 60 and 6000 nl. The dMDA reactions presented in this paper are 6 nl in volume, while our microfluidic single-cell MDA reactions are typically 60 nl in volume, and 6 μl represents the smallest MDA reactions typically set up manually or with fluid-handling robots. Using the commercial enzyme preparations, all of the 6 μl MDA reactions will be contaminated, while more than half of the 60 nl MDA reactions could be contaminant-free, depending on the manufacturer chosen and the particular enzyme lot. We and others have made the disconcerting observation that contaminants in the commercial MDA reagents are not limited to the expression host (presumably an E. coli B strain), but are rather drawn from an eclectic, albeit stereotyped group of bacterial species, making informatic subtraction of contaminating sequences an unattractive solution.
Table 1.
Contaminating DNA fragment number as function of enzyme batch and MDA reaction volume, based on median fragment values shown in Figure 3B and the assumption that all reagents other than the enzyme are free of contaminates
| Frag/6 nl MDA reaction | Frag/60 nl MDA reaction | Frag/6 μl MDA reaction | |
|---|---|---|---|
| Qiagena | 0.040 | 0.398 | 39.8 |
| GEa | 0.044 | 0.444 | 44.4 |
| Epicentreb | 0.023 | 0.233 | 23.3 |
| High-purityb | 0.000 | 0.002 | 0.24 |
With the high-purity enzyme, even microliter-scale MDA reactions free of contamination are possible. The commercial suppliers of MDA reagents typically specify 10 ng as the minimum quantity of template material. This limitation is not imposed by the sensitivity of MDA, but rather by the competing amplification of contaminants that reduce the amplification yield from the intended template material. The ability to amplify smaller amounts of template contaminant-free is a critical capability for single-cell genomics and other applications where a small amount of template material needs to be specifically amplified in a sequence-independent manner.
In the 12.765 digital array, with a clean ϕ29 DNAP preparation, the dMDA limit of detection is below 1 fragment per μl and the limit of quantification (10 positives per d12 panel) is two fragments per microliter (∼1 fg/ml for 2 kb fragments) or about one bacterial GE per microliter given uniformly-sized fragments. The fact that we see a somewhat larger number of spots than expected for uniform fragments indicates that smaller fragments exist in our E. coli genomic sample, that dMDA can detect these smaller fragments with high efficiency, and that the practical quantification limit for highly fragmented bacterial DNA is much better than one GE per microliter. The LOD and LOQ values can be pushed even lower by using more than one panel per sample on the 12.765 chips or by increasing the assay volume in a different microfluidic device or emulsion-based platform. Assuring a low reagent background, including low contamination of the DNA polymerase, is key to realize the improved sensitivity in a larger assay volume. Conversely, shrinking the compartment volume is called for in dMDA quantification of nucleic acids at higher concentrations without dilution.
The LOQ <1 fg/ml demonstrated here for dMDA compares favorably with chemical, immunologic and PCR-based assays for organismal DNA. With respect to PCR detection of DNA fragments, the factor by which dMDA is expected to outperform dPCR depends on several features of the analyte, including PCR target locus density, the DNA fragment size and the template quality. For example, taking bacterial DNA in 4 kb fragments, with a PCR target locus density of 1 per Megabyte (typical of ssu rRNA genes), under idealized circumstances, there exist 250 analyte molecules for dMDA for each PCR-active analyte molecule, implying two to three orders of magnitude better sensitivity for the MDA-based assay. However, our real-world data show an even greater performance advantage for dMDA (Figure 2). This is due to the fact that dMDA comes closer to its theoretical potential than does dPCR on the sheared E. coli test sample. First, the shearing operation creates double-stranded breaks at random locations in the template DNA, reducing the fraction of fragments carrying the intact target locus for PCR. Second, it is likely that nicks are introduced by the harsh shearing condition in addition to double-stranded breaks. Since we carried out no steps to repair the template and diluted the template at low ionic strength, the presence of nicks is likely to reduce further the fraction of PCR-amplifiable fragments. The requirements for intact target loci and longer priming sites render PCR-based quantification more sensitive to template quality than MDA-based assays.
Digital MDA also provides new insights into the MDA process itself. For instance, we can test hypotheses about the source(s) of background amplification in microliter-scale MDA reactions. The digital nature of the dMDA results (i.e. bimodal distribution of spot intensities) and template concentration-dependence in observed in experiments with E. coli genomic DNA fragments strongly implicate high molecular weight contaminants, but not primer-primer interactions, as the source of background amplification. The bimodal distribution of spot intensities cannot be explained by varying combinations of random primer sequences in different nanoliter MDA reactions. For perfectly random hexamers at 50 μM, the 6 nl wells in the 12.765 digital array each contain 150 billion primer molecules. Since there are 4096 6-mer sequence variants, each sequence is represented by nearly 50 million copies in each reaction well. According to random sampling statistics, the relative variation in concentration of sequence variants from well to well is <0.02%. Such small differences in representation are unlikely to explain the observed variation of SYBR green fluorescence intensity in the dMDA assay. The fact that the background level of SYBR green in negative spots does not increase over the course of the reaction is also inconsistent with the generation of high molecular weight product from primers alone in the GE and Qiagen reaction mixes. Thus, we are able to show that the background amplification indeed arises from high molecular weight contaminants and is not intrinsic to the MDA reagents.
This realization, in combination with the application of dMDA as a means of counting nucleic acid fragments in a sequence-independent manner, opens up new possibilities for the ultra-sensitive detection of DNA in various contexts. Often, in assays for the presence of microbes, ssu rRNA-encoding DNA serves as a proxy analyte for the organisms themselves. Digital MDA promises to improve the sensitivity of such assays by orders of magnitude in contexts where the loss of sequence-specificity can be tolerated or constitutes an advantage. The dMDA assay extends several features of the dPCR assay to MDA, including absolute quantification that requires no standard curve. Digital MDA demonstrates superior sensitivity to dPCR in this application because every MDA-active DNA fragment is detected, rather than some fraction containing a particular, intact, sequence locus (which may be a tiny fraction). Digital MDA of whole genomes or chromosomes from cells represents the ultimate goal of sample preparation for single-cell genomics. Capabilities for cell lysis and product recovery need to be integrated to access this application. Forthcoming innovations in hardware platforms intended for digital PCR will improve performance (especially sample throughput and dynamic range) while driving down costs for both digital PCR and digital MDA.
The ability to conduct contaminant-free MDA has important implications. For instance, positive and negative amplification results are more obviously apparent; small quantities of template DNA can be amplified with high fractional yield; and the reaction products can be analyzed without the false-positive signals or interfering sequences that arise from contaminants. In addition, there may be benefits of compartmentalization for certain studies that rely on MDA for preparative amplifications. MDA sequence and length bias may be reduced in metagenomic or library-based applications by eliminating competition among template molecules for amplification reagents. For example, a high-amplification-efficiency template molecule is limited to the reagents inside its microchamber, while a low-amplification-efficiency template molecule can be given extra time to catch up as it utilizes a privileged supply of reagents inside its own microchamber. Emulsion dMDA may prove to be another effective way to access the advantages of compartmentalized MDA.
Here, we introduced a new method, dMDA, for quantitatively enumerating the number of high molecular weight DNA fragments in a sample. We demonstrated the dMDA limit of quantification at 1 fg/ml for 2 kb fragments or less than one (fragmented) bacterial GE per microliter and identified a straightforward development path toward even better sensitivity. Digital MDA allows the enumeration of DNA fragments and other MDA-active DNA templates (such as plasmids and single- or double-stranded minicircles) on an absolute basis, without a standard curve. Furthermore, the digital nature of the microfluidic MDA results is consistent with high molecular weight DNA contaminants as the sole source of the so-called ‘template-independent’ MDA background. This method has applications in quality control of pharmaceuticals, biological and chemical reagents (especially DNA-modifying enzymes and assay components), as well as in characterization/quantification of DNA libraries, water quality testing, surveillance of industrial reactors, counter-bioterrorism, forensics, space exploration and astrobiology. Digital MDA is well-suited for analysis at the site of sample collection due to the low power requirements for isothermal incubation and straightforward interpretation of the raw data.
We also overproduced and purified a tagged ϕ29 DNAP with extremely low levels of nucleic acid contamination. Such a clean preparation of a strand-displacing polymerase is required to realize the low dMDA LOQ and LOD values quoted above. We characterized contamination in commercial lots of ϕ29 DNAP, finding levels sufficient to interfere with sensitive applications (such as single cell genomics or quantification of DNA fragments at concentrations <10 fg/ml) in every lot from the three vendors tested. High contaminant levels were also observed in most lots of MDA reaction buffers. We recommend that suppliers of reagents for ultrasensitive molecular biology applications use dMDA to quantify the concentration of contaminating DNA fragments and report the detected level in units of contaminating fragments per unit enzyme activity and fragments per reaction microliter to the end user. No contamination was detected above the dMDA LOD in the high-purity ϕ29 DNAP sample prepared in-house.
FUNDING
The National Institutes of Health funded this study through a Director's Pioneer Award (DP1OD000251 to S.R.Q.) and an R01 (5R01HG004863-02). The Howard Hughes Medical Intitute funded the open access charge. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Conflict of interest statement. This work has potential commercial applications, and Stanford University may file a patent application based on the results presented here. Furthermore, we demonstrate a new method using an instrument manufactured by Fluidigm, a company with which one of us (S.R.Q.) has financial relationships.
ACKNOWLEDGEMENTS
The authors thank Geoffrey Schiebinger for assistance with implementation of an earlier form of quantitative MDA, Joanna Tsai and Anastasia Potanina for assistance in carrying out digital MDA assays, and anonymous reviewers for insightful comments that helped improve this article.
REFERENCES
- 1.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dietmaier W, Hartmann A, Wallinger S, Heinmoller E, Kerner T, Endl E, Jauch K, Hofstadter F, Ruschoff J. Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am. J. Pathol. 1999;154:83. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Telenius H, Carter N, Bebb C. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. 1992;89:5847. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lao K, Xu N, Straus N. Whole genome amplification using single-primer PCR. Biotechnol. J. 2008;3:378–382. doi: 10.1002/biot.200700253. [DOI] [PubMed] [Google Scholar]
- 6.Pinard R, De Winter A, Sarkis G, Gerstein M, Tartaro K, Plant R, Egholm M, Rothberg J, Leamon J. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorensen K, Turteltaub K, Vrankovich G, Williams J, Christian A. Whole-genome amplification of DNA from residual cells left by incidental contact. Anal. Biochem. 2004;324:312–314. doi: 10.1016/j.ab.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL, Beeson KY, Goldberg SM, Quake SR. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl Acad. Sci. USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang H, Sung Y, Kim K, Roh S, Kim M, Jeon C, Bae J. Development of microbial genome-probing microarrays using digital multiple displacement amplification of uncultivated microbial single cells. Environ. Sci. Technol. 2008;42:6058–6064. doi: 10.1021/es8006029. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigue S, Malmstrom RR, Berlin AM, Birren BW, Henn MR, Chisholm SW. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, Lapidus A, Wu D, McCutcheon JP, McDonald BR, et al. One bacterial cell, one complete genome. PLoS ONE. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulakakis N, Parmakelis A, Lymberakis P, Mylonas M, Zouros E, Reese D, Glaberman S, Caccone A. Ancient DNA forces reconsideration of evolutionary history of Mediterranean pygmy elephantids. Biol. Lett. 2006;2:451. doi: 10.1098/rsbl.2006.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handyside A, Robinson M, Simpson R, Omar M, Shaw M, Grudzinskas J, Rutherford A. Isothermal whole genome amplification from single and small numbers of cells: a new era for preimplantation genetic diagnosis of inherited disease. Mol. Hum. Reprod. 2004;10:767. doi: 10.1093/molehr/gah101. [DOI] [PubMed] [Google Scholar]
- 15.Pan X, Urban A, Palejev D, Schulz V, Grubert F, Hu Y, Snyder M, Weissman S. A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc. Natl Acad. Sci. 2008;105:15499. doi: 10.1073/pnas.0808028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhang D, Li W, Chen J, Peng Y, Cao W. A novel real-time quantitative PCR method using attached universal template probe. Nucleic Acids Res. 2003;31:e123. doi: 10.1093/nar/gng123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White RA, 3rd, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane D, Pace B, Olsen G, Stahl D, Sogin M, Pace N. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl Acad. Sci. USA. 1985;82:6955. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sykes P, Neoh S, Brisco M, Hughes E, Condon J, Morley A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 20.Kalinina O, Lebedeva I, Brown J, Silver J. Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res. 1997;25:1999. doi: 10.1093/nar/25.10.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogelstein B, Kinzler K. Digital PCR. Proc. Natl Acad. Sci. USA. 1999;96:9236. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]