Examining the global distribution of dominant archaeal populations in soil (original) (raw)
Abstract
Archaea, primarily Crenarchaeota, are common in soil; however, the structure of soil archaeal communities and the factors regulating their diversity and abundance remain poorly understood. Here, we used barcoded pyrosequencing to comprehensively survey archaeal and bacterial communities in 146 soils, representing a multitude of soil and ecosystem types from across the globe. Relative archaeal abundance, the percentage of all 16S rRNA gene sequences recovered that were archaeal, averaged 2% across all soils and ranged from 0% to >10% in individual soils. Soil C:N ratio was the only factor consistently correlated with archaeal relative abundances, being higher in soils with lower C:N ratios. Soil archaea communities were dominated by just two phylotypes from a constrained clade within the Crenarchaeota, which together accounted for >70% of all archaeal sequences obtained in the survey. As one of these phylotypes was closely related to a previously identified putative ammonia oxidizer, we sampled from two long-term nitrogen (N) addition experiments to determine if this taxon responds to experimental manipulations of N availability. Contrary to expectations, the abundance of this dominant taxon, as well as archaea overall, tended to decline with increasing N. This trend was coupled with a concurrent increase in known N-oxidizing bacteria, suggesting competitive interactions between these groups.
Keywords: archaea, Crenarchaeota, microbial ecology, pyrosequencing, soil
Introduction
Since the formal recognition of the archaeal domain around two decades ago (Woese et al., 1990), knowledge of the biology, diversity and ecology of this microbial group has grown considerably. Archaea have been found in a wide variety of habitats including hydrothermal vents (for example, Ehrhardt et al., 2007), marine waters (for example, DeLong, 1992), hypersaline sediments (for example, Demergasso et al., 2004), freshwater sediments (for example, Schleper et al., 1997) and soil environments (for example, Bintrim et al., 1997; Buckley et al., 1998; Oline et al., 2006). These advances have largely been fueled by the advent of PCR-based molecular techniques, which have permitted the detection of archaea in samples without cultivation and have improved our knowledge of this domain of life. Recent progress has included the recognition of novel archaeal lineages, the development of new functional markers and probes, the cultivation of over 50 different archaeal strains, and the sequencing of several archaeal genomes, including that of an ammonia-oxidizing group 1 archaeon (see Schleper et al., 2005; Walker et al., 2010).
Although our knowledge of the group is expanding, we still have an inadequate understanding of terrestrial archaea. Previous surveys of soil archaea have typically examined a limited number of samples or sites, often focusing on only a single category of soils, such as agricultural soils (Buckley et al., 1998; Furlong et al., 2002; Gattinger et al., 2007) or soils from extreme environments including deserts and recently deglaciated sites (Nicol et al., 2006; Pointing et al., 2009; Soule et al., 2009). The most comprehensive survey of archaeal populations to date (Auguet et al., 2009) provided valuable insight into broad-scale ecological patterns exhibited by the archaeal domain in general. That investigation, however, did not specifically focus on terrestrial archaea, nor did it examine in any detail the variables that might affect archaeal abundance in soils. The picture emerging from these, and other, studies is that soils are typically dominated by a few groups of Crenarchaeota (Auguet et al., 2009), containing members that are possibly key players in soil nitrification (Leininger et al., 2006; Nicol and Schleper, 2006).
There has been much speculation regarding the factors that influence soil-dwelling archaeal communities, their diversity, relative abundance and the functional roles that they have in terrestrial environments. Recent investigations have reached varying conclusions about the environmental variables, which structure archaeal communities, suggesting that these communities can be influenced by pH (for example, Nicol et al., 2008), elevation (for example, Zhang et al., 2009) or climate and vegetation cover (for example, Angel et al., 2009). Similarly, there are numerous lines of evidence suggesting that archaea may function as important soil nitrifiers (Treusch et al., 2005; Leininger et al., 2006; Nicol and Schleper, 2006), however, studies that have focused specifically on this question (for example, Nicol et al., 2004; Le Roux et al., 2008; Tourna et al., 2008; Di et al., 2009; Jia and Conrad, 2009; Offre et al., 2009; Schauss et al., 2009), have produced divergent results regarding the overall importance of archaeal-mediated ammonia oxidization in soil.
It is important to note, however, that inferences in the above mentioned studies have typically been drawn from either a relatively limited number of soils or from soils collected across a single environmental gradient. With the ongoing development of high-throughput sequencing technologies, we can now address these knowledge gaps by comprehensively characterizing microbial communities in large numbers of individual soils to examine global trends for soil microbes. Recent studies have demonstrated the value of using these ‘next generation' techniques in identifying broad ecological patterns that govern the structure and diversity of soil bacteria (Jones et al., 2009; Lauber et al., 2009).
Here, we utilized barcoded pyrosequencing targeting the 16S ribosomal RNA (rRNA) gene to consistently and thoroughly survey a large number of soils (nearly 150 samples, resulting in over 150 000 archaeal and bacterial 16S sequences, with over 2500 of these corresponding to archaea), which represent soils from a wide variety of ecosystem types (from Antarctic dry valleys to tropical forests of South America). From the pyrosequencing data, we directly determined the relative abundance of archaea as well as the general structure of dominant archaeal populations in soil, and investigated how these factors may be influenced by soil and site characteristics. Contained within our data set are specific sample subsets (for example, nitrogen (N) addition plots, latitudinal transects within a biome) that permit more focused examinations of how different environmental factors may influence the relative abundances of soil archaeal taxa in situ. As the one of the more wide-ranging examinations of the global-scale patterns exhibited by soil archaea to date, this work addresses the following questions: (i) What are the dominant archaeal taxa in soil? (ii) How does the relative abundance and occurrence of dominant soil archaea vary across a wide range of biomes? (iii) Do key soil archaea inhabit a particular niche, or set of niches, which are predictable based on soil edaphic or site characteristics? (iv) How do dominant archaeal populations in soil respond to inorganic N amendments in situ?
Materials and methods
Collection site characterization, sampling, and isolation of soil DNA
Soil samples were collected from 146 sites across North and South America and Antarctica. These sites represent a broad variety of ecosystem, climate and soil types. We also sampled soils across a latitudinal gradient of native tallgrass prairie sites within the United States. Finally, we included samples from two long-term N fertilization experiments located at US Long-Term Ecological Research sites with plots receiving 0, 100 and 280–290 kg N ha−1 year−1: Cedar Creek (CC in MN, USA—Experimental N Gradient Exp 001; for example, see Tilman, 1987) and Kellogg Biological Station (KBS in MI, USA—N Fertility Gradient; for example, see Buckley et al., 1998). Data on the location, climate and vegetation cover for each site included in this study are provided in Supplementary Table S1. The methods used to collect edaphic and environmental data have been described previously (Fierer and Jackson, 2006).
The soil collection protocol followed that of Fierer and Jackson (2006). Briefly, mineral soil samples were gathered from relatively undisturbed sites (with the exception of the experimental plots at CC and KBS) around the time of the peak growing season for vascular plants (with the exception of the Antarctic sites, which were basically plant-free). As our objective was to examine the variability in archaeal communities across a wide range of sites, rather than an examination of the spatial and temporal variability within individual plots, DNA was extracted and amplified from a single composited soil sample from each site. Approximately 10 g of soil was removed from the sample and homogenized under liquid N2 using a mortar and pestle. DNA was then extracted from a 0.25 g subsample using a procedure described previously (Lauber et al., 2009).
PCR-amplification of bacterial/archaeal rRNA genes and barcoded pyrosequencing
Preparation of extracted DNAs for pyrosequencing followed the protocol described by Fierer et al. (2008). Briefly, the method includes targeted amplification of a portion of the 16S small-subunit ribosomal gene, triplicate PCR-product pooling (per sample) to mitigate reaction-level PCR-biases, and Roche 454 pyrosequencing. PCR amplification used the primers F515 (5′-GTGCCAGCMGCCGCGGTAA-3′) and R806 (5′-GGACTACVSGGGTATCTAAT-3′). This primer set was developed for this project and was designed to be universal for nearly all bacterial and archaeal taxa. We have demonstrated in silico that this primer set should amplify 16S rRNA genes from a broad range of archaeal and bacterial groups with few biases or excluded taxa (see Supplementary Figures S1 and S2). In spite of the short-read lengths (∼250 bp), this targeted gene region should also provide sufficient resolution for the accurate taxonomic classification of microbial sequences (Liu et al., 2007). The F515 primer included a Roche 454-A pyrosequencing adapter (Roche Applied Science, Branford, CT, USA) and a ‘GT' linker sequence, although R806 incorporated a 12-bp barcode sequence (unique to each individual sample), a ‘GG' linker, and a Roche 454-B sequencing adapter.
PCR reactions were performed in 25 μl reactions, each containing 2 μl (15 μ concentration) of forward and reverse primers, 10 μl of 5Prime Hot MasterMix (Eppendorf-5Prime Inc., Gaithersburg, MD, USA) and 1 μl of genomic community DNA as a template; using the following cycling parameters: 35 cycles (95 °C, 30 s; 50 °C, 1 min; 72 °C, 1 min) after an initial denaturation 95 °C, 3 min. Pooled triplicate reactions were purified using the UltraClean PCR clean-up kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) and then quantified using PicoGreen dsDNA assay (Invitrogen, Carlsbad, CA, USA). A single composite sample was produced that contained barcoded PCR product, normalized in equimolar amounts to produce equivalent sequencing depth from all samples. The sample was sent for sequencing at EnGenCore (University of South Carolina) on a Roche GS-FLX 454 automated pyrosequencer.
Sequence processing, assigning taxonomic identity, abundance and phylogeny
Before analyses, raw sequence data generated from the 454-sequencing runs were processed using QIIME (Caporaso et al., 2010). In brief, quality sequences were binned into phylotypes (⩾97% similarity) using Cd-hit (Li and Godzik, 2006) and grouped by samples according to their unique 12-bp barcode. Phylotypes were assigned an identity based on comparisons with sequences in the Ribosomal Database Project (Cole et al., 2005). In order to assess the phylogenetic placement of archaeal sequences obtained from soil samples, a composite alignment was created by first assembling representative sequences of each archaeal phylotype identified. These were then aligned using the NAST alignment function of the GreenGenes public database (http://greengenes.lbl.gov), applying the PH Lane Mask. The sequence alignment was then used to construct bootstrapped neighbor-joining phylogenetic trees, recommended for inferring phylogenies for large sets of sequences (Price et al., 2009), in the MEGA 4 software package (Tamura et al., 2007).
We defined relative archaeal abundances according to the total number of individual archaeal sequences (each corresponding to a specific phylotype) obtained from each soil sample (determined by the sequence barcode that corresponded to a specific sample). From these data, the relative archaeal abundance for each soil was calculated as a percentage: the number of archaeal sequences divided by the total number of 16S rRNA gene sequences (archaeal+bacterial) obtained from each soil sample, and multiplied by 100.
Clone libraries of archaeal 16S rRNA genes
In order to independently verify our pyrosequencing results and to obtain longer sequence reads, we constructed clone libraries from a subset of our soils. For these libraries, we used the archaeal-specific primers A2Fb (López-García et al., 2002) and U1406R (Reysenbach and Pace, 1994) to produce archaeal PCR-amplicons. These primers have been shown to effectively amplify a broad range of archaeal groups when using an optimized cycling protocol (Baker et al., 2003), which we used here. One clone library was constructed for each of three composited soil samples using the pooled products from triplicate PCR reactions. Clone libraries were then constructed using a TOPO TA Cloning Kit (Invitrogen) according to the manufacturer's specifications. The cloned amplicons were sequenced in the forward and reverse directions at a commercial facility. Contiguous sequences were assembled from the sequences obtained, checked for quality, and then binned into phylotypes (⩾97% similarity) using FastGroupII (Yu et al., 2006). A second alignment was then produced for these sequences, and phylogeny was assessed using the methods described above.
Statistical analyses
Parametric Pearson's correlation coefficients were calculated to examine trends between relative archaeal abundances and site/soil characteristics or relative bacterial abundances (calculated in the same manner as for archaea, see above). Archaeal relative abundance data were transformed (Log (x+1)) when required in order to satisfy the assumption of normality. All correlations (including tests for significance) and multiple regression analyses as well as analysis of variance or Kruskal–Wallis tests (including the Levene's test to verify homoscedasticity) were performed using R statistical software (http://www.r-project.org/).
Results and discussion
Dominant archaea recovered from soil samples
A total of 159 714 high-quality sequences (∼250 bp on average) were obtained from the 146 soil samples in the pyrosequencing run, at an average of 1094 sequences per soil and with coverage ranging from 813 to 1497 reads per sample. With this level of coverage, the full extent of microbial diversity has not been surveyed; however, previous work (Fierer et al., 2008; Jones et al., 2009; Lauber et al., 2009; Miller et al., 2009) has shown that patterns of beta diversity and overall taxon relative abundances of dominant lineages can be accurately inferred with this depth of sequencing. Of these reads, 159 618 (99%) could be confidently identified, the majority to the level of family or genus. Across all samples, we identified over 15 000 phylotypes (defined at the 97% sequence similarity level). Here, we largely restrict our analyses to the archaeal sequences, determining the relative abundance of archaea (number of archaeal sequences as a percentage of the total number for bacteria and archaea) to gain insight into factors that influence dominant populations across these soils.
Of the total number of sequences obtained, 2672 (1.7% of all sequences) represented lineages from the domain Archaea, and of the 146 soils sampled, archaeal populations were large enough to be detected by our methods in 127 (87% of the samples) soils. The phylogenetic placement of the archaeal phylotypes represented by these sequences is shown in Figure 1. As only the most abundant archaea were recovered by our method, it should be stressed that the phylogenetic tree represents relationships among the most dominant archaea and does not depict the full extent of archaeal diversity in these soils, which is potentially much greater (Fierer et al., 2007). The majority of the archaeal phylotypes (90.9% of all archaeal sequences) were contained within the group 1.1b crenarchaeotal clade (Jurgens et al., 2000), which has been shown previously to be dominant in soils (Auguet et al., 2009).
Figure 1.
Neighbor-joining tree based on the alignment of 16S rRNA gene sequences (∼250-bp long) showing the relationship between archaeal phylotypes (PT) recovered from samples of 146 soils by pyrosequencing. The dominant soil Crenarchaeota (DSC1 and DSC2) are indicated along with a basic classification for clades within the Archaea. Sequences of representative archaeal isolates (and clone 54d9) have been included and their GenBank accession numbers are given. For simplicity, some well-supported clades have been collapsed at their nodes. The tree is rooted with a bacterial phylotype (PT 01742) recovered in our study and consensus bootstrap confidence levels are indicated if >60%.
Interestingly, our soil samples were overwhelmingly dominated by just a few of the group 1.1b crenarchaeotal phylotypes (see Supplementary Figure S3). One phylotype (defined at the ⩾97% similarity level) was quite abundant, having been detected in 73 (50%) of the soils sampled and representing approximately 46% of all archaeal sequences recovered across the 146 soils. This phylotype is designated here as ‘dominant soil crenarchaeota number 1′ (DSC1). A second phylotype was more frequently encountered, having been found in 92 (63%) of the soils, but less abundant than DSC1, representing 27% of all archaeal sequences in these soils. This phylotype is designated here as ‘dominant soil crenarchaeota number 2′ (DSC2). In total, 101 soils (79.5% of samples with detectable populations of archaea) contained either DSC1 or DSC2; however, in only 60 of these soils (59.4%) could both dominant phylotypes be found together. Collectively, these two individual phylotypes dominated the archaeal communities, accounting for >70% of the archaeal sequences overall. DSC1 and DSC2 were not closely related to the only known group 1.1b crenarchaeotal isolate Candidatus Nitrososphaera gargensis (from hot springs; Hatzenpichler et al., 2008); however, they did form a tight clade (Figure 1) with the uncultured soil clone ‘54d9' (a large genomic fragment obtained from an soil fosmid library that included the entire 16S/23S rRNA gene; Ochsenreiter et al., 2003; Treusch et al., 2005).
These results were independently verified with archaeal clone libraries of 92 sequences constructed from a subset of three soils (samples AR3, CF1 and CO3). The short sequence reads (∼250 bp) representing DSC1 and DSC2 obtained by pyrosequencing could also be confidently placed within the alignment (nearly identical in this overlapping segment) of the considerably longer (∼1300 bp) clone library sequences. Thus, the presence of phylotypes of DSC1 and DSC2 within the clone library sequences was confirmed. Longer clone library sequence reads representing the DSC1 phylotype were also identical to the soil clone 54d9 (see Supplementary Figure S4) at the ⩾97% similarity level. The general archaeal community structure revealed by these clone libraries also mirrored that recovered by pyrosequencing (see Supplementary Figure S4 and Figure 2). For example, the DSC1 phylotype represented 83% of the clone library sequences for sample AR3 (see Supplementary Figure S4), which reflected the high relative abundance of this phylotype indicated by pyrosequencing for the same sample (see AR3 in Figure 2). Similarly, both clone library and pyrosequencing suggested the dominance of crenarcheotal sequences representing phylotypes other than DSC1 and DSC2 in sample CF1. Finally, both methods revealed the more equitable distribution of DSC1, DSC2 and other crenarcheotal phlotypes in sample CO3.
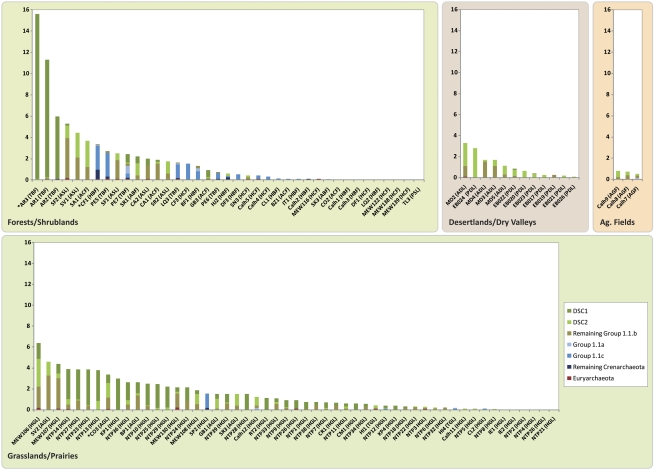
Figure 2.
Relative archaeal abundances (on the y axis as archaeal % of all 16S rRNA gene sequences in each sample) grouped by general biome for each of the non-experimental soil samples (those excluding experimental N addition sites at CC and KBS; *soil samples used for clone libraries). The portions represented by the dominant soil Crenarchaeota (DSC1 and DSC2) as well as for remaining Crenarchaeotal and Euryarchaeotal phylotypes are indicated. More specific biome vegetation/climate classes are given on the x axis, as is the site of origin for each sample. AGF, agricultural field; ACF, arid/semi-arid conifer forest; ABF, arid/semi-arid broadleaf forest; ADL, arid/semi-arid desert land; AGL, arid/semi-arid grassland; ASL, arid/semi-arid shrubland; HCF, humid conifer forest; HBF, humid broadleaf forest; HGL, humid grassland/prairie; PDL, polar desert land; TBF, tropical broadleaf forest; TGL, tropical grassland.
Our finding that archaea are nearly ubiquitous in soil agrees with other studies, which have identified archaeal DNA in a wide variety of soils (for example, Leininger et al., 2006; Oline et al., 2006; Auguet et al., 2009), including even the extreme environment of Antarctic dry valleys (see Cary et al., 2010). Although the dominance of the crenarchaeotal group 1.1b in terrestrial systems has been noted previously (Buckley et al., 1998; Ochsenreiter et al., 2003; Auguet et al., 2009), to the best of our knowledge this is the first study to show the extensive dominance of just two phylotypes, within a very phylogenetically restricted group 1.1b clade, from archaeal communities across a broad range of soils and environments. The close relationship of these phylotypes, especially for DSC1, to the clone 54d9 (recovered from a calcareous grassland soil; Ochsenreiter et al., 2003) suggests a potential functional role as important soil ammonia oxidizers, as this clone was shown to contain genes encoding ammonia monooxygenase (Amo)-related proteins (Treusch et al., 2005).
Although our primer set should amplify 16S rRNA genes from a broad variety of archaeal groups with few biases against specific taxa (see Supplementary Figures S1 and S2), archaea from groups outside of group 1.1b Crenarchaeota were poorly represented in these soils. For example, only 6% of all archaeal sequences represented group 1.1c, which has been found to be abundant in forest soils as well as freshwater systems (Ochsenreiter et al., 2003). Other crenarchaeotal groups (for example, group 1.1a) were recovered here; however, they were represented by low sequence numbers in only a few soil samples. Similarly, euryarchaeotal sequences were very rare in our data set having been recovered in fewer than 20 soil samples and representing only 1.5% of all archaeal sequences. A greater diversity and abundance of these archaeal groups may actually be present in our samples but they could not be detected with the methods employed here given that we only averaged 1094 sequence reads per sample. Although limitations are inherent when using molecular techniques for microbial community surveys (Bent and Forney, 2008), the further development of ‘next generation' sequencing techniques (for example Caporaso et al., in press) will improve our ability to characterize the full extent of archaeal diversity and abundance in soil.
Archaeal relative abundance and correlation analysis for non-experimental soils
Our pyrosequencing-based survey recovered detectable archaeal populations in a high percentage (87%) of the soils examined, with the relative abundance of archaea averaging nearly 2% across all soils. In those soils wherein archaea were detected, relative archaeal abundances ranged from 0.08% to 15.6%, with just five soil samples having over 5% archaea (Figure 2). Of these five, three samples were from wet tropical forests in Argentina. The remaining two soils were from a high-elevation meadow in Colorado and a semi-arid shrubland in New Mexico, USA. There was a high degree of variability in relative archaeal abundances and no single vegetation or biome type consistently had higher soil relative archaeal abundances than any other (Figure 2). This intra-biome variability is evident across the native tallgrass prairie soils (designated in Figure 2 by ‘NTP'), samples collected from a latitudinal gradient within a single biome and vegetation type, wherein archaeal relative abundances ranged from among the highest levels (3.9%) to individual samples in which archaeal populations were not detected. If we examine only oxisols from tropical forests in South America (AR1–3, PE5–7 in Figure 2), we find some samples dominated by DSC1, and others by group 1.1c. This high degree of variability highlights the risks associated with using smaller sample sets to reach conclusions regarding the environmental factors regulating archaeal relative abundances in soil. For example, if the Argentinean soils had been the only tropical forest soils included in our sample set, we would have mistakenly concluded that archaea are far more abundant in tropical soils than in those from other biomes.
Many of the soil or site characteristics that were correlated with relative archaeal abundances differed, some diametrically opposing, when examining grassland and forest/shrubland soils separately (see Table 1), which suggests vegetation type modulates the relationship between relative archaeal abundances and environmental factors. Particular groups of aquatic archaea are known to dominate specific niches defined by discreet environmental parameters (for example, non-saline vs saline habitats) perhaps representing habitat filtering among various archaeal groups (Auguet et al., 2009). For soil archaea we did see some evidence for this phenomenon; however, it was reflected only in the general trends observed (see Figure 2). For example, group 1.1c crenarchaeota were found almost exclusively in forest/shrubland soils (present in 42% of samples with detectable archaeal populations), typically when DSC1 and DSC2 were not present, whereas group 1.1c were basically absent from other soils from non-forested sites (present in only 7% of samples with detectable archaeal populations). Similarly, the dominant phylotype DSC1 tended to prefer grassland/prairie soils (present in 73% of samples with detectable archaeal populations) over forest/shrubland soils (present in 28% of samples with detectable archaeal populations). On the other hand, DSC2 had a more equal distribution between forest/shrubland soils (present in 53% of samples with detectable archaeal populations) and grassland/prairie (present in 66% of samples with detectable archaeal populations).
Table 1. Correlations between relative archaeal abundances (as archaeal percentage of all 16S rRNA gene sequences, Log (x+1) transformed) and soil/site characteristics for non-experimental (CC and KBS sites excluded) soil samples.
| | Correlation coefficients | | | | | ------------------------------ | ------------------------------- | -------------------------------- | ----------- | | | All soilsa | Tallgrass prairieb | Forests/shrublandsc | | | Edaphic properties | | | | | C:N ratio | −0.43*** | −0.55** | −0.59*** | | Total organic carbon | −0.23* | 0.22 | −0.27 | | Total nitrogen | −0.10 | 0.32 | −0.10 | | pH | 0.19* | 0.19 | 0.36* | | % Silt+clay | 0.17 | −0.09 | 0.37* | | | | | | | | Site characteristics | | | | | Latitude (degrees, minutes) | −0.09 | 0.38* | −0.61*** | | Mean annual precipitation (mm) | 0.002 | −0.59*** | 0.09 | | Mean annual temperature (°C) | 0.21* | −0.39* | 0.46*** |
Soil C:N ratio was the only measured variable that was consistently correlated with relative archaeal abundances across all data sets, a result that parallels previous research showing that C:N ratios were strongly related to the composition of soil archaeal communities at the landscape scale (Nielsen et al., 2010). The pattern that relative archaeal abundances were typically higher in soils with lower C:N ratios was highly significant in all cases (Table 1). However, we note that soil C:N ratio was not the only variable correlated with archaeal relative abundances and C:N ratio alone only explained a portion of the variability in relative archaeal abundance across all samples or sample subsets. As with nearly all studies that utilize broad biogeographical surveys, it is difficult to ascertain which specific environmental variables are driving the observed biological patterns because many of the soil and site characteristics are unavoidably correlated with one another. As C:N ratios provide a relative index of soil nutrient status, this correlation may suggest the response of archaeal communities to soil carbon inputs; however, it is equally probable that these populations are being influenced by an increasing availability of N. We examined N availability in more detail with our experimental N-amended plots of CC and KBS.
Archaeal relative abundance and correlation analysis for N-amended soils
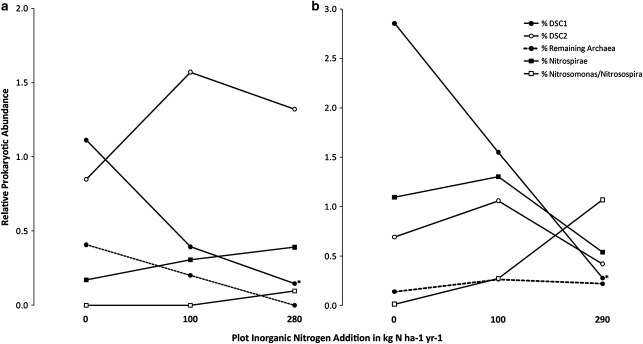
Considering the potential important role of archaea in soil nitrification, suggested by the close relation of our dominant archaeal phylotypes to a putative ammonia oxidizer (clone 54d9) and previous studies (for example, Leininger et al., 2006; Nicol and Schleper, 2006), we specifically examined the link between relative archaeal abundances and the level of soil N in situ. Samples collected from two long-term N fertilization experiments were analyzed to determine how prolonged N fertilization affects archaeal populations in soils from these sites. In these soils, we found that the sizes of the extractable inorganic N pools (NH4+ and NO3−) were both significantly correlated with relative archaeal abundances (_r_=−0.39, _P_=0.023 and _r_=−0.42, _P_=0.015, respectively). This correspondence was particularly apparent in the mean relative abundance values for archaeal groups, including DSC1 and DSC2, plotted in Figure 3 for both N amendment experiments (exact plot means and s.d. are given in Supplementary Table S2). Significant trends (P<0.05) of diminishing relative abundance with higher N levels were observed for DSC1 (but not DSC2) at both the native grassland site (CC) and KBS, which were coupled with a general trend of an increase relative abundances for putative groups of nitrifying bacteria between the control plots and those plots receiving the highest N amendment rates.
Figure 3.
Relative abundances (group % of all 16S rRNA gene sequences in each sample) for archaea and specific groups of bacteria across soils from medium (∼100 kg ha−1 year−1) and high (∼300 kg ha−1 year−1) experimental N addition as well as control plots at CC (a) and KBS (b). The portions represented by the dominant soil crenarchaeota (DSC1 and DSC2) and the remaining Archaea as well as Nitrosomonas/Nitrosospira and Nitrospirae bacterial groups are indicated (actual mean values and s.d. are reported in Supplementary Table S2). Significant difference (P<0.5) in relative abundances between the control and N addition plots is indicated with an asterisk (*).
As 54d9 (a putative ammonia oxidizer) was most closely related to DSC1, it is interesting to note that in the N-amended soils of CC and KBS the relative abundance of DSC1 sharply declines with heavy N application as Nitrosomonas/Nitrosospira and Nitrospirae tended to become more abundant (Figure 3). This pattern suggests that these soil-dwelling archaea are inhibited by high levels of available N, perhaps because of competitive interactions with bacterial nitrifiers. The cultivated marine ammonia-oxidizing archaeon, Nitosopumilus maritimus, has an extremely high substrate affinity and is thought to be adapted to nutrient limited conditions (Martens-Habbena et al., 2009). These investigators have also suggested that ammonia oxidation kinetics may have an important role in shaping ammonia-oxidizing microbial communities, with archaeal ammonia oxidizers more abundant in soils with lower levels of available N, and bacterial ammonia oxidizers outcompeting their archaeal counterparts and becoming more dominant in soils with higher levels of available N. Jia and Conrad (2009) as well as Di et al. (2009) have also shown that bacterial ammonia oxidizers functionally dominate over their archaeal complement in soils, which have high levels of available inorganic N. Although our results are not definitive proof that archaea in these soils are actively oxidizing ammonia, they are congruent with the concept of ‘niche separation' between ammonia-oxidizing archaea and bacteria. The recent sequencing of the Cenarchaeum symbiosum and Nitrosopumilus maritimus genomes, however, suggests these organisms may also be capable of some level of mixotrophy (Hallam et al., 2006; Walker et al., 2010), and additional research will be required in this area before these dynamics can be more fully resolved.
Conclusions
This survey demonstrates that there is a high degree of variability in the general structure and relative abundance of the dominant archaeal communities inhabiting soils on the global scale, underscoring the importance of examining a broad array of soils before attempting to draw conclusions about the ecological characteristics of archaea, or any other soil microbial taxa. Some general trends of a preference of particular soil archaeal groups for specific habitat types were observed, which supports the concept of niche partitioning for terrestrial archaeal groups. Across the non-experimental soils examined here, we found that soil C:N ratio was the single best predictor of archaeal relative abundances. Perhaps more surprisingly, we found these soils were basically dominated by just two phylotypes within the crenarchaeotal group 1.1b. This finding, only alluded to in previous studies, has important implications for future studies in this field as it suggests that archaea inhabit a far more restricted ecological niche in soils than bacteria. Although these dominant archaeal taxa, particularly DSC1, may have key roles in the soil N cycling, surveys such as this cannot confirm this functional role for soil archaea. We do show, however, that the dominance of certain soil archaea (DSC1) is diminished under conditions of high inorganic N availability, perhaps because of competitive interactions with nitrifying bacteria. The differing responses of DSC1 and DSC2 within soils, linked to both edaphic and environmental factors, suggest very different roles for these dominant terrestrial archaea and present an interesting avenue for future research. Metagenomic surveys targeting those soils wherein these taxa are relatively abundant, coupled with efforts to cultivate these microbes should significantly add to our knowledge of this prominent, but still poorly understood, group of soil-dwelling archaea.
Acknowledgments
We thank David Tilman, Jay Lennon, Zach Aanderud, and the personnel at Cedar Creek Ecosystem Science Reserve and the Kellogg Biological Station for facilitating sample collection from these sites. We also thank Rebecca McCulley, Diana Wall, and the large number of other collaborators who helped us collect soil samples at the various sites. We acknowledge Dan Arp, Peter Bottomley, Eric Triplett, Ian Clark, Penny Hirsch and Steve McGrath for their collaboration and valuable comments on this paper. Members of the Fierer lab, particularly Chris Lauber, Kathryn Eilers and Kelly Ramirez helped with the laboratory analyses, sample processing and paper preparation. We also appreciated additional comments on the paper by Albert Barberán. Joe Jones, from EnGenCore, supervised the pyrosequencing. This research was funded by grants to RK and NF from the Bill and Melinda Gates Foundation, Crohn's and Colitis Foundation of America, National Institutes of Health, National Science Foundation and the US Department of Agriculture.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary Table S2
References
- Angel R, Soares MIM, Ungar ED, Gillor O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 2009;4:553–563. doi: 10.1038/ismej.2009.136. [DOI] [PubMed] [Google Scholar]
- Auguet J-C, Barberan A, Casamayor EO. Global ecological patterns in uncultured archaea. ISME J. 2009;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Bent SJ, Forney LJ. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2008;2:689–695. doi: 10.1038/ismej.2008.44. [DOI] [PubMed] [Google Scholar]
- Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM. Molecular phylogeny of archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DH, Graber JR, Schmidt TM. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl Environ Microbiol. 1998;64:4333–4339. doi: 10.1128/aem.64.11.4333-4339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone C, Turnbaugh P, et al. 2010Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample Proc Natl Acad Sci USAdoi: 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed]
- Cary SC, McDonald IR, Barrett JE, Cowan DA. On the rocks: the microbiology of Antarctic dry valley soils. Nat Rev Microbiol. 2010;8:129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, et al. The ribosomal database project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demergasso C, Casamayor EO, Chong G, Galleguillos P, Escudero L, Pedrós-Alió C. Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. FEMS Microbiol Ecol. 2004;48:57–69. doi: 10.1016/j.femsec.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- Ehrhardt CJ, Haymon RM, Lamontagne MG, Holden PA. Evidence for hydrothermal archaea within the basaltic flanks of the East Pacific rise. Environ Microbiol. 2007;9:900–912. doi: 10.1111/j.1462-2920.2006.01211.x. [DOI] [PubMed] [Google Scholar]
- Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R, et al. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol. 2007;73:7059–7066. doi: 10.1128/AEM.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong MA, Singleton DR, Coleman DC, Whitman WB. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl Environ Microbiol. 2002;68:1265–1279. doi: 10.1128/AEM.68.3.1265-1279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinger A, Höfle MG, Schloter M, Embacher A, Böhme F, Munch JC, et al. Traditional cattle manure application determines abundance, diversity and activity of methanogenic archaea in arable European soil. Environ Microbiol. 2007;9:612–624. doi: 10.1111/j.1462-2920.2006.01181.x. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Conrad R. Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009;3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G, Glöckner F-O, Amann R, Saano A, Montonen L, Likolammi M, et al. Identification of novel archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization1. FEMS Microbiol Ecol. 2000;34:45–56. doi: 10.1111/j.1574-6941.2000.tb00753.x. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux X, Poly F, Currey P, Commeaux C, Hai B, Nicol GW, et al. Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure. ISME J. 2008;2:221–232. doi: 10.1038/ismej.2007.109. [DOI] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Gaill F, Moreira D. Wide bacterial diversity associated with tubes of the vent worm Riftia pachyptila. Environ Microbiol. 2002;4:204–215. doi: 10.1046/j.1462-2920.2002.00286.x. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Miller SR, Strong AL, Jones KL, Ungerer MC. Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Appl Environ Microbiol. 2009;75:4565–4572. doi: 10.1128/AEM.02792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Schleper C. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle. Trends Microbiol. 2006;14:207–212. doi: 10.1016/j.tim.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Tscherko D, Chang L, Hammesfahr U, Prosser JI. Crenarchaeal community assembly and microdiversity in developing soils at two sites associated with deglaciation. Environ Microbiol. 2006;8:1382–1393. doi: 10.1111/j.1462-2920.2006.01031.x. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Webster G, Glover LA, Prosser JI. Differential response of archaeal and bacterial communities to nitrogen inputs and pH changes in upland pasture rhizosphere soil. Environ Microbiol. 2004;6:861–867. doi: 10.1111/j.1462-2920.2004.00627.x. [DOI] [PubMed] [Google Scholar]
- Nielsen UN, Osler GHR, Campbell CD, Burslem DFRP, Van Der Wal R. The influence of vegetation type, soil properties and precipitation on the composition of soil mite and microbial communities at the landscape scale. J Biogeogr. 2010;37:1317–1328. [Google Scholar]
- Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- Oline DK, Schmidt SK, Grant MC. Biogeography and landscape-scale diversity of the dominant Crenarchaeota of soil. Microbial Ecol. 2006;52:480–490. doi: 10.1007/s00248-006-9101-5. [DOI] [PubMed] [Google Scholar]
- Pointing SB, Chan Y, Lacap DC, Lau MCY, Jurgens JA, Farrell RL. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA. 2009;106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A-L, Pace NR. Archaea—A Laboratory Manual: Thermophiles. Cold Spring Harbor Laboratory Press: Woodbury, NY; 1994. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction; pp. 101–105. [Google Scholar]
- Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, et al. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol. 2009;11:446–456. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- Schleper C, Holben W, Klenk H. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- Soule T, Anderson IJ, Johnson SL, Bates ST, Garcia-Pichel F. Archaeal populations in biological soil crusts from arid lands in North America. Soil Biol Biochem. 2009;41:2069–2074. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. Mega4: molecular evolutionary genetics analysis (MEGA) software, version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tilman D. Secondary succession and the pattern of plant dominance along experimental nitrogen gradients. Ecol Monographs. 1987;57:189–214. [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk H-P, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Breitbart M, McNairnie P, Rohwer F. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinform. 2006;7:57. doi: 10.1186/1471-2105-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-M, Wang M, Prosser JI, Zheng Y-M, He J-Z. Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol Ecol. 2009;70:208–217. doi: 10.1111/j.1574-6941.2009.00775.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary Table S2