Revisiting the host as a growth medium (original) (raw)
. Author manuscript; available in PMC: 2011 Jun 15.
Published in final edited form as: Nat Rev Microbiol. 2008 Sep;6(9):657–666. doi: 10.1038/nrmicro1955
Abstract
The ability of the human body to play host to bacterial pathogens has been studied for more than 200 years. Successful pathogenesis relies on the ability to acquire the nutrients that are necessary for growth and survival, yet relatively little is understood about the in vivo physiology and metabolism of most human pathogens. This Review discusses how in vivo carbon sources can affect disease and highlights the concept that carbon metabolic pathways provide viable targets for antibiotic development.
Billions of dollars are spent annually on research into, and treatment of, bacterial infections, and it is likely that over the course of our lifetimes each of us will be treated for a bacterial infection. As antibiotic resistance continues to increase, it is imperative that researchers not only continue to develop conventional antimicrobials but also pursue more unconventional targets. One under-exploited therapeutic opportunity is based on the simple premise that for a bacterium to cause an infection, it must obtain the nutrients that are necessary for replication from the infection site. This is not a new concept, as Louis Pasteur developed a model that described the body as a culture vessel in the late 1870s as a means of describing immunity1. Subsequent work expanded this ‘culture-vessel model’ as an incomplete explanation for bacterial host and tissue tropism2,3. More recently, E. D. Garber presented a paper entitled ‘The host as a growth medium’ in 1960 (REF. 4), in which he proposed the fundamental importance of understanding the physiology of the bacterium in the infection site. Over the intervening 50 years, a wealth of literature has been generated that details the response of both hosts and pathogens to infection. However, relatively few studies have examined the fundamental question: what makes the human body a good growth medium for bacterial pathogens? In this Review, we outline how the host growth environment affects disease and discuss the potential for targeting metabolic pathways for therapeutic development.
Interest in the host as a growth environment has resurfaced in recent years. More than just a culture vessel, the human body is now considered a chemostat, in which the nutrients that are crucial for bacterial growth are replenished over time. Bacterial pathogens are adept at localizing to and responding to specific nutritional cues within host microenvironments (TABLE 1), and growth within these microenvironments requires specific metabolic pathways. Indeed, some of the first antimicrobial therapeutics targeted bacterial metabolism (BOX 1). More recently, inactivation of lactic acid production in the caries-causing bacterium Streptococcus mutans has demonstrated therapeutic promise5. Treating infections with defined dietary regimens, such as low-iron diets, has also shown limited success. Iron sequestration is a primary means of defence used by the human body during infection, as most bacterial pathogens require iron for growth6. As such, dietary supplementation with iron is correlated with increased incidence of human Mycobacterium tuberculosis infection as well as increased mortality7,8. Conversely, iron chelator therapy and dietary iron restriction diminishes M. tuberculosis growth in animal models, and these treatments have been proposed for human use9. Iron chelator therapy has also shown promise in individuals with malaria10. The rationale is that through dietary restriction one can starve a pathogen of iron, an essential limiting nutrient in the human host6.
Table 1.
Impact of carbon substrate on host colonization and disease
| Bacterium | Location | Carbon substrate | Impact |
|---|---|---|---|
| Aggregatibacter actinomycetemcomitans | Oral cavity | Lactic acid | Persistence and growth |
| Escherichia coli | Intestine | Intestinal mucus sugars | Establishment of infection |
| Legionella pneumophila | Macrophage | Threonine | Differentiation signal |
| Listeria monocytogenes | Macrophage | Hexose phosphates, lipoate | Persistence and growth |
| Mycobacterium tuberculosis | Macrophage | Cholesterol | Immune manipulation |
| Neisseria meningitidis | Bloodstream, cerebrospinal fluid | Lactic acid | Molecular mimicry |
| Pseudomonas aeruginosa | Lung | Aromatic amino acids | Increased production of virulence factors |
| Uropathogenic E. coli | Urinary tract | d-Serine | Tissue localization and growth |
Box 1 Targeting bacterial metabolism: a historical perspective.
One of the oldest antibiotics has its roots in the textile industry100. In 1908, Paul Gelmo synthesized _p_-aminobenzenesulphonamide (sulphanilamide), which was later discovered to make superior textile dyes when mixed with azo dyes. In the 1930s one of these dyes was found to have antimicrobial properties. A French patent in 1934 and a German patent in 1935 were obtained for the newly named Prontosil, which was soon made available for therapeutic use. Prontosil proved to be an effective antimicrobial agent, although turning the patient red was an unfortunate side effect. It was only months later that a team in Paris discovered that the sulphanilamide component of Prontosil was the active portion of the compound, and was, conveniently, colourless. This led to widespread use throughout Europe. The drug did not gain popularity in the United States until late 1936, when the president’s son, Franklin D. Roosevelt Jr, was treated successfully for a streptococcal infection. The sulfa drug family went on to play a major part in reducing casualties during the Second World War.
It is now known that sulfa drugs interfere with the folic acid biosynthesis pathway101, a pathway that is essential for bacterial survival. Since the discovery of sulfa drugs, the development of new antibiotics, along with an increasing level of bacterial resistance, has led to a decrease in their use. Nevertheless, sulfa drugs are still reliable treatment options for bacterial infections today.
This Review cannot encompass every instance in which the nutritional content of the infection site has been linked to disease. Therefore, we will focus on four general virulence-related phenotypes that are known to be affected by the growth environment: evasion of the host immune system; tissue tropism; niche specialization and resource partitioning; and cell–cell communication.
Immune evasion
A healthy immune system has the remarkable ability to quickly and effectively identify and eradicate disease-causing bacteria from the host. However, some pathogenic bacteria have developed ways to thwart the host attack and proliferate in vivo. Here, we discuss two bacterial species in which specific carbon catabolic pathways are metabolically coupled to immune subversion.
Neisseria meningitidis
Neisseria meningitidis, a Gram-negative commensal of the human oropharynx, can cause septicaemia by crossing the mucosal barrier and entering the bloodstream or meningitis by crossing the blood–brain barrier into the cerebrospinal fluid11. In each case, N. meningitidis uses molecular mimicry to survive exposure to the innate immune system. The ability of the immune system to distinguish foreign cells from self is mediated in part by sialic acid, which coats the surface of human cells and acts as a ‘self signal’. N. meningitidis exploits this feature by decorating its outer surface with sialic acid residues, thus masking itself from human defences12,13. Molecular mimicry is a key component of N. meningitidis pathogenesis and mutants that are unable to sialylate their outer surface are highly attenuated14.
The in vivo nutritional environment has an impact on N. meningitidis host cell mimicry. N. meningitidis can use lactate and glucose as carbon and energy sources15, and lactate and glucose are present in nasopharyngeal tissue, serum and cerebrospinal fluid at 1–2 mM and 6–10 mM levels, respectively16,17. Interestingly, although both of these carbon sources are present in vivo, N. meningitidis catabolizes lactate at a faster rate than it does glucose16, and mutants that are deficient for lactate transport are defective colonizers of nasopharyngeal tissue17. Why might a bacterium preferentially metabolize a more oxidized substrate such as lactate over glucose? One likely explanation is that intermediates of lactate consumption feed directly into the sialylation pathway16, thus enhancing sialic acid biosynthesis. This, in turn, leads to increased sialylation of the N. meningitidis outer membrane (FIG. 1). As expected, an N. meningitidis strain that is unable to transport lactate (Δ_lctP_) is highly deficient for sialic acid modification of the outer membrane and is more susceptible to complement-mediated killing16. Thus, for N. meningitidis, catabolism of a preferred carbon source in vivo is coupled to a unique immune-evasion strategy.
Figure 1. Molecular mimicry by Neisseria meningitidis.
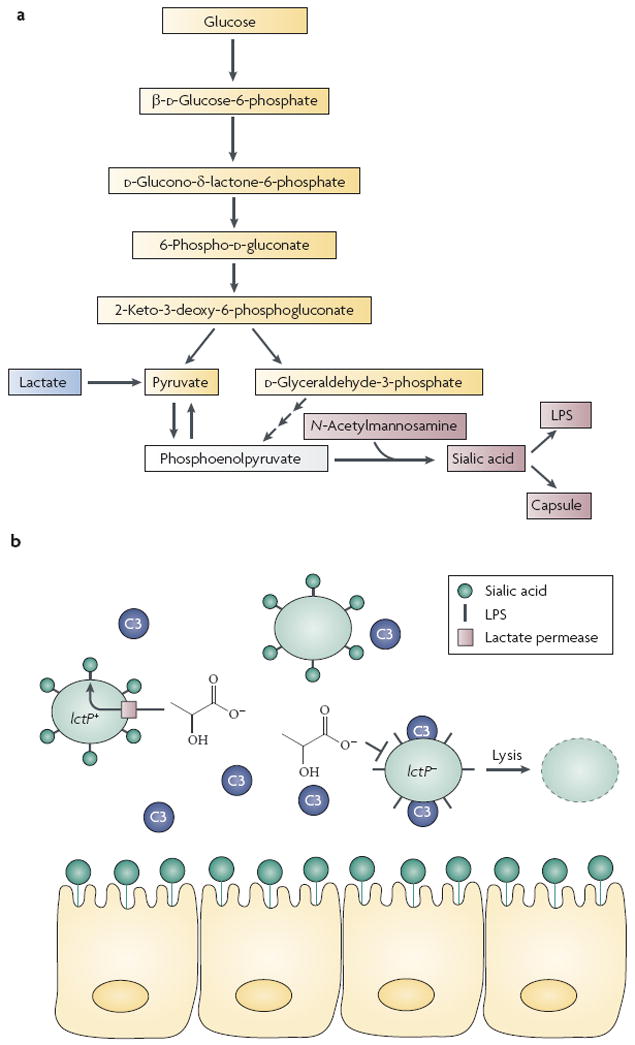
a Glucose catabolism in N. meningitidis proceeds by the Entner–Doudoroff pathway and lactate catabolism feeds directly into the sialic acid pathway16. Note the relative number of metabolic steps from glucose to phosphoenolpyruvate compared with that from lactate to phosphoenolpyruvate. b Sialylated lipopolysaccharide (LPS) on the N. meningitidis surface mimics the surface of eukaryotic cells, preventing deposition of the complement molecule C3. Inactivation of the lactate permease gene lctP results in C3-mediated cell lysis16.
Mycobacterium tuberculosis
M. tuberculosis, the causative agent of tuberculosis, infects millions of people worldwide and is a significant health concern in our increasingly globalized society. Upon colonization of the lung, this intracellular respiratory pathogen is engulfed by pulmonary macrophages and then manipulates the host immune response using elaborate mechanisms, many of which are not fully understood18. Ultimately, host cell manipulation results in the creation of an M. tuberculosis niche — a granuloma19 — and chronic infection.
It has been proposed that M. tuberculosis uses fatty acids as carbon sources during chronic infection. Evidence supporting this hypothesis includes stimulation of M. tuberculosis respiration by fatty acids20, survival of glycolytic enzyme mutants in murine infection models21 and a requirement for isocitrate lyase enzymes (which are involved in the glyoxylate shunt) for intracellular growth and virulence22,23. Significantly, treatment with an isocitrate lyase inhibitor blocks M. tuberculosis growth in vitro with fatty acids as well as in vivo growth in murine macrophages22, suggesting that a similar approach could be used to develop human tuberculosis therapies.
M. tuberculosis carbon catabolism and virulence have been linked by the flux of a key fatty-acid intermediate within the cell24. During infection, M. tuberculosis secretes polyketide lipids that interact with components of the host immune system. These lipids are required for immune evasion and pathogenesis25,26 and are often termed virulence lipids. Interestingly, catabolism of the small fatty acid propionate by M. tuberculosis leads to higher production of virulence lipids than observed for growth on glucose or acetate24. It has been proposed that high intracellular levels of methyl-malonyl-CoA, an intermediate in propionate catabolism and a biosynthetic precursor of polyketide lipids, mediates this phenotype. The model dictates that during growth on fatty acids such as propionate, intracellular levels of methyl-malonyl-CoA increase, thereby increasing the available substrate for the biosynthesis of polyketide lipids. Consequently, these virulence lipids are produced at higher levels during growth on fatty acids.
More recently, it was reported that M. tuberculosis uses cholesterol as a sole carbon source and a transport system that is important for cholesterol uptake was identified27. An M. tuberculosis mutant deficient for cholesterol uptake was impaired for replication in interferon-γ (IFN-γ)-activated macrophages but not resting macrophages, suggesting that cholesterol metabolism is specifically required for growth in the nutrient-restrictive intracellular conditions that are induced by IFN-γ (BOX 2). In addition, M. tuberculosis was found to co-localize with high-cholesterol regions of IFN-γ-activated macrophages. Significantly, oxidation of the cholesterol side chain yields propionate28,29, and labelled carbon in the cholesterol side chain is incorporated into M. tuberculosis virulence lipids27. These exciting results provide a direct link between M. tuberculosis in vivo carbon catabolism and the production of important immune-evasion factors (FIG. 2).
Box 2 Two can play this game: IFN-γ and intracellular nutrient depletion.
The immunomodulatory compound interferon-γ (IFN-γ) has a crucial role in human immune defence. This chemokine is produced by a host of immune cells in response to pathogen invasion and is a key factor in promoting the antimicrobial activities of macrophages and other cell types102. In fact, for intracellular pathogens such as Mycobacterium tuberculosis and Listeria monocytogenes, in vivo control of bacterial growth and host survival is dependent on IFN-γ production103-105.
Arguably, the best-known antimicrobial activity of IFN-γ-stimulated macrophages is the production of reactive nitrogen species such as nitric oxide. However, this activity is not responsible for all of the growth-inhibitory properties of IFN-γ-stimulated cells. Over two decades ago it was discovered that stimulation of human fibroblasts with IFN-γ induces intracellular tryptophan degradation and concomitantly suppresses growth of the intracellular eukaryotic parasite Toxoplasma gondii106. Mechanistically, IFN-γ stimulation induces production of indoleamine-2,3-dioxygenase107, an enzyme that degrades tryptophan to kynurenine and _N_-formylkynurenine108. Because T. gondii is a tryptophan auxotroph and must scavenge tryptophan from the host during infection106,109, indoleamine-2,3-dioxygenase induction leads to T. gondii tryptophan starvation and growth suppression. In vivo tryptophan starvation has been observed for other pathogens including Rickettsia conorii110 and group B streptococci111. Thus, cellular stimulation by IFN-γ promotes in vivo carbon substrate (tryptophan) deprivation as a growth control mechanism for intracellular pathogens.
The effect of IFN-γ on access to in vivo carbon substrates might go beyond tryptophan depletion. For example, in resting macrophages, M. tuberculosis resides in a maturation-arrested phagosomal compartment that has access to endosomal components such as glycosphingolipids112, which are potential in vivo carbon sources. However, IFN-γ stimulation of macrophages promotes maturation and autophagy of this compartment113, probably restricting access to host-derived nutrients. In turn, it has been proposed that IFN-γ-dependent mechanisms of in vivo nutrient control, beyond tryptophan degradation, are at play during intracellular infection114.
Figure 2. Mycobacterium tuberculosis cholesterol catabolism and virulence factor production.
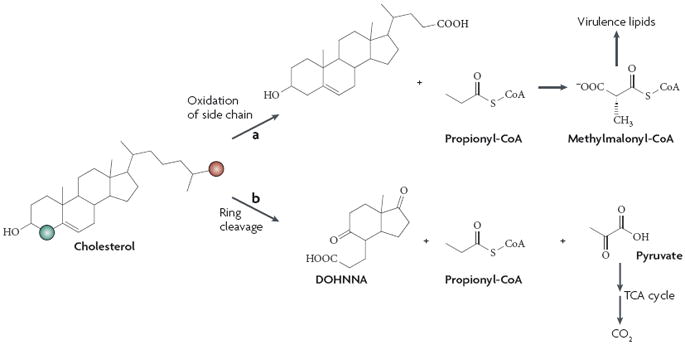
A proposed pathway for cholesterol catabolism with relevant gene products has recently been published for the soil bacterium Rhodococcus sp. strain RHA1 (Ref. 115). M. tuberculosis possesses homologues for almost all of the genes involved in cholesterol degradation115, several of which are important for intracellular growth116. However, definitive M. tuberculosis use of cholesterol as a sole source of carbon and energy was reported only recently, and in this study radioactive labelling was used to demonstrate the differential fates of cholesterol carbons27. A labelled side-chain carbon (shown in red) was incorporated into the virulence lipid phthiocerol dimycocerosate27, presumably by conversion of side-chain-derived propionyl-CoA to methylmalonyl-CoA, a precursor to phthiocerol dimycocerosate and other virulence lipids24. Successive oxidation of the remaining side chain produces an acetyl-CoA and an additional propionyl-CoA moiety (not shown). Conversely, a labelled ring carbon (shown in green) was released as CO2, indicating that this carbon enters the tricarboxylic acid (TCA) cycle and is mineralized27. Some aspects of M. tuberculosis cholesterol catabolism remain unclear, including the fates of the degradation product 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid (DOHNAA)115 and propionyl-CoA moieties produced through ring cleavage and successive side chain degradation.
Tissue tropism
Primarily on the basis of work by Pasteur1, bacterial tissue tropism was originally proposed to rely solely on the nutritional conditions within specific host tissues. Of course this is not the case, and tissue localization and specificity are often determined by devoted adhesins such as external appendages (pili and fimbriae) or outer membrane proteins30. The molecular interactions between the bacterial and host cell outer surfaces have long been a topic of intense interest; consequently, most tissue tropism studies have focused on interactions between host tissues and bacterial surface components. However, and perhaps not surprisingly, for some bacterial pathogens the nutritional environment at the infection site also influences tissue tropism.
Escherichia coli
Escherichia coli is a pervasive microorganism. A commensal in the human intestinal tract, non-pathogenic E. coli is the workhorse of molecular biology and a model system for microbial geneticists and physiologists. By contrast, its pathogenic counterparts colonize multiple infection sites, generate steep health-care costs and can be fatal31,32. In general, pathogenic E. coli strains have acquired mobile genetic elements that carry novel virulence determinants such as new toxins and adhesins33. Two well-known examples of pathogenic E. coli are enterohaemorrhagic (EHEC) and uropathogenic E. coli (UPEC). EHEC, an intestinal pathogen that causes severe diarrhoea, has achieved notoriety as a result of highly publicized food-borne outbreaks involving contaminated beef and vegetable products. UPEC is the predominant cause of uncomplicated urinary tract infections34; treatment of these infections in women had an estimated cost of ~$1.5 billion in 1995 in the USA alone35. Curiously, although UPEC strains benignly inhabit the intestine, they colonize and cause disease in sites outside the intestine, a virulence trait that is not shared by EHEC strains.
Why does UPEC cause infections outside the intestine whereas EHEC presumably does not? The host nutritional environment might have a role. The amino acid d-serine is present in human urine36 at levels that are potentially toxic to E. coli37; however, some E. coli strains are able to use d-serine as a carbon, nitrogen and energy source. Comparative genomic analyses of E. coli strains demonstrated that UPEC strains have the genes that are necessary for d-serine catabolism but that in EHEC these are partially absent and are replaced by sucrose utilization genes38. Correspondingly, d-serine catabolism is observed in the majority of UPEC clinical isolates but rarely in EHEC isolates38. Finally, the genes that are required for d-serine catabolism are induced during human in vivo growth of an asymptomatic bacteriuria E. coli strain39, suggesting that d-serine metabolism is important during urinary tract colonization and infection. These results suggest that EHEC lacks the metabolic capacity to replicate and/or survive in human urine, a capacity that UPEC has retained, and it is this metabolic capability that defines pathogenesis in the urinary tract. Additionally, sucrose catabolism (in lieu of d-serine catabolism) in diarrhoeagenic E. coli isolates40 suggests that this metabolism confers an advantage in the intestinal growth environment.
Brucella abortus
Spontaneous abortions of cattle caused by the intracellular pathogen Brucella abortus are a serious concern to the agriculture industry. Although several industrialized nations have launched historically successful vaccination campaigns against B. abortus, these infections still pose a threat in developing countries41. An archetypal zoonotic pathogen, B. abortus can also cause disease in humans, and countries that lack effective animal disease-control programmes generally have higher rates of B. abortus infection41,42.
B. abortus tissue tropism during bovine infection is influenced by nutrition. The naturally occurring sugar alcohol erythritol is the preferred carbon source for B. abortus43. Extracts derived from ground bovine tissues have been used as in vitro B. abortus growth media, and experiments using these media have shown that B. abortus growth is promoted by the high levels of erythritol in bovine fetal tissues44. Furthermore, injection of calves with erythritol enhances experimental B. abortus infection44. Interestingly, expression of erythritol catabolism genes in the closely related human pathogen Brucella suis increases during intracellular growth in macrophages45, suggesting that erythritol catabolism (or catabolism of structural analogues) might be conserved during human infection.
Niche specialization and resource partitioning
Bacterial adaptations to in vivo growth have been well documented. Niche specialization refers to the ability of an organism to occupy a specific environment, for example, the adaptation of some bacteria to life in an intracellular compartment. Resource partitioning proposes that bacterial species living within a mixed community can avoid competition by using a carbon source that is not preferred by other members of the community. In this section, we discuss four examples of niche specialization and resource partitioning in the human body: Legionella pneumophila and Listeria monocytogenes in macrophages, Aggregatibacter actinomycetemcomitans in the oral cavity and E. coli in the intestine.
Legionella pneumophila
Legionellosis, commonly known as Legionnaire’s disease, is caused by the intracellular pathogen L. pneumophila. This Gram-negative bacterium is found in aquatic environments where it can be internalized and survive digestion by bacteriovorus amoebae46. L. pneumophila resides within intracellular compartments of the amoeba, and infected amoebae serve as an important environmental reservoir for this bacterium. After internalization, L. pneumophila undergoes a two-step life cycle whereby it replicates internally (replication phase) and eventually kills the host, returning to the aquatic environment (transmission phase)46. Each phase has a characteristic gene expression profile that is controlled by conditions inside the host cell.
When aerosolized and inhaled, L. pneumophila can reach the alveoli of the human lung and be internalized by macrophages, in which it undergoes a life cycle similar to that observed in amoebae46,47. Normally, bacteria contained within the phagosome would be destroyed upon phagolysosomal fusion; however, L. pneumophila has evolved a unique means of avoiding destruction by coordinating the construction of a specialized compartment that is derived from the host endoplasmic reticulum. This compartment is rich in nutrients and stimulates the bacteria to enter the replication phase. Once nutrients become limiting for growth, L. pneumophila changes its gene-expression profile and enters the transmission phase.
What triggers the switch from transmission to replication? L. pneumophila uses amino acids as carbon growth substrates, and recent studies suggest that the availability of a specific amino acid, threonine, could be the differentiation switch. Mutants that are unable to acquire threonine can survive within a macrophage but fail to differentiate and do not enter the replication phase. Thus, threonine can serve as both a nutrient source and a differentiation signal during infection. Without threonine acquisition, L. pneumophila is unable to replicate, escape from the cell and infect other cells48. Therapeutics targeting this pathway could aid in treatment of legionellosis, a disease that affects up to 18,000 people in the United States each year. It is unknown why threonine in particular acts as a differentiation switch, although it is hypothesized that L. pneumophila senses the presence of six essential amino acids (threonine, arginine, cysteine, methionine, serine and valine) before entering the replication phase48.
Listeria monocytogenes
L. monocytogenes is a Grampositive, facultative intracellular bacterium that is responsible for the food-borne illness listeriosis in humans. During infection of a human host, L. monocytogenes is internalized by phagocytic host cells where it temporarily resides within phagocytic vacuoles. Normally, the phagocytic vacuole would fuse with a lysosome and kill the bacterium; however, upon entry into the phagocytic vacuole, L. monocytogenes produces specific factors that allow it to escape the vacuole and enter the cytoplasm. This escape is controlled by the global virulence regulator PrfA, which is activated upon entry into the host cytosol49,50. Once inside the cytoplasm, the bacteria undergo replication and subsequently use host actin to spread to neighbouring cells51.
Unlike non-pathogenic species of Listeria, pathogenic species such as L. monocytogenes use glucose phosphates as carbon sources52. Interestingly, transcription of the hexose phosphate transporter gene (hpt) is dependent upon PrfA53. Mutants lacking Hpt can escape the phagocytic vacuole, but are deficient for replication within the cytoplasm53,54. By using a substrate that is available in the host cytoplasm and controlling its consumption with the global virulence regulator PrfA, L. monocytogenes effectively couples virulence factor production to specific catabolic pathways.
During residence in the host cytoplasm, L. monocytogenes must also scavenge a number of amino acids and vitamins, including the cofactor lipoate. In vivo most lipoate is associated with host proteins and is therefore not freely accessible. L. monocytogenes possesses two lipoate protein ligases, LplA1 and LplA2. Interestingly, LplA1 is functionally specialized to acquire lipoate from hostderived lipoyl peptides, which are present at low concentrations in the host cytoplasm, whereas LplA2 scavenges free lipoate55. Inactivation of LplA1 significantly attenuates L. monocytogenes intracellular growth and virulence56, indicating that the ability to scavenge lipoate directly from host proteins is required for successful pathogenesis. These two examples demonstrate that L. monocytogenes has undergone multiple metabolic adaptations to life inside the host cell that allow for efficient replication and dissemination of the bacterium.
Aggregatibacter actinomycetemcomitans
Periodontitis and heart disease have been linked to the presence of A. actinomycetemcomitans, a Gram-negative bacterium that is found exclusively in the mammalian oral cavity57-59. More specifically, A. actinomycetemcomitans resides in the gingival crevice, the area around the tooth that is bounded by the tooth surface on one side and the epithelium lining the gingiva on the other, in a complex bacterial consortium that includes lactate-producing Streptococcus spp.60,61. A. actinomycetemcomitans has a relatively slow growth rate compared with many other species within the oral cavity, which could limit its competitiveness. However, A. actinomycetemcomitans has evolved a novel mechanism of carbon resource partitioning whereby lactate is preferentially consumed over sugars. This preference exists despite higher growth yields and a faster doubling time with the sugars glucose and fructose. It has been demonstrated that the addition of lactate to A. actinomycetemcomitans cultures rapidly inhibits glucose transport and metabolism. The current model is that high intracellular levels of pyruvate resulting from lactate consumption inhibit glucose use; however, a detailed mechanism has yet to be elucidated.62. Lactate is produced at high levels in the oral cavity, primarily by Streptococcus spp., and A. actinomycetemcomitans consumes lactate produced by Streptococcus spp. during in vitro co-culture. These results indicate that by using lactate, A. actinomycetemcomitans avoids competition with faster growing species in the oral cavity, thereby enabling it to establish residence within the competitive environment of the human mouth (FIG. 3).
Figure 3. Resource partitioning by oral bacteria.
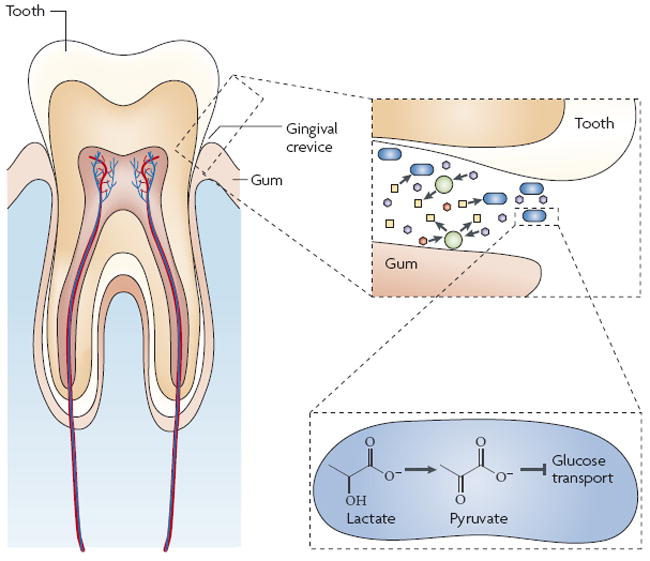
The figure shows a view of the microbial community that inhabits the gingival crevice, which is the space between the tooth and gum. Gingival crevicular fluid, which is similar in composition to serum, contains millimolar concentrations of glucose and lactate117 and micromolar concentrations of other phosphotransferase system (PTS) sugars118. Streptococcus species (green circles) within the gingival crevice rapidly consume sugars (hexagons) and produce lactate (yellow squares). Despite constitutive expression of PTS sugar and lactate catabolic genes, Aggregatibacter actinomycetemcomitans (blue rods) preferentially uses lactate62. Lactate metabolism by the lactate dehydrogenase enzyme results in a rapid increase in intracellular pyruvate levels which is hypothesized to inhibit glucose transport or metabolism by A. actinomycetemcomitans through an unknown post-transcriptional mechanism62.
Escherichia coli
How are resources partitioned in the human intestine, a diverse polymicrobial environment that can support the growth of billions of bacteria63? For the study of intestinal pathogens such as EHEC, this question is highly relevant. Once ingested by a potential human host, EHEC must successfully colonize the host intestinal mucosa to cause disease. Colonization, however, has been hypothesized to rely on the availability of an open niche (the nutrient-niche hypothesis64,65). As the colon contains high levels of commensal E. coli, invading EHEC must either directly compete with commensal E. coli for carbon substrates or, according to the nutrient-niche hypothesis, use substrates that are not consumed by commensal strains. The in vivo hierarchy of carbon substrate preference has been determined for commensal E. coli using murine intestinal mucus as an in vitro growth substrate64. Interestingly, when compared with the carbon substrate preference of EHEC, important differences are observed66, lending credence to the nutrient-niche hypothesis. However, both E. coli strains undergo simultaneous catabolism during growth with mixed carbon substrates64,66, suggesting that a distinct nutrient niche (that is, a niche based on the availability of a single carbon substrate) might not exist. What is clear is that carbon preference is an important component of EHEC-mediated disease and elucidation of the nature of the interactions between commensal and pathogenic E. coli might provide new therapeutic strategies to specifically target pathogenic strains.
Bacterial communication
It is now commonly accepted that bacteria are capable of intraspecies communication using diverse chemical ‘languages’ in a process that is termed quorum sensing. The growing body of research on bacterial cell–cell communication indicates that producing and responding to specific signals allow bacterial populations to coordinate their gene expression and thus their group behaviours. Quorum signals have diverse ecological roles, going beyond the intraspecies level to participate in interspecies67,68 and even interdomain communications69,70. The collective work on bacterial communication to date suggests that bacteria participate in dynamic ‘conversations’ with their microbial neighbours and eukaryotic hosts that intimately influence their physiology. Signal production by bacterial populations can be heavily influenced by nutrients in the growth environment, and for pathogenic bacteria this environment is the host. In this section, we review two well-studied pathogenic relationships in which cell–cell communication, carbon metabolism and pathogenesis intersect.
Agrobacterium tumefaciens
The Gram-negative α-proteobacterium Agrobacterium tumefaciens uses a remarkable mechanism to elicit tumour formation in plants. This soil bacterium, which is recruited by phenolic compounds and sugars that are released by wounded plant tissues71-73, enters compromised plants and effects DnA transfer to plant cell nuclei through illegitimate recombination74,75. The transferred DNA, called T-DNA, originates from the Ti (tumour-inducing) plasmids that are carried by A. tumefaciens. The transfer of T-DNA from the bacterial cell to the host plant has revolutionized plant molecular biology and genetic engineering76,77. In nature, this process is believed to confer a major growth advantage for A. tumefaciens, as T-DNA carries oncogenic genes that promote plant cell proliferation78 as well as genes that direct the plant to synthesize opines79. Opines are carbon substrates that are catabolized by agrobacterial species but not by most other bacteria80,81. This phenomenon has been referred to as the ‘opine concept’ hypothesis, and in this way A. tumefaciens induces formation of an ecological niche that selects for its own growth82.
Opines themselves fulfil a twofold purpose, serving as both carbon growth substrates and as inducers of A. tumefaciens quorum signal production. Similarly to many Gram-negative bacteria, A. tumefaciens produces an acyl-homoserine lactone (AHL) communication signal that is used for density-dependent gene regulation83-85. In A. tumefaciens, AHL signalling promotes the conjugal transfer of Ti plasmids to other, potentially avirulent A. tumefaciens cells in the environment83. Thus, host-derived opines promote genetic diversity among A. tumefaciens strains by indirectly inducing the conjugative transfer of the Ti plasmid86. It was recently demonstrated that A. tumefaciens can transfer T-DNA to unwounded plants and elicit plant opine production without tumour formation87, reminiscent of a commensal relationship. This work suggests that the outcomes of _Agrobacterium_–host interactions could be more complex, with tumour formation being only one of several outcomes. It remains to be seen how this non-tumorigenic interaction will affect agrobacterial plasmid transfer and genetic diversity in the rhizosphere.
Pseudomonas aeruginosa
The heritable disease cystic fibrosis (CF) is a well-studied model for chronic bacterial infections. Non-CF airways are generally sterile, but inadequate ion transport, combined with defective mucociliary clearance, is thought to predispose CF airways to persistent microbial colonization88. Numerous bacterial species are commonly present in the CF lung89; however, the Gram-negative opportunistic pathogen Pseudomonas aeruginosa often predominates. Within the CF lung, P. aeruginosa grows within the copious sputum (mucus), where it can reach densities of 109 bacteria per ml of sputum90,91. P. aeruginosa infections within the CF lung are highly refractory to antimicrobial treatment and are often maintained throughout the life of the individual.
P. aeruginosa produces an arsenal of extracellular factors that are crucial for colonization and competition with other bacterial species. The production of these factors is regulated by several cell–cell communication signals, including 2-heptyl-3-hydroxy-4-quinolone, termed the Pseudomonas quinolone signal (PQS)92,93. Outer membrane vesicles released by P. aeruginosa disseminate the hydrophobic PQS and deliver antimicrobial quinolone compounds to potential competitors94-96. Interestingly, P. aeruginosa initiates PQS production at lower cell densities during in vitro growth in CF sputum than in common laboratory medium97. Consequently, P. aeruginosa exhibits higher production of virulence factors and antimicrobial factors during growth in CF sputum than during growth in normal laboratory media. Use of a synthetic defined medium to nutritionally model CF sputum demonstrated that aromatic amino acids present in CF sputum serve as cues to modulate P. aeruginosa PQS production98. PQS and aromatic amino acids share biosynthetic precursors, and it is hypothesized that high levels of aromatic amino acids in CF sputum allow these precursors to be assimilated into PQS, thereby enhancing PQS production in CF sputum (FIG. 4a). Thus, it appears that increased flux of metabolic intermediates to PQS — mediated by the presence of aromatic amino acid cues in the CF lung — might enhance the virulence and competitiveness of P. aeruginosa in this environment98. These results also suggest that development of therapeutics that decrease the levels of these amino acids in the CF lung might affect the ability of P. aeruginosa to colonize and initiate disease (FIG. 4b).
Figure 4. Cell–cell communication, carbon metabolism and pathogenesis in Pseudomonas aeruginosa.
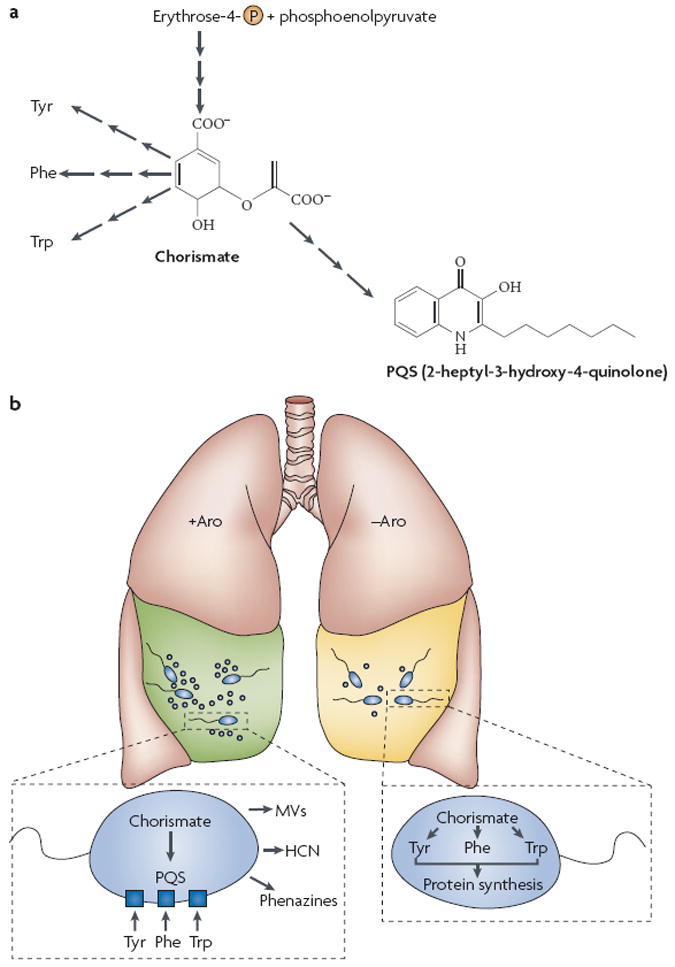
a A simplified view of the fates of chorismate in P. aeruginosa. Chorismate, the biosynthetic precursor of the aromatic amino acids tyrosine (Tyr), phenylalanine (Phe) and tryptophan (Trp), is synthesized from the central metabolites erythrose-4-phosphate and phosphoenolpyruvate. Chorismate synthesis is a highly regulated process in Escherichia coli119 but might be constitutive in P. aeruginosa120. In addition to the aromatic amino acids, chorismate is also the biosynthetic precursor of 2-heptyl-3-hydroxy-4-quinolone, a cell–cell signal that is unique to P. aeruginosa (the Pseudomonas quinolone signal; PQS)121. b A model for the impact of aroxmatic amino acids on P. aeruginosa physiology during cystic fibrosis (CF) lung infection. High levels of aromatic amino acids are present in CF sputum98. P. aeruginosa scavenges these amino acids for protein synthesis, thereby diverting intracellular chorismate pools to PQS biosynthesis. PQS induces virulence factor production including the blue-green phenazine pigment pyocyanin92,121, hydrogen cyanide (HCN)121 and toxin-loaded membrane vesicles (MVs)94 (+Aro). In the second scenario, depletion of aromatic amino acids from the CF lung (potentially by some novel therapeutic), forces P. aeruginosa to use intracellular chorismate pools to support protein biosynthesis (−Aro). In turn, production of PQS and PQS-regulated virulence factors decreases98.
Conclusions and future directions
What is the future of antimicrobial development? Using genetic, proteomic and bioinformatic techniques in serovars of Salmonella enterica, Becker et al. recently illuminated genetic and metabolic redundancies that could hinder efforts to target S. enterica metabolism99. In fact, based on their data the authors concluded that most broad-spectrum antimicrobial targets have already been exploited. As disheartening as these results are, it is likely that the future of antimicrobial development lies in novel narrow-spectrum antibiotics and combination therapies, particularly in light of widespread gene-based resistance to many broad-spectrum compounds. By understanding the basic physiology of pathogens during residence in the human body, several novel avenues of drug and treatment development can be explored, including targeting specific enzymes within a bacterial species (such as the isocitrate lyase enzymes of M. tuberculosis), specific manipulation of the in vivo growth environment (such as targeted destruction of aromatic amino acids in CF sputum), and modification of host diet (such as iron restriction during infection). Although not discussed at length here, the growing body of work on microbial community dynamics in host environments such as the intestine might also lead to appropriate dietary modifications and probiotic prophylactics and treatments. Regardless of the approach used, clever strategies will be necessary to prevent, treat and cure microbial infections in the future.
It is clear that there is interest in revisiting the host as a growth medium. This is most evident from the renewed interest in examining the in vivo carbon preference of extracellular and intracellular pathogens. Unfortunately, the growth conditions in many infection sites are poorly defined, making nutritional modelling of these sites difficult. Perhaps as a result of this, the preferred in vivo carbon source of many pathogens is unknown. This observation is troubling as it suggests that, in general, we still do not understand the basic physiology of bacterial pathogens in vivo. Using _in vivo_-relevant growth substrates for in vitro studies provides a simple and versatile method for examining nutritional effects on bacterial physiology. The use of relevant nutritional substrates has led to significant breakthroughs in the study of several pathogens, including UPEC (human urine), P. aeruginosa (lung mucus) and commensal and enterohaemorrhagic E. coli strains (intestinal mucus). These in vitro studies allow for the development of molecular models that can be assessed in more challenging in vivo systems. Ultimately, understanding the nutritional environment of the infection will provide important information for understanding the basis of infection.
Acknowledgments
We would like to acknowledge members of the Whiteley laboratory for critical discussion and review of this manuscript.
Glossary
Molecular mimicry
Chemical imitation of a host structural component by a foreign body.
Granuloma
A tissue lesion that occurs when macrophages are unable to clear foreign substances from the body. The centre of a granuloma contains a high density of macrophages (sometimes necrotic), which are surrounded by multiple immune cell types, including polymorphonuclear leukocytes and fibroblasts.
Glyoxylate shunt
An anaplerotic pathway used to bypass CO2-generating reactions of the tricarboxylic acid cycle to conserve carbon for intermediate biosynthesis; required for growth on fatty acids or acetate.
Polyketide
A member of a diverse class of secondary metabolites formed by condensation of small fatty acid groups. Polyketides are produced by bacteria, archaea and eukaryotes and can function as natural antimicrobials.
Phagosome
An intracellular vacuole that is formed by phagocytosis of extracellular components. Phagocytic cells, such as macrophages, are involved in bacterial clearance during infection. Properly trafficked phagosomes fuse with lysosomes — compartments that are filled with degradative enzymes.
Cofactor
A non-proteinaceous chemical compound that is required for enzyme catalysis.
Illegitimate recombination
A type of genetic recombination that occurs in spite of little homology between nucleotide sequences.
References
- 1.Pasteur L, Theorie des Germes La. Comptes Rendus l’Academie des Sciences. 1878;86:1037–1043. [Google Scholar]
- 2.McFarland J. A Text-Book upon the Pathogenic Bacteria. W B Saunders; Philadelphia: 1900. [Google Scholar]
- 3.Zinsser H. Infection and Resistance. Macmillan; New York: 1918. [Google Scholar]
- 4.Garber ED. The host as a growth medium. Ann NY Acad Sci. 1960;88:1187–1194. doi: 10.1111/j.1749-6632.1960.tb20108.x. [DOI] [PubMed] [Google Scholar]
- 5.Hillman JD, et al. Construction and characterization of an effector strain of Streptococcus mutans for replacement therapy of dental caries. Infect Immun. 2000;68:543–549. doi: 10.1128/iai.68.2.543-549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg ED. Iron and susceptibility to infectious disease. Science. 1974;184:952–956. doi: 10.1126/science.184.4140.952. [DOI] [PubMed] [Google Scholar]
- 7.Gordeuk VR, McLaren CE, MacPhail AP, Deichsel G, Bothwell TH. Associations of iron overload in Africa with hepatocellular carcinoma and tuberculosis: Strachan’s 1929 thesis revisited. Blood. 1996;87:3470–3476. [PubMed] [Google Scholar]
- 8.Gangaidzo IT, et al. Association of pulmonary tuberculosis with increased dietary iron. J Infect Dis. 2001;184:936–939. doi: 10.1086/323203. [DOI] [PubMed] [Google Scholar]
- 9.Polla BS. Therapy by taking away: the case of iron. Biochem Pharmacol. 1999;57:1345–1349. doi: 10.1016/s0006-2952(99)00017-9. [DOI] [PubMed] [Google Scholar]
- 10.Mabeza GF, Loyevsky M, Gordeuk VR, Weiss G. Iron chelation therapy for malaria: a review. Pharmacol Ther. 1999;81:53–75. doi: 10.1016/s0163-7258(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 11.van Deuren M, Brandtzaeg P, van der Meer JW. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin Microbiol Rev. 2000;13:144–166. doi: 10.1128/cmr.13.1.144-166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGee DJ, Rest RF. Regulation of gonococcal sialyltransferase, lipooligosaccharide, and serum resistance by glucose, pyruvate, and lactate. Infect Immun. 1996;64:4630–4637. doi: 10.1128/iai.64.11.4630-4637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons NJ, et al. Lactic acid is the factor in blood cell extracts which enhances the ability of CMP-NANA to sialylate gonococcal lipopolysaccharide and induce serum resistance. Microb Pathog. 1996;20:87–100. doi: 10.1006/mpat.1996.0008. [DOI] [PubMed] [Google Scholar]
- 14.Vogel U, et al. Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis. Infect Immun. 1997;65:4022–4029. doi: 10.1128/iai.65.10.4022-4029.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leighton MP, Kelly DJ, Williamson MP, Shaw JG. An NMR and enzyme study of the carbon metabolism of Neisseria meningitidis. Microbiology. 2001;147:1473–1482. doi: 10.1099/00221287-147-6-1473. [DOI] [PubMed] [Google Scholar]
- 16.Exley RM, et al. Available carbon source influences the resistance of Neisseria meningitidis against complement. J Exp Med. 2005;201:1637–1645. doi: 10.1084/jem.20041548. Using cerebrospinal fluid as an in vitro growth substrate and in vivo animal studies, the authors find that the ability of N. meningitidis to camouflage itself from host defences is linked to in vivo carbon acquisition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Exley RM, et al. Neisseria meningitidis lactate permease is required for nasopharyngeal colonization. Infect Immun. 2005;73:5762–5766. doi: 10.1128/IAI.73.9.5762-5766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell DG, Mwandumba HC, Rhoades EE. Mycobacterium and the coat of many lipids. J Cell Biol. 2002;158:421–426. doi: 10.1083/jcb.200205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders BM, Cooper AM. Restraining mycobacteria: role of granulomas in mycobacterial infections. Immunol Cell Biol. 2000;78:334–341. doi: 10.1046/j.1440-1711.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- 20.Bloch H, Segal W. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J Bacteriol. 1956;72:132–141. doi: 10.1128/jb.72.2.132-141.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nature Med. 2005;11:638–644. doi: 10.1038/nm1252. This study demonstrates that a metabolic inhibitor can effectively control M. tuberculosis intracellular growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz-Elias EJ, McKinney JD. Carbon metabolism of intracellular bacteria. Cell Microbiol. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- 24.Jain M, et al. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci USA. 2007;104:5133–5138. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 26.Rousseau C, et al. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell Microbiol. 2004;6:277–287. doi: 10.1046/j.1462-5822.2004.00368.x. [DOI] [PubMed] [Google Scholar]
- 27.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sih CJ, Tai HH, Tsong YY, Lee SS, Coombe RG. Mechanisms of steroid oxidation by microorganisms. XIV. Pathway of cholesterol side-chain degradation. Biochemistry. 1968;7:808–818. doi: 10.1021/bi00842a039. [DOI] [PubMed] [Google Scholar]
- 29.Sih CJ, Wang KC, Tai HH. Mechanisms of steroid oxidation by microorganisms. 13. C22 acid intermediates in the degradation of the cholesterol side chain. Biochemistry. 1968;7:796–807. doi: 10.1021/bi00842a038. [DOI] [PubMed] [Google Scholar]
- 30.Niemann HH, Schubert WD, Heinz DW. Adhesins and invasins of pathogenic bacteria: a structural view. Microbes Infect. 2004;6:101–112. doi: 10.1016/j.micinf.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Spears KJ, Roe AJ, Gally DL. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol Lett. 2006;255:187–202. doi: 10.1111/j.1574-6968.2006.00119.x. [DOI] [PubMed] [Google Scholar]
- 32.Vallance BA, Chan C, Robertson ML, Finlay BB. Enteropathogenic and enterohemorrhagic Escherichia coli infections: emerging themes in pathogenesis and prevention. Can J Gastroenterol. 2002;16:771–778. doi: 10.1155/2002/410980. [DOI] [PubMed] [Google Scholar]
- 33.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronald A. The etiology of urinary tract infection: traditional and emerging pathogens. Am J Med. 2002;113(Suppl. 1A):14S–19S. doi: 10.1016/s0002-9343(02)01055-0. [DOI] [PubMed] [Google Scholar]
- 35.Foxman B, Barlow R, D’Arcy H, Gillespie B, Sobel JD. Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol. 2000;10:509–515. doi: 10.1016/s1047-2797(00)00072-7. [DOI] [PubMed] [Google Scholar]
- 36.van de Merbel NC, et al. Determination of d- and l-amino acids in biological samples by two-dimensional column liquid chromatography. Chromatographia. 1995;41:6–14. [Google Scholar]
- 37.Cosloy SD, McFall E. Metabolism of d-serine in Escherichia coli K-12: mechanism of growth inhibition. J Bacteriol. 1973;114:685–694. doi: 10.1128/jb.114.2.685-694.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roesch PL, et al. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol Microbiol. 2003;49:55–67. doi: 10.1046/j.1365-2958.2003.03543.x. [DOI] [PubMed] [Google Scholar]
- 39.Roos V, Klemm P. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect Immun. 2006;74:3565–3575. doi: 10.1128/IAI.01959-05. First transcriptome analysis of E. coli harvested from the human urinary tract. This study used in vitro growth in human urine as well as laboratory medium as experimental controls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moritz RL, Welch RA. The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA. J Clin Microbiol. 2006;44:4038–4048. doi: 10.1128/JCM.01172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinsstag J, et al. Human benefits of animal interventions for zoonosis control. Emerg Infect Dis. 2007;13:527–531. doi: 10.3201/eid1304.060381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth F, et al. Human health benefits from livestock vaccination for brucellosis: case study. Bull World Health Organ. 2003;81:867–876. [PMC free article] [PubMed] [Google Scholar]
- 43.Essenberg RC, Seshadri R, Nelson K, Paulsen I. Sugar metabolism by Brucellae. Vet Microbiol. 2002;90:249–261. doi: 10.1016/s0378-1135(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 44.Smith H, et al. Foetal erythritol: a cause of the localization of Brucella abortus in bovine contagious abortion. Nature. 1962;193:47–49. doi: 10.1038/193047a0. In a novel approach to studying infectious abortions in cattle, the investigators used filtered tissue extracts from pregnant cattle as B. abortus growth media. [DOI] [PubMed] [Google Scholar]
- 45.Kohler S, et al. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci USA. 2002;99:15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson MS, Hammer BK. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol. 2000;54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- 47.Nash TW, Libby DM, Horwitz MA. Interaction between the Legionnaires’ disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J Clin Invest. 1984;74:771–782. doi: 10.1172/JCI111493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer JD, Bachman MA, Swanson MS. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc Natl Acad Sci USA. 2005;102:9924–9929. doi: 10.1073/pnas.0502767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakraborty T, et al. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leimeister-Wachter M, Haffner C, Domann E, Goebel W, Chakraborty T. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc Natl Acad Sci USA. 1990;87:8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cossart P, Lecuit M. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 1998;17:3797–3806. doi: 10.1093/emboj/17.14.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripio MT, Brehm K, Lara M, Suarez M, Vazquez-Boland JA. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J Bacteriol. 1997;179:7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chico-Calero I, et al. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci USA. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goetz M, et al. Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc Natl Acad Sci USA. 2001;98:12221–12226. doi: 10.1073/pnas.211106398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keeney KM, Stuckey JA, O’Riordan MX. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol Microbiol. 2007;66:758–770. doi: 10.1111/j.1365-2958.2007.05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Riordan M, Moors MA, Portnoy DA. Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- 57.Norskov-Lauritsen N, Kilian M. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int J Syst Evol Microbiol. 2006;56:2135–2146. doi: 10.1099/ijs.0.64207-0. [DOI] [PubMed] [Google Scholar]
- 58.Slots J, Reynolds HS, Genco RJ. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun. 1980;29:1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 60.Dzink JL, Tanner AC, Haffajee AD, Socransky SS. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648–659. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 61.Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189:6407–6414. doi: 10.1128/JB.00554-07. A unique mechanism, inducer exclusion, was elucidated that is potentially used to regulate in vivo carbon substrate utilization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laux DC, Cohen PS, Conway T. In: Colonization of Mucosal Surfaces. Nataro JP, editor. ASM Press; Washington DC: 2005. pp. 199–212. [Google Scholar]
- 64.Chang DE, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. Used intestinal mucus and mouse model studies to investigate the in vivo carbon substrate preference hierarchy of non-pathogenic E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fabich AJ, et al. Comparative carbon nutrition of pathogenic and commensal Escherichia coli in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDougald D, Srinivasan S, Rice SA, Kjelleberg S. Signal-mediated cross-talk regulates stress adaptation in Vibrio species. Microbiology. 2003;149:1923–1933. doi: 10.1099/mic.0.26321-0. [DOI] [PubMed] [Google Scholar]
- 68.Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shiner EK, et al. Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell Microbiol. 2006;8:1601–1610. doi: 10.1111/j.1462-5822.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 70.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria–host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ankenbauer RG, Nester EW. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990;172:6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cangelosi GA, Ankenbauer RG, Nester EW. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci USA. 1990;87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stachel SE, Messens E, Van Montagu M, Zambryski P. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature. 1985;318:624–629. [Google Scholar]
- 74.Gheysen G, Villarroel R, Van Montagu M. Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev. 1991;5:287–297. doi: 10.1101/gad.5.2.287. [DOI] [PubMed] [Google Scholar]
- 75.Mayerhofer R, et al. T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 1991;10:697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caplan A, et al. Introduction of genetic material into plant cells. Science. 1983;222:815–821. doi: 10.1126/science.222.4625.815. [DOI] [PubMed] [Google Scholar]
- 77.Horsch RB, et al. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- 78.Akiyoshi DE, et al. Cytokinin/auxin balance in crown gall tumors is regulated by specific loci in the T-DNA. Proc Natl Acad Sci USA. 1983;80:407–411. doi: 10.1073/pnas.80.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Komro CT, DiRita VJ, Gelvin SB, Kemp JD. Site-specific mutagenesis in the TR-DNA region of octopine-type Ti plasmids. Plant Mol Biol. 1985;4:253–263. doi: 10.1007/BF02418244. [DOI] [PubMed] [Google Scholar]
- 80.Moore LW, Chilton WS, Canfield ML. Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl Environ Microbiol. 1997;63:201–207. doi: 10.1128/aem.63.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oger PM, Mansouri H, Nesme X, Dessaux Y. Engineering root exudation of Lotus toward the production of two novel carbon compounds leads to the selection of distinct microbial populations in the rhizosphere. Microb Ecol. 2004;47:96–103. doi: 10.1007/s00248-003-2012-9. [DOI] [PubMed] [Google Scholar]
- 82.Guyon P, Chilton MD, Petit A, Tempe J. Agropine in “null-type” crown gall tumors: Evidence for generality of the opine concept. Proc Natl Acad Sci USA. 1980;77:2693–2697. doi: 10.1073/pnas.77.5.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fuqua WC, Winans SC. A LuxR–LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hwang I, et al. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang L, Murphy PJ, Kerr A, Tate ME. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 86.Petit A, et al. Substrate induction of conjugative activity of Agrobacterium tumefaciens Ti plasmids. Nature. 1978;271:570–572. [Google Scholar]
- 87.Brencic A, Angert ER, Winans SC. Unwounded plants elicit Agrobacterium vir gene induction and T-DNA transfer: transformed plant cells produce opines yet are tumour free. Mol Microbiol. 2005;57:1522–1531. doi: 10.1111/j.1365-2958.2005.04763.x. [DOI] [PubMed] [Google Scholar]
- 88.Chmiel JF, Davis PB. State of the art: why do the lungs of patients with cystic fibrosis become infected and why can’t they clear the infection? Respir Res. 2003;4:8. doi: 10.1186/1465-9921-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rogers GB, et al. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoiby N. Pseudomonas in cystic fibrosis: past, present, and future. Cystic Fibrosis Trust; Berlin, Germany: 1998. [Google Scholar]
- 91.Ohman DE, Chakrabarty AM. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun. 1982;37:662–669. doi: 10.1128/iai.37.2.662-669.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Diggle SP, et al. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 93.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 95.Mashburn-Warren LM, Whiteley M. In: Chemical Communication among Bacteria. Winans SC, Bassler BL, editors. ASM Press; Washington DC: 2008. pp. 333–344. [Google Scholar]
- 96.Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 97.Palmer KL, Mashburn LM, Singh PK, Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. Outlined the development of a defined growth medium designed to mimic cystic fibrosis sputum, allowing for mechanistic studies of P. aeruginosa responses to specific in vivo nutrients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Becker D, et al. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 100.Lesch JE. The First Miracle Drugs: How the Sulfa Drugs Transformed Medicine. Oxford University Press; Oxford; New York: 2007. [Google Scholar]
- 101.Brown GM. The biosynthesis of folic acid. II. Inhibition by sulfonamides. J Biol Chem. 1962;237:536–540. [PubMed] [Google Scholar]
- 102.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 103.Cooper AM, et al. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Flynn JL, et al. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buchmeier NA, Schreiber RD. Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc Natl Acad Sci USA. 1985;82:7404–7408. doi: 10.1073/pnas.82.21.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pfefferkorn ER. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci USA. 1984;81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yoshida R, Imanishi J, Oku T, Kishida T, Hayaishi O. Induction of pulmonary indoleamine 2,3-dioxygenase by interferon. Proc Natl Acad Sci USA. 1981;78:129–132. doi: 10.1073/pnas.78.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Takikawa O, Yoshida R, Kido R, Hayaishi O. Tryptophan degradation in mice initiated by indoleamine 2,3-dioxygenase. J Biol Chem. 1986;261:3648–3653. [PubMed] [Google Scholar]
- 109.Sibley LD, Messina M, Niesman IR. Stable DNA transformation in the obligate intracellular parasite Toxoplasma gondii by complementation of tryptophan auxotrophy. Proc Natl Acad Sci USA. 1994;91:5508–5512. doi: 10.1073/pnas.91.12.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feng HM, Walker DH. Mechanisms of intracellular killing of Rickettsia conorii in infected human endothelial cells, hepatocytes, and macrophages. Infect Immun. 2000;68:6729–6736. doi: 10.1128/iai.68.12.6729-6736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.MacKenzie CR, Hadding U, Daubener W. Interferon-gamma-induced activation of indoleamine 2,3-dioxygenase in cord blood monocyte-derived macrophages inhibits the growth of group B streptococci. J Infect Dis. 1998;178:875–878. doi: 10.1086/515347. [DOI] [PubMed] [Google Scholar]
- 112.Russell DG, Dant J, Sturgill-Koszycki S. Mycobacterium avium- and Mycobacterium tuberculosis-containing vacuoles are dynamic, fusion-competent vesicles that are accessible to glycosphingolipids from the host cell plasmalemma. J Immunol. 1996;156:4764–4773. [PubMed] [Google Scholar]
- 113.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 114.Appelberg R. Macrophage nutriprive antimicrobial mechanisms. J Leukoc Biol. 2006;79:1117–1128. doi: 10.1189/jlb.0206079. [DOI] [PubMed] [Google Scholar]
- 115.Van der Geize R, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nuttall FQ, Khan MA, Gannon MC. Peripheral glucose appearance rate following fructose ingestion in normal subjects. Metabolism. 2000;49:1565–1571. doi: 10.1053/meta.2000.18553. [DOI] [PubMed] [Google Scholar]
- 118.Soyama K. Enzymatic determination of d-mannose in serum. Clin Chem. 1984;30:293–294. [PubMed] [Google Scholar]
- 119.Pittard AJ. Escherichia coli and Salmonella. American Society for Microbiology; Washington DC: 1996. pp. 458–484. [Google Scholar]
- 120.Song J, Jensen RA. PhhR, a divergently transcribed activator of the phenylalanine hydroxylase gene cluster of Pseudomonas aeruginosa. Mol Microbiol. 1996;22:497–507. doi: 10.1046/j.1365-2958.1996.00131.x. [DOI] [PubMed] [Google Scholar]
- 121.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]