The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology (original) (raw)
Abstract
Thaumarchaeota range among the most abundant archaea on Earth. Initially classified as ‘mesophilic Crenarchaeota’, comparative genomics has recently revealed that they form a separate and deep-branching phylum within the Archaea. This novel phylum comprises in 16S rRNA gene trees not only all known archaeal ammonia oxidizers but also several clusters of environmental sequences representing microorganisms with unknown energy metabolism. Ecophysiological studies of ammonia-oxidizing Thaumarchaeota suggest adaptation to low ammonia concentrations and an autotrophic or possibly mixotrophic lifestyle. Extrapolating from the wide substrate range of copper-containing membrane-bound monooxygenases, to which the thaumarchaeal ammonia monooxygenases belong, the use of substrates other than ammonia for generating energy by some members of the Thaumarchaeota seems likely.
Introduction
Aerobic ammonia oxidation, the first and rate-limiting step in nitrification, is the only biological process converting reduced to oxidized inorganic nitrogen species on Earth [1]. For over 100 years, this process was thought to be mediated by autotrophic Beta-proteobacteria and Gamma-proteobacteria (AOB) [2] occasionally supported by heterotrophic nitrifiers in soil environments [3]. However, in situ measurements of nitrification in marine and terrestrial environments showed that ammonia oxidation often proceeds at substrate concentrations significantly below the growth threshold of cultured AOB (e.g. [4]) indicating the presence of unknown nitrifiers. The recent discovery of homologs of ammonia monooxygenase genes in archaea [5–7] and the cultivation of autotrophic ammonia-oxidizing archaea (AOA) [8–11] revealed that an additional group of microorganisms is able to catalyze this process. The widespread distribution of putative archaeal ammonia monooxygenase (amo) genes and their numerical dominance over their bacterial counterparts in most marine and terrestrial environments suggested that AOA play a major role in global nitrification [12–15], but our understanding of their evolutionary history and metabolic repertoire is still in its infancy.
From mesophilic Crenarchaeota to Thaumarchaeota
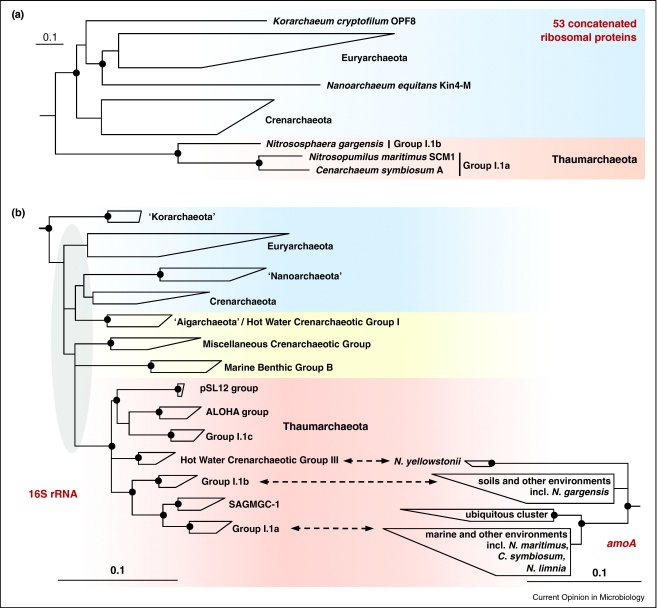
In 1992, Jed Fuhrman's team and Ed DeLong reported the discovery of a novel clade of archaeal 16S rRNA sequences from ocean surface waters, which formed a mesophilic sister group to the hyperthermohilic Crenarchaeota [16,17]. When it became apparent that this novel group contained AOA, these organisms were consequently also referred to as mesophilic Crenarchaeota. This perception was questioned by phylogenetic analysis of the first available genome sequence of a putative AOA, the sponge symbiont Candidatus Cenarchaeum symbiosum. When Brochier-Armanet and colleagues analyzed a concatenated data set of 53 ribosomal proteins common to Archaea and Eukarya, they made the surprising observation that C. symbiosum branched off before the separation of Crenarchaeota and Euryarchaeota. Based on this phylogenetic analysis, on gene presence/absence data, and on the diversity and wide distribution of AOA, they proposed that that these organisms belong to the phylum Thaumarchaeota [18••]. Recently, this analysis was extended to the ammonia-oxidizing Candidatus Nitrosopumilus maritimus, a marine group I.1a representative, and Candidatus Nitrososphaera gargensis, a soil group I.1b representative enriched from a hot spring. In this study, phylogenetic analysis of concatenated ribosomal proteins (Figure 1a) and several other marker genes as well as presence/absence patterns of information processing machineries in Archaea strongly supported the assignment of AOA to the deep-branching phylum Thaumarchaeota [19•]. Consistent with this finding, comparative genomics revealed that 6 conserved signature indels and >250 proteins are unique to the thaumarchaeota C. symbiosum and N. pumilus and are not found in Crenarchaeota [20]. Additional support for the phylum Thaumarchaeota stems from comparative analysis of fosmid clones obtained from different deep-sea locations. Among 200 phylogenetic trees of protein families present in thaumarchaeotal fosmids from these sites, Thaumarchaeota sequences branched as separate cluster distinct from hyperthermophilic Crenarchaeota and Euryarchaeota in 162 phylogenetic trees [21]. Independent from genomic data, the presence of the lipid crenarchaeol in all analyzed AOA [9,22–24] is consistent with a separate placement of these organisms in the archaeal tree as this lipid has so far not been found in any other bacterium or archaeon. Thus, it seems likely that this membrane lipid, which may now be more appropriately termed thaumarchaeol, is an invention of an early thaumarchaote and represents a signature lipid for this phylum.
Figure 1.
Phylogeny of ammonia-oxidizing Thaumarchaeota. (a) Schematic phylogeny of Archaea redrawn after a rooted maximum likelihood tree of 53 concatenated ribosomal proteins of Archaea (4853 deduced amino acid positions), with permission from Spang et al. [19•]. The scale bar represents 10% estimated sequence divergence. (b) Majority consensus trees based on the 16S rRNA gene (1067 nucleic acid positions conserved in >50% of all Archaea) and archaeal amoA gene (592 nucleic acid positions) as inferred by maximum likelihood, distance, and maximum parsimony methods. ‘Nanoarchaeota’ have been shown to represent a fast evolving lineage of the Euryarchaeota (reviewed by Brochier-Armanet et al. in this issue and (a)) and are misplaced in 16S rRNA-based trees as a sister group of the Crenarchaeota. The shadowed area highlights a region of the 16S rRNA tree with unstable branching order and low bootstrap support. The bars represent 10% Jukes-Cantor corrected sequence divergence. Dots (●) indicate in all trees bootstrap support above 80% as inferred by maximum likelihood. Abbreviations: N. yellowstonii, Nitrosocaldus yellowstonii; N. gargensis, Nitrososphaera gargensis; N. maritimus, Nitrosopumilus maritimus; C. symbiosum, Cenarchaeum symbiosum; N. limnia, Nitrosoarchaeum limnia, SAGMCG-1, South African Gold Mine Crenarchaeotic Group 1.
Revisiting the phylogenetic placement of Thaumarchaeota in 16S rRNA-based trees also reveals a clear separation from Crenarchaeota and Euryarchaeota (Figure 1b). A number of environmentally retrieved clone groups consisting of the SAGMGC-1 group (subsurface mine), group I.1c (acidic soils), ALOHA group (open ocean), pSL12 group (hot spring), and the HWCGIII/Nitrosocaldus group (hot springs/hydrothermal vents) form a monophyletic cluster with known Thaumarchaota. Since this cluster is supported by all treeing methods and has a bootstrap value of 100% (Figure 1b), its representatives very likely all belong to the phylum Thaumarchaeota and at least some of them might be AOA. Supporting this hypothesis, a good correlation between copy numbers of archaeal amoA (coding for the α-subunit of ammonia monooxygenase) and 16S rRNA genes of the ALOHA group has been observed in the North Pacific [25]. It will be fascinating to see whether all Thaumarchaeota have the capability to perform ammonia oxidation or whether certain members use a different energy metabolism. Just recently two giant thaumarchaeota, Candidatus Giganthauma karukerense and Candidatus Giganthauma insulaporcus, were characterized by molecular methods but all attempts to amplify archaeal amoA genes failed [26•]. However, this could also be caused by primer bias as has been previously recognized for archaeal _amoA_-targeted surveys in deep ocean waters [27,28].
Currently, the MCG (Miscellaneous Crenarchaeotic Group), MBGB (Marine Benthic Group B), and HWCGI (Hot Water Crenarchaeotic Group I) clusters have no clear affiliation to any of the established archaeal phyla and show an unstable branching order when 16S rRNA-based trees inferred with different treeing methods are compared (Figure 1b). Little is known about these organisms but recently the first genome of a representative of the HWCGI cluster, that of Candidatus Caldiarchaeum subterraneum, was found to be distinct from other archaeal phyla including genes encoding a ubiquitin-like protein modifier system that was so far only found in eukaryotes. As a consequence, the lineage ‘Aigarchaeota’ was proposed [29••]. However, a comparative genome analysis by Brochier-Armanet and colleagues revealed some typical thaumarchaeal features in C. subterraneum and thus places it at the base of Thaumarchaeota in protein trees (for details see Brochier-Armanet et al., this issue). With the availability of more genomes within this and related lineages, comparative genomics will show whether ‘Aigarchaeota’ represent a new archaeal phylum or will be classified as deep-branching members of the Crenarchaeota or Thaumarchaeota.
The phylogenetic structure of AOA can also be analyzed by the functional marker gene amoA, which is found in all ammonia-oxidizing microorganisms. The presence of AOA within Group I.1a and Group I.1b Thaumarchaeota as well as within the Thaumarchaeota-group HWCGIII/Nitrosocaldus is mirrored in the respective amoA phylogeny (Figure 1b). In addition, a fourth _amoA_-cluster with no established link to a thaumarchaeotal lineage in the 16S rRNA-based tree became apparent during the accumulation of environmental amoA sequences within the last few years. Since amoA sequences from a wide range of habitats (including various marine, terrestrial, and hot water environments) are affiliated with this lineage, we have named it the ‘ubiquitous cluster’. It is tempting to speculate that this cluster represents so-far unrecognized AOA within the SAGMGC-1, group I.1c, ALOHA, or pSL12 cluster.
Emerging ecophysiology of Thaumarchaeota
Almost every study that investigates ammonia-oxidizing Thaumarchaeota uses the amoA gene to explore their diversity and abundance with the implicit assumption that all _amoA_-carrying archaea are oxidizing ammonia. However, of the >10,000 deposited archaeal amoA sequences, thus far only four have been directly linked to archaeal strains for which experimental evidence of ammonia oxidation exists [8–11]. Although phylogenetically closely related enzymes often perform the same function, it deserves consideration that the family of copper-containing membrane-bound monooxgenases (CuMMO), to which archaeal ammonia monooxygenases belong, has a wide substrate range. In addition to ammonia [ammonia monooxygenase (AMO) in β-Proteobacteria, γ-Proteobacteria, and Thaumarchaeota] [30], this includes methane [particulate methane monooxygenase (pMMO) in α-Proteobacteria, γ-Proteobacteria, Verucomicrobia, and Candidatus _Methylomirabilis oxyfera_] [31,32], and short-chained alkanes [particulate butane monooxygenase (pBMO) in the Gram-positive Nocardioides strain CF8] [33••]. In addition, non-specific substrate catabolism such as oxidation of chlorinated ethenes and aromatic hydrocarbons has been observed with some members of this enzyme family [34,35], clearly indicating substrate promiscuity. Therefore, it has been suggested that not necessarily the type of CuMMO but rather the downstream enzyme machinery defines the energy metabolism of a microorganism [36]. For example, the γ-Proteobacterium AOB Nitrosococcus oceani can oxidize methane but lacks all subsequent enzymes to gain energy by methane oxidation [37]. Likewise, co-oxidation of ammonia by methane oxidizing bacteria does not support their growth [38]. Furthermore, it is interesting to note that γ-proteobacterial AMOs are more closely related to γ-proteobacterial pMMOs than to β-proteobacterial AMOs and have a near equal substrate specificity for ammonia and methane [39]. Consequently, the mere presence of an _amoA_-like gene, transcript, or protein is insufficient to infer that the respective organism is oxidizing ammonia.
Currently, it is not clear whether AOA are strict autotrophs or also assimilate organic substrates. For N. maritimus, autotrophy has been shown [8] and for N. gargensis CO2-fixation has been experimentally demonstrated [10]. Incorporation of labeled bicarbonate into lipids, proteins, and cells of marine thaumarchaeota [27,40,41] are consistent with autotrophy, which is enabled by a modified 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle for CO2-fixation as found in known AOA genomes and in marine thaumarchaeal fosmids [7,42•,43,44]. However, analysis of the C. symbiosum and N. maritimus genomes as well as of thaumarchaeal fosmids from bathypelagic plankton also has revealed the presence of a TCA cycle (possibly incomplete) and of potential transporters for organic substances such as amino acids, oligopeptides, and glycerol [42•,43,45]. Thus, mixotrophic or even heterotrophic growth of marine Thaumarchaeota as supported by other isotope labeling studies and natural distribution of radiocarbon in archaeal membrane lipids [27,46,47] can to date not be excluded. Furthermore, it has been suggested that parts of the HP/HB cycle can serve to co-assimilate organic compounds including, for example, 3-hydroxypropionate, an intermediate in the metabolism of the ubiquitous marine osmoprotectant dimethylsulphoniopropionate [48•]. For soil environments, 13CO2-stable isotope probing revealed ammonia oxidizing activity of members of group I.1a as well as I.1b Thaumarchaeota [49•,50,51•] indicating an autotrophic or mixotrophic lifestyle. Two of these studies found label incorporation into genes or transcripts of the 4-hydroxybutyryl-CoA-dehydratase [51•] or acetyl-CoA-propionyl-CoA-carboxylase [49•], respectively, with both enzymes being involved in the CO2-fixing HP/HB cycle [48•]. However, growth of soil AOA with no concomitant incorporation of 13CO2 has been also observed when nitrification was inhibited [52] indicating that at least some soil AOA can grow heterotrophically. For comparison, heterotrophic growth of Crenarchaeota that possess the HP/HB cycle is known for Sulfolobus solfataricus and Metallosphaera sedula with the latter being able to switch between an autotrophic and heterotrophic lifestyle [48•,53].
The question under which conditions AOA or AOB dominate ammonia oxidation is currently attracting a lot of attention. For ammonia oxidation by the group I.1a Thaumarchaeote N. maritimus, an extremely low substrate threshold (<10 nM total NH4+ + NH3, representing the detection limit of the used method) and apparent _K_m-value (133 nM) were determined with the latter being very similar to _in situ_ nitrification measurements made in oligotrophic oceans [54••]. Adaptation to low ammonium concentrations has also been reported for the thermophilic group I.1b Thaumarchaeote _N. gargensis_ [10], indicating a widespread distribution of oligotrophic ammonia oxidation within the Thaumarchaeota. In comparison, minimum total ammonium concentrations required for growth of cultured AOB are 100-fold higher (>1 μM near neutral pH) with _K_m-values ranging from 46 to 1780 μM total ammonium [54••,55]. Thus, a dominating activity of AOA in the large water bodies of oligotrophic oceans is highly likely with AOB being restricted to organic-matter rich particles and coastal environments with higher nutrient loads [54••]. Measured apparent _K_m-values for soils range from 2 to 42 μM total ammonium [54••,55] and may therefore be influenced by both AOA and AOB. In general, activity of soil AOA was seen when total ammonia concentrations were below 15 μg NH4+-N (g dw. soil)−1 [30,49•] whereas AOB responded to high ammonia concentrations [>100 μg NH4+-N (g dw. soil)−1] [49•,50,52,56]. In addition, the form of supplied nitrogen might also play a critical role: AOA activity was seen when N was supplied as mineralized organic N derived from composted manure or soil organic matter and AOB-dominated activity was seen with ammonia from inorganic fertilizer (reviewed in [30]).
Based on genome analyses of N. maritimus and C. symbiosum and due to the fact that AOA do not contain a homologue of the bacterial hydroxylamine oxidoreductase, a mechanism for ammonia oxidation distinctly different from that of AOB has been proposed. Here, ammonia is not oxidized via hydroxylamine (NH2OH) as in AOB but rather via nitroxyl (HNO) to nitrite [42•], which possibly involves only 0.5 O2 per NH3 oxidized (proposed by Martin Klotz (Louisville) [30]). This hypothesized lower oxygen demand could explain why AOA are found not only in fully areated soils and oxic marine waters but also in suboxic marine waters, sediments, and oxygen-depleted hot springs [30,57]. In oxygen gradients of marine sediments and in the stratified water body of the Black Sea different AOA ecotypes were found to reside at different oxygen concentrations [58,59]. AOA can also be found over a wide range of pH, temperature, salinity, and phosphate concentrations with some AOA being adapted to sulfidic environments, which extends the potential range of AOA niche differentiation to a multitude of environmental factors (reviewed in [30,57]).
Conclusion and outlook
Until recently, methanogenic euryarchaeota were the only known archaea of global relevance for element cycling. This perception changed with the discovery of ammonia-oxidizing archaea, which belong to the newly recognized archaeal phylum Thaumarchaeota and contribute significantly to the global N-cycle and C-cycle. Their shear abundance in the ocean (up to 20% of all bacteria and archaea [60]) and extremely low substrate threshold for total ammonium provide compelling evidence for their role as dominant ammonia oxidizers in the open ocean, where they also contribute to primary production by their autotrophic (or possibly partly mixotrophic) lifestyle. The dominance of AOA over AOB in many terrestrial environments cannot be so easily explained. Low _K_m-values of unfertilized soils for ammonia oxidation [54••,55] might point to a contribution of certain AOA ecotypes to nitrification, especially under low ammonia availability. On the other hand, it is well possible that some soil thaumarchaeotes use other substrates than ammonia for energy generation and are heterotrophs or that they switch to ammonia oxidation only under certain environmental conditions. Future research is needed to investigate the ecophysiology of thaumarchaeota in greater detail. Further dissection of the ecological interplay of AOA groups among themselves and with AOB is urgently required and might reveal that AOA exhibit a similar type of niche partitioning as found for different nitrite oxidizers. Here, Nitrobacter spp. are known to dominate nutrient rich and oxygen saturated environments whereas Nitrospira spp. prefer low nutrient and microoxic sites [61], with different Nitrospira lineages adapted to different nitrite concentrations [62].
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
- • of special interest
- •• of outstanding interest
Acknowledgements
We would like to thank Roland Hatzenpichler, Alexander Galushko, Anja Spang, and Pierre Offre for helpful discussions and David Berry for proof-reading the manuscript. We apologize to all authors whose work could not be cited due to the limited amount of allowed references. This research was financially supported by the Austrian Science Fund (P23117, MP).
References
- 1.Gruber N., Galloway J.N. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 2.Purkhold U., Pommerening-Roser A., Juretschko S., Schmid M.C., Koops H.P., Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Boer W., Kowalchuk G.A. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- 4.Prosser J.I. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 5.Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D.Y., Paulsen I., Nelson K.E., Nelson W. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 6.Treusch A.H., Leininger S., Kletzin A., Schuster S.C., Klenk H.-P., Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 7.Hallam S.J., Mincer T.J., Schleper C., Preston C.M., Roberts K., Richardson P.M., DeLong E.F. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:2412–12412. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Könneke M., Bernhard A.E., de la Torre J.R., Walker C.B., Waterbury J.B., Stahl D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 9.de la Torre J.R., Walker C.B., Ingalls A.E., Konneke M., Stahl D.A. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 10.Hatzenpichler R., Lebedeva E.V., Spieck E., Stoecker K., Richter A., Daims H., Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A. 2008:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blainey P.C., Mosier A.C., Potanina A., Francis C.A., Quake S.R. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis C.A., Roberts K.J., Beman J.M., Santoro A.E., Oakley B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wuchter C., Abbas B., Coolen M.J.L., Herfort L., van Bleijswijk J., Timmers P., Strous M., Teira E., Herndl G.J., Middelburg J.J. Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leininger S., Urich T., Schloter M., Schwark L., Qi J., Nicol G.W., Prosser J.I., Schuster S.C., Schleper C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C.L., Ye Q., Huang Z.Y., Li W.J., Chen J.Q., Song Z.Q., Zhao W.D., Bagwell C., Inskeep W.P., Ross C. Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol. 2008;74:6417–6426. doi: 10.1128/AEM.00843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuhrman J.A., McCallum K., Davis A.A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 17.Delong E.F. Archaea in coastal marine environments. Proc Natl Acad Sci U S A. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Brochier-Armanet C., Boussau B., Gribaldo S., Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]; First suggestion that AOA might be sufficiently different from other archaea to warrant the proposal of a new phylum, based on detailed phylogenetic and comparative genomic data.
- 19•.Spang A., Hatzenpichler R., Brochier-Armanet C., Rattei T., Tischler P., Spieck E., Streit W., Stahl D.A., Wagner M., Schleper C. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]; Extended study with group I.1a and I.1b AOA that supports the deep-branching archaeal phylum Thaumarchaeota. The presence/absence analysis of genes revealed among other things that Thaumarchaeota encode homologues of both archaeal cell-division systems (from Euryarchaeota and Crenarchaeota) and might thus reflect the ancestral distribution of those genes in the last archaeal common ancestor.
- 20.Gupta R., Shami A. Molecular signatures for the Crenarchaeota and the Thaumarchaeota. Antonie van Leeuwenhoek. 2011;99:133–157. doi: 10.1007/s10482-010-9488-3. [DOI] [PubMed] [Google Scholar]
- 21.Brochier-Armanet C., Deschamps P., Lopez-Garcia P., Zivanovic Y., Rodriguez-Valera F., Moreira D. Complete-fosmid and fosmid-end sequences reveal frequent horizontal gene transfers in marine uncultured planktonic archaea. ISME J. 2011 doi: 10.1038/ismej.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitcher A., Rychlik N., Hopmans E.C., Spieck E., Rijpstra W.I.C., Ossebaar J., Schouten S., Wagner M., Damsté J.S.S. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic Group I.1b Archaeon. ISME J. 2010;4:542–552. doi: 10.1038/ismej.2009.138. [DOI] [PubMed] [Google Scholar]
- 23.Schouten S., Hopmans E.C., Baas M., Boumann H., Standfest S., Konneke M., Stahl D.A., Damsté J.S.S. Intact membrane lipids of “Candidatus Nitrosopumilus maritimus” a cultivated representative of the cosmopolitan mesophilic group I crenarchaeota. Appl Environ Microbiol. 2008;74:2433–2440. doi: 10.1128/AEM.01709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damsté J.S.S., Schouten S., Hopmans E.C., van Duin A.C.T., Geenevasen J.A.J. Crenarchaeol: the characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota. J Lipid Res. 2002;43:1641–1651. doi: 10.1194/jlr.m200148-jlr200. [DOI] [PubMed] [Google Scholar]
- 25.Mincer T.J., Church M.J., Taylor L.T., Preston C., Karl D.M., DeLong E.F. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 26•.Muller F., Brissac T., Le Bris N., Felbeck H., Gros O. First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol. 2010;12:2371–2383. doi: 10.1111/j.1462-2920.2010.02309.x. [DOI] [PubMed] [Google Scholar]; This study reports on the discovery of two marine thaumarchaeal species from shallow waters that consist of multiple giant cells.One species was shown to be coated with sulfur-oxidizing γ-Proteobacteria indicating that some thaumarchaeotes might form symbioses with bacteria.
- 27.Herndl G.J., Reinthaler T., Teira E., van Aken H., Veth C., Pernthaler A., Pernthaler J. Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konstantinidis K.T., Braff J., Karl D.M., DeLong E.F. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific Subtropical Gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Nunoura T., Takaki Y., Kakuta J., Nishi S., Sugahara J., Kazama H., Chee G.-J., Hattori M., Kanai A., Atomi H. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the first metagenome of a representative of the HWCGI cluster that forms a sister group to Thaumarchaeota and hyperthermophilic Crenarchaeota in 16S rRNA trees. Candidatus Caldiarchaeum subterraneum is apparently unable to oxidize ammonium for energy generation but might grow chemolithotrophically with carbon monoxide. As the composite genome was found to be clearly distinct from other archaea the new lineage ‘Aigarchaeota’ was proposed.
- 30.Schleper C., Nicol G.W. Ammonia-oxidising archaea — physiology, ecology and evolution. Adv Microb Physiol. 2010;57:1–41. doi: 10.1016/B978-0-12-381045-8.00001-1. [DOI] [PubMed] [Google Scholar]
- 31.Semrau J.D., DiSpirito A.A., Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 32.Ettwig K.F., Butler M.K., Le Paslier D., Pelletier E., Mangenot S., Kuypers M.M.M., Schreiber F., Dutilh B.E., Zedelius J., de Beer D. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 33••.Sayavedra-Soto L.A., Hamamura N., Liu C.-W., Kimbrel J.A., Chang J.H., Arp D.J. The membrane-associated monooxygenase in the butane-oxidizing Gram-positive bacterium Nocardioides sp. strain CF8 is a novel member of the AMO/PMO family. Environ Microbiol Rep. 2011 doi: 10.1111/j.1758-2229.2010.00239.x. [DOI] [PubMed] [Google Scholar]; This study extends the specific substrate range of copper-containing membrane-bound monooxygenases to short-chained alkanes (C2–C10) and shows that this enzyme family also exists in Gram-positive Actinobacteria.
- 34.Chang S.W., Hyman M.R., Williamson K.J. Cooxidation of naphthalene and other polycyclic aromatic hydrocarbons by the nitrifying bacterium, Nitrosomonas europaea. Biodegradation. 2002;13:373–381. doi: 10.1023/a:1022811430030. [DOI] [PubMed] [Google Scholar]
- 35.Yoon S., Im J., Bandow N., DiSpirito A.A., Semrau J.D. Constitutive expression of pMMO by Methylocystis strain SB2 when grown on multi-carbon substrates: implications for biodegradation of chlorinated ethenes. Environ Microbiol Rep. 2011;3:182–188. doi: 10.1111/j.1758-2229.2010.00205.x. [DOI] [PubMed] [Google Scholar]
- 36.Tavormina P.L., Orphan V.J., Kalyuzhnaya M.G., Jetten M.S.M., Klotz M.G. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3:91–100. doi: 10.1111/j.1758-2229.2010.00192.x. [DOI] [PubMed] [Google Scholar]
- 37.Klotz M.G., Arp D.J., Chain P.S.G., El-Sheikh A.F., Hauser L.J., Hommes N.G., Larimer F.W., Malfatti S.A., Norton J.M., Poret-Peterson A.T. Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl Environ Microbiol. 2006;72:6299–6315. doi: 10.1128/AEM.00463-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trotsenko Y.A., Murrell J.C. Metabolic aspects of aerobic obligate methanotrophy. In: Allen I., Laskin S.S., Geoffrey M.G., editors. Advances in Applied Microbiology. Academic Press; 2008. pp. 183–229. [DOI] [PubMed] [Google Scholar]
- 39.Lontoh S., DiSpirito A.A., Krema C.L., Whittaker M.R., Hooper A.B., Semrau J.D. Differential inhibition in vivo of ammonia monooxygenase, soluble methane monooxygenase and membrane-associated methane monooxygenase by phenylacetylene. Environ Microbiol. 2000;2:485–494. doi: 10.1046/j.1462-2920.2000.00130.x. [DOI] [PubMed] [Google Scholar]
- 40.Wuchter C., Schouten S., Boschker H.T.S., Damsté J.S.S. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett. 2003;219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 41.Yakimov M.M., Cono V.L., Smedile F., DeLuca T.H., Juarez S., Ciordia S., Fernandez M., Albar J.P., Ferrer M., Golyshin P.N. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea) ISME J. 2011 doi: 10.1038/ismej.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Walker C.B., de la Torre J.R., Klotz M.G., Urakawa H., Pinel N., Arp D.J., Brochier-Armanet C., Chain P.S.G., Chan P.P., Gollabgir A. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci U S A. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Based on the genome analysis of Candidatus Nitrosopumilus maritimus and on the fact that known AOA do not contain a homolog of the bacterial hydroxylamine oxidoreductase, a mechanism for ammonia oxidation distinctly different from that of AOB was proposed. Here, ammonia is not oxidized via hydroxylamine such as in AOB but possibly via nitroxyl to nitrite.
- 43.Martin-Cuadrado A.-B., Rodriguez-Valera F., Moreira D., Alba J.C., Ivars-Martinez E., Henn M.R., Talla E., Lopez-Garcia P. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008;2:865–886. doi: 10.1038/ismej.2008.40. [DOI] [PubMed] [Google Scholar]
- 44.La Cono V., Smedile F., Ferrer M., Golyshin P.N., Giuliano L., Yakimov M.M. Genomic signatures of fifth autotrophic carbon assimilation pathway in bathypelagic Crenarchaeota. Microb Biotechnol. 2010;3:595–606. doi: 10.1111/j.1751-7915.2010.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hallam S., Mincer T., Schleper C., Preston C., Roberts K., Richardson P., DeLong E. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine crenarchaeota. PLoS Biol. 2006;4:2412–12412. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingalls A.E., Shah S.R., Hansman R.L., Aluwihare L.I., Santos G.M., Druffel E.R.M., Pearson A. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci U S A. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouverney C.C., Fuhrman J.A. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Berg I.A., Kockelkorn D., Ramos-Vera W.H., Say R.F., Zarzycki J., Hügler M., Alber B.E., Fuchs G. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]; This review gives a detailed overview on the different carbon fixation pathways found in archaea. Of special interest is the recently discovered hydroxypropionate/hydroxybutyrate cycle found in thermophilic Crenarchaeota and its modified version proposed for ammonia-oxidizing Thaumarchaeota. The fundamental difference of this cycle to most other archaeal CO2-fixation pathways is the fact that its enzymes tolerate oxygen.
- 49•.Pratscher J., Dumont M.G., Conrad R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci U S A. 2011;108:4170–4175. doi: 10.1073/pnas.1010981108. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study gives support that members of the very abundant soil group I.1b Thaumarchaeota are actually involved in ammonia oxidation in soils. In addition, this study is one of the first that shows that mRNA-stable isotope probing can be conducted in soil environments.
- 50.Xia W., Zhang C., Zeng X., Feng Y., Weng J., Lin X., Zhu J., Xiong Z., Xu J., Cai Z. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 2011 doi: 10.1038/ismej.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Zhang L.-M., Offre P.R., He J.-Z., Verhamme D.T., Nicol G.W., Prosser J.I. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci U S A. 2010;107:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows for the first time ammonia-oxidizing activity of group I.1a Thaumarchaeota in soils by 13CO2-stable isotope probing and gives support for an autotrophic lifestyle by label incorporation into the gene of a key enzyme of the hydroxypropionate/hydroxybutyrate cycle.
- 52.Jia Z., Conrad R. Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 53.Huber H, Prangishvili D. Sulfolobales. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.H., Stackebrandt E., editors. edn 3. vol 3. Springer; 2006. pp. 23–51. (Prokaryotes: A Handbook on the Biology of Bacteria). [Google Scholar]
- 54••.Martens-Habbena W., Berube P.M., Urakawa H., de la Torre J.R., Stahl D.A. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]; This very exciting study on the physiology of the marine isolate Nitrosopumilus maritimus showed an extremely low half-saturation constant and substrate threshold for total ammonium, which gave strong support that AOA dominate ammonia oxidation in oligotrophic oceans.
- 55.Koper T.E., Stark J.M., Habteselassie M.Y., Norton J.M. Nitrification exhibits Haldane kinetics in an agricultural soil treated with ammonium sulfate or dairy-waste compost. FEMS Microbiol Ecol. 2010;74:316–322. doi: 10.1111/j.1574-6941.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- 56.Di H.J., Cameron K.C., Shen J.P., Winefield C.S., O’Callaghan M., Bowatte S., He J.Z. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol. 2010;72:386–394. doi: 10.1111/j.1574-6941.2010.00861.x. [DOI] [PubMed] [Google Scholar]
- 57.Erguder T.H., Boon N., Wittebolle L., Marzorati M., Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 58.Coolen M.J.L., Abbas B., van Bleijswijk J., Hopmans E.C., Kuypers M.M.M., Wakeham S.G., Damste J.S.S. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol. 2007;9:1001–1016. doi: 10.1111/j.1462-2920.2006.01227.x. [DOI] [PubMed] [Google Scholar]
- 59.Durbin A.M., Teske A. Sediment-associated microdiversity within the Marine Group I Crenarchaeota. Environ Microb Rep. 2010;2:693–703. doi: 10.1111/j.1758-2229.2010.00163.x. [DOI] [PubMed] [Google Scholar]
- 60.Karner M.B., DeLong E.F., Karl D.M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 61.Schramm A., De Beer D., Gieseke A., Amann R. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ Microbiol. 2000;2:680–686. doi: 10.1046/j.1462-2920.2000.00150.x. [DOI] [PubMed] [Google Scholar]
- 62.Maixner F., Noguera D.R., Anneser B., Stoecker K., Wegl G., Wagner M., Daims H. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol. 2006;8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]