HIF induces human embryonic stem cell markers in cancer cells (original) (raw)
. Author manuscript; available in PMC: 2012 Jul 1.
Abstract
Low oxygen levels have shown to promote self-renewal in many stem cells. In tumors, hypoxia is associated with aggressive disease course and poor clinical outcomes. Furthermore, many aggressive tumors have shown to display gene expression signatures characteristic of human embryonic stem cells (hESC). We now tested whether hypoxia might be responsible for the hESC signature observed in aggressive tumors. We show that hypoxia, through hypoxia inducible factor (HIF), can induce a hESC-like transcriptional program, including the iPSC inducers, OCT4, NANOG, SOX2, KLF4, cMYC and miRNA-302 in eleven cancer cell lines (from prostate, brain, kidney, cervix, lung, colon, liver and breast tumors). Further, non-degradable forms of HIFα, combined with the traditional iPSC inducers are highly efficient in generating A549 iPSC-like colonies that have high tumorigenic capacity. To test potential correlation between iPSC inducers and HIF expression in primary tumors, we analyzed primary prostate tumors and found a significant correlation between NANOG-, OCT4- and HIF1α-positive regions. Further, NANOG and OCT4 expression positively correlated with increased prostate tumor Gleason score. In primary glioma-derived CD133 negative cells neurospheres and hESC markers were induced in hypoxia but not in normoxia. Together, these findings suggest that HIF targets may act as key inducers of a dynamic state of stemness in pathological conditions.
Keywords: cancer, hESC, HIF, hypoxia, stemness
INTRODUCTION
Stem cells grow in a specialized microenvironment, the stem cell niche, which can regulate the balance between self-renewal, differentiation and stem cell quiescence. Several factors are important within the niche, including cell interactions and environmental factors (1). Recent evidence suggests that many stem cells are also localized in areas with, and benefit from low oxygen, supporting the hypothesis that hypoxia might be important for the undifferentiated phenotype of stem/precursor cells (1, 2).
Many solid tumors contain poorly vascularized regions that are severely hypoxic (3) and contribute to cancer progression by activating transcription factors that promote cell survival, tumor angiogenesis and metastasis (2, 3). Tumor hypoxia is also associated with resistance to radiation and chemotherapy (4). It is therefore not surprising that tumor hypoxia is associated with a more aggressive disease course and poor clinical outcomes (4).
A small proportion of cancer cells exhibit stem cell properties (5). These cells, which have been considered as “tumor initiating cells” or “cancer stem cells”, show the ability of self-renewal and multi-potential differentiation, and have the ability to initiate and propagate tumors (5). Recent findings suggest that these tumorigenic cells are rather common in some cancers (6), suggesting that stem-like cell state could be more dynamic than previously thought. It has been suggested that hypoxia could contribute to the formation of a cancer stem cell niche within the tumor (1).
Low oxygen levels stabilize Hypoxia Inducible Factor (HIF) alpha subunits and thereby induce the transcriptional activity of the heterodimers comprising an α subunit and a β subunit (HIFβ/ARNT) that can bind to hypoxia response elements (HRE) and activate multiple genes, including cMYC and OCT4 (2, 3, 7, 8). In some cells HIF-signaling is also known to regulate cellular metabolism by up-regulating the expression of the glycolytic genes and down-regulating mitochondrial activity by transactivating PDK1, a repressor of Pyruvate dehydrogenase and up-regulating miR-210, a suppressor of the iron-sulfur cluster assembly protein (ISCU) (9). Under normoxic conditions, HIFα undergoes prolyl hydroxylation, binds to an ubiquitin E3-ligase, the Von Hippel-Lindau (VHL) protein and undergoes polyubiquitination dependent rapid proteasomal degradation. Other means of HIF regulation have also been described, including regulation through enzymes involved in the Krebs cycle and miRNA or HDAC-dependent regulation (10–13).
Some aggressive cancers and cancer stem cells display gene expression signatures characteristic of ESCs (14–16). However, whether Oct4 is expressed and function in cancer cells is not yet clear (17, 18). It has been proposed that HIFs play a role in tumor aggressiveness, and that the HIF-target, miR-210, could be a circulating biomarker for certain tumors (19, 20). Whether hypoxia and, specifically, active HIFs are responsible for the hESC signature observed in aggressive tumors is, however, unknown.
Data presented here show that HIF induces hESC markers, including the critical iPSC inducers, OCT4, SOX2, NANOG, MYC and miR-302 in cancer cells. Further, HIF, combined with the traditional iPSC inducers is efficient in generating iPSC-like colonies that are highly tumorigenic. In prostate tumor specimens HIF1α co-localizes with hESC markers, NANOG and OCT4 and expression of these stem cell markers positively correlates with high Gleason score, indication of prostate tumor aggressiveness. Furthermore, primary non stem glioma cells are able to form neurospheres that up-regulate hESC markers in hypoxic but not in normoxic conditions.
MATERIALS AND METHODS
Cells, tissue culture and hypoxia induction
Human embryonic stem cells (hESC) lines were obtained from Wicell Research Institute (Madison, WI, USA) and cultured as previously described (21, 22).
HCT116, HT29, DLD1 and RKO (colorectal carcinoma), HeLa and ME180 (cervical carcinoma), A549 and H1299 (lung carcinoma), MCF7 (breast carcinoma), U251 (glioma), Hep3B and HuH7 (hepatocarcinoma) cells were from the American Type Culture Collection (Rockville, MD). 786-O cells transfected either with an empty vector or wild-type VHL (23), and HCT116 Dicer hypomorph line (HCT116 Dcr-, 24) were obtained from Dr. Kaelin and Dr. Vogelstein, respectively.
Cultures enriched for or depleted of glioma stem cells (GSCs) were isolated from primary human brain tumor specimens as previously described in accordance with a Duke University Institutional Review Board approved protocol concurrent with the national regulatory standards with patients signing for informed consent (25). CD133+ cells were designated as GSCs whereas CD133− cells were used as non-stem glioma cells.
Hypoxia induction: cells were cultured in multi-gas incubators (Sanyo, San Diego, CA). Nitrogen gas was supplied to the chambers in order to induce a controlled reduced percentage of oxygen. For normoxia, cells were cultured in incubators containing 5% CO2 and atmospheric concentration of O2, approximately 20% O2.
Overexpression of Oct4, Sox2, Nanog and Lin28 in human lung adenocarcinoma cell line
A549 cells were first infected in 6-well plates by retroviruses with non degradable forms of HIF1α and HIF2α (23) or an empty vector control, and then infected 6 days later by lentiviruses expressing Oct4/Sox2, and Nanog/Lin28 (OSLN, 26). Viral transductions were performed in the presence of polybrene 4 μg/ml (Sigma-Aldrich). Following 24h, cells were trypsinized, transferred to 10-cm dishes and cultured in DMEM/10% FBS. iPSC-like colonies were picked at day 12 post-infection based on hESC like colony morphology. The picked colonies were subsequently expanded and maintained on irradiated mouse embryonic fibroblasts in hESC media. They were then stained for alkaline phosphatase (AP) activity using a Black Alkaline Phosphatase Substrate Kit II (Vector Laboratories, Burlingame, CA, USA). See supplemental information for details on xenograft formation.
Reporter constructs
Oct4-GFP construct
A 3.9kb OCT4 promoter region-GFP fusion construct was linearized using Apal I restriction enzyme and transfected into cells using lipofectamine 2000 as previously described (27).
HIF sensor
We constructed a HIF-sensor lentiviral vector expressing an enhanced yellow florescent protein (eYFP) under the regulation of six tandem repeats of HIF-binding sites (CGTGTACGTG), followed by a minimal human thymidine kinase (TK) promoter (Zhou et al, in preparation).
miR-302-cluster promoter luciferase reporter
The miR-302-cluster promoter-pGL3 enhancer vector, contains a 3.9-kb miR-302-cluster promoter (as described in (28)) inserted between the KpnI and XhoI sites of pGL3 enhancer vector (Promega, Madison, WI).
siRNA and plasmid transfection
siRNA transfections were performed as described previously (24).
Cells were transfected with non-degradable forms of HIF1α, HIF2α (23) or empty vector control using Lipofectamine 2000. Over-expression of HIFs was confirmed by qPCR and Western blot 2 to 3 days after transfection. Cells expressing the Oct4-GFP construct were seeded in chamber slides 24 hours before transfection. Cells were fixed 48 hours post-transfection with 4% PFA and incubated with 1 μg/ml of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Sigma-Aldrich). GFP and DAPI expression were scored by confocal microscopy (SPE5, Leica).
Luciferase assay, bisulfate sequencing, flow cytometry and immunoblot analysis
Standard protocols were used. Please see details in the Supplemental information.
Gene expression (mRNA and miRNA) analyses
mRNA microarray analysis was performed as described previously (22). Gene expression data analysis was done with the Rosetta Resolver gene expression analysis software (version 7.1 Rosetta Biosoftware, Seattle, WA). The miRNA levels were determined using a quantitative primer-extension PCR assay (29) or miRNA microarrays. For the microRNA qPCR analysis, Ct values were converted to copy numbers by comparison to standard curves generated with single-stranded mature miRNAs and were expressed as copies/10 pg input RNA (approximately equivalent to copies/cell).
mRNA and miRNA qRT-PCR
Total RNA was extracted using TriZol (Invitrogen) and treated with DNase (Ambion, Austin, TX). RNA abundance was determined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). After reverse transcription reaction using Omniscript RT kit (Qiagen, Valencia, CA), quantitative PCR was performed using SybrGreen master mix (Applied Biosystems, Carlsbad, CA) or, TaqMan assay (Applied Biosystems) on Applied Biosystems 7300 Real-time PCR cycler. All data were normalized to 28S or β-ACTIN transcript levels. The gene primers used are listed in Suppl. Table VI. TaqMan miRNA assays (Applied Biosystems) were used for miRNA qPCR according to the manufacturer’s protocol using RNU66 snoRNA as a loading control.
Immunohistochemistry on patient samples
Prostate
This study was carried out under approval by the University of Washington IRB for the research use of excess tissue from surgeries. Frozen blocks of prostate tumor tissue were obtained through the UWMC Division of Urology. Serial sections were stained with anti-HIF1α, anti-NANOG and anti-OCT4 antibodies (Suppl. Table VII) using a protocol previously described (30). Gleason pattern of prostate tumors was given by a pathologist and CD10, CD107b and CD104 (BD Biosciences) antibodies were used to stain tumor specimens (30). Prostate cancer glands were scored for the presence of HIF1α and NANOG staining. Please see supplemental information for details. For each patients, local Gleason grading was performed as described previously (31) in individual microscopic areas (100X magnification). Particularly, Gleason 5 was assigned when single round and undifferentiated epithelial cells were observed without any trace of gland formation.
GBM
Formalin fixed and paraffin embedded blocks of GBM (glioblastoma multiforme) were obtained throught the UWMC Division of Neuropathology with IRB approval. Serial sections were processed from neuropathologically verified representative sections of grade IV glioblastoma according to a previously described protocol (32) and stained with HIF1α and NANOG antibodies (Suppl. Table VII).
Statistical analysis
P values were calculated using Student’s t-test. *, P< 0.05 and **, P< 0.01. Bars show SEM for 3 or 4 separate experiments.
RESULTS
Hypoxia induces stem cell markers in cancer cell lines
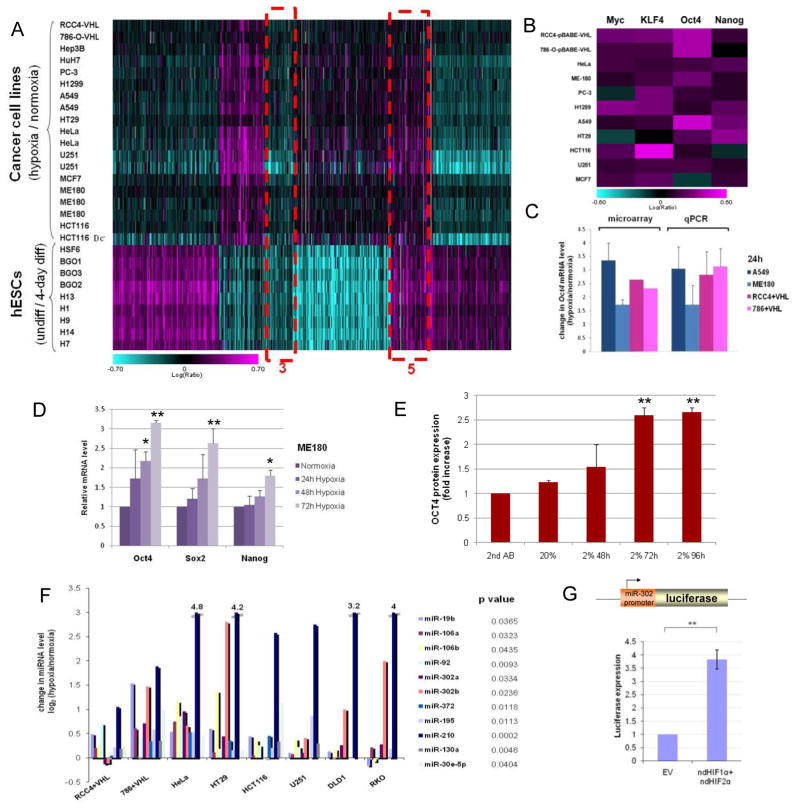
We analyzed the common gene expression signature of 11 different cancer cell lines under hypoxic conditions (2% O2) and compared it to the hESC expression pattern (9 hESC lines; 22). A significant overlap was observed between the differentially expressed mRNAs in these two groups (Fig. 1, cluster 5; Suppl. Fig. 1; Suppl. Table I). Many of the overlapping mRNAs corresponded to genes involved in stemness and in the reprogramming of fibroblasts to hESC-like cells, iPSCs (Fig. 1B; 26, 33). Among the hypoxia up-regulated genes were cMYC, KLF4, OCT4 and NANOG (Fig. 1B). The hypoxia induced OCT4 up-regulation was validated in multiple cancer cell lines using qPCR (Fig. 1C). Multiple primers were utilized to confirm that the isoform of OCT4 shown to be involved in stemness (OCT4-A) was up-regulated in these hypoxic cancer cell lines (Suppl. Fig. 2A–B; 18). Oct4 was previously shown to be a transcriptional target of HIF2α (7). Overall, the HIF target genes and genes enriched in tumors were observed in the overlap between hypoxic cancer cell and hESC signatures (Suppl. Fig. 1A, Suppl. Tables I and II; brain, prostate and kidney tumors; Suppl. Table III). Particularly, genes expressed in Wilms tumors were found in the hypoxic cancer cell and hESC overlap (Suppl. Fig. 1B–C). Wilms tumor and hESC similarity has previously been identified on epigenetic level (34). The data presented suggest that HIF activity regulates these processes.
Figure 1. Hypoxia induces hESC markers among cancer cells.
(A) mRNA profiling of hESC and hypoxic cancer cell lines (2% O2 24h) shown in a heatmap representation of gene expression changes. The color bar represents log10 expression ratio −0.7 (teal) to +0.7 (magenta) (P<0.01 in at least 9 experiments). Outlined in dotted red are the overlap between these two profiling experiments. Cluster #5 (common up-regulated genes) contains OCT4 and other stem cell markers. (B) iPSC inducers are upregulated in cancer cells in response to hypoxia. The color bar represents log10 expression ratio (hypoxia relative to normoxia) −0.6 (teal) to +0.6 (magenta). (C) qPCR validation of OCT4 expression in various cancer cell lines after 24h in hypoxia (2% O2). (D) Stem cell markers are up-regulated by hypoxia over time in ME180 cells (qPCR analysis). (E) OCT4 protein is up-regulated by hypoxia in ME180 cells (flow cytometry analysis). Incubation with the secondary antibody (2nd AB) alone was used as a negative control. (F) hESC enriched miRNAs are up-regulated by hypoxia treatment (24h in 2% O2) in multiple tumor cell lines (qPCR analysis). p values were calculated using paired t-test to measure the difference between the microRNA level under hypoxia and normoxia conditions in all cell lines. (G) The miR-302 promoter is responsive to over-expression of non-degradable forms of HIFα in HeLa cells.
In ME180, while SOX2, a well established stem cell marker (26, 33), was not among the group of hypoxia induced mRNAs at 24h, it was significantly induced at later time points (Fig. 1D). Moreover OCT4, SOX2 and NANOG remained up-regulated in these cells grown in hypoxia compared to normoxia (Fig. 1D). Since multiple pseudogenes for OCT4 exist, we tested whether the true OCT4 promoter was responsive to hypoxia in cancer cells by stably integrating Oct4-GFP reporter in ME180 and U251 lines (ME180-Oct4GFP and U251-Oct4GFP). When these lines were incubated for two days in 2% O2, 6- and 3-fold increase of GFP-positive cells was observed, respectively (Suppl. Fig. 3A–B, Fig. 2C). We also confirmed induction of OCT4 protein in ME180 cells by flow cytometry (Fig. 1E). The OCT4 antibody used in these experiments could detect a nuclear signal in tumor tissues (Fig. 4Q, Suppl. Fig. 10–11). Although in some situations and cell types hypoxia is shown to affect cell division and/or cell death (3), no discernible changes in these processes were observed in hypoxia (2% O2) for the cell lines analyzed (Suppl. Fig.3C). Thus, the increased expression of OCT4, NANOG and SOX2 was unlikely due to altered cell division or survival.
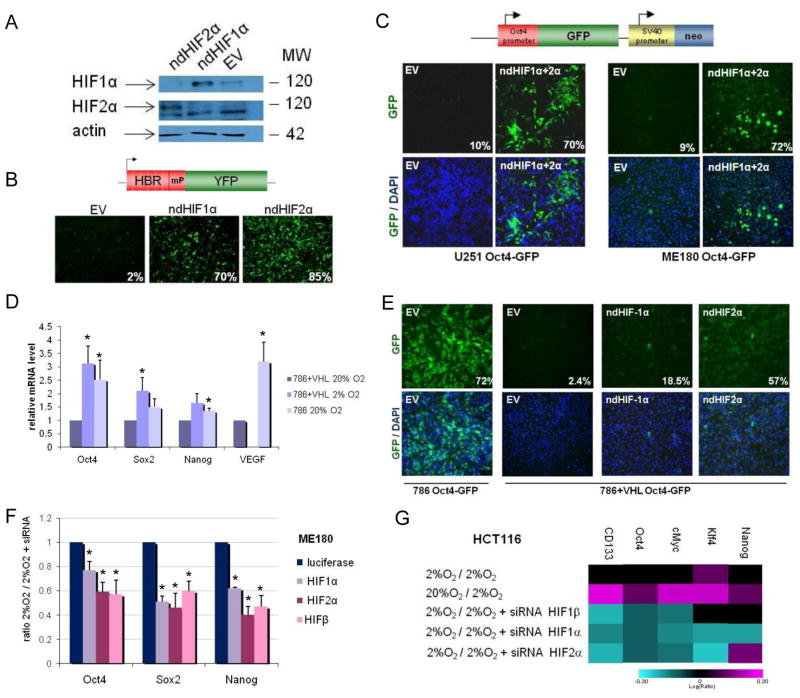
Figure 2. Dependence of HIF for hypoxia-induced expression of stem cell markers.
(A) Overexpression of non-degradable (nd) forms of HIFα under normoxia was assessed by Western blot analysis in ME180 cells 3 days after transfection with either an empty vector control (EV) or ndHIF1α or ndHIF2α. (B) The HIF-sensor containing 6 repeats of the HIF binding site (HBR) upstream of a minimal promoter (mP) and the yellow fluorescence protein (YFP) coding gene (upper panel) is upregulated by overexpression of ndHIF1α or ndHIF2α. (C) OCT4 promoter-GFP reporter is activated in ME180 Oct4-GFP and U251 Oct4-GFP cells two days after transfection with ndHIF1α and ndHIF2α but not with EV. Percentage of GFP-positive cells are indicated. (D) Expression of stem cell markers in renal carcinoma cells expressing either no functional VHL (qPCR; 786-0-pBABE = 786) or a functional VHL (786-0-pBABE-VHL = 786+VHL). (E) Oct4-GFP is induced in 786 but not in 786+VHL cells after transfection with ndHIF1α or ndHIF2α. Percentages of GFP-positive cells are indicated. (F–G) Hypoxia-induced expression of OCT4, NANOG and SOX2 is dependent on both HIF1α and HIF2α in ME180 (F) and HCT116 cells (G). The cells were transfected with siRNA directed against HIF1α, HIF2α or HIFβ. 24h later, the cells were cultured in normoxia or hypoxia (2% O2) for 24h before qPCR (F) or microarray (G) analysis. The color bar represents log10 expression ratio (hypoxia vs. normoxia) −0.3 (teal) to +0.3 (magenta) (G).
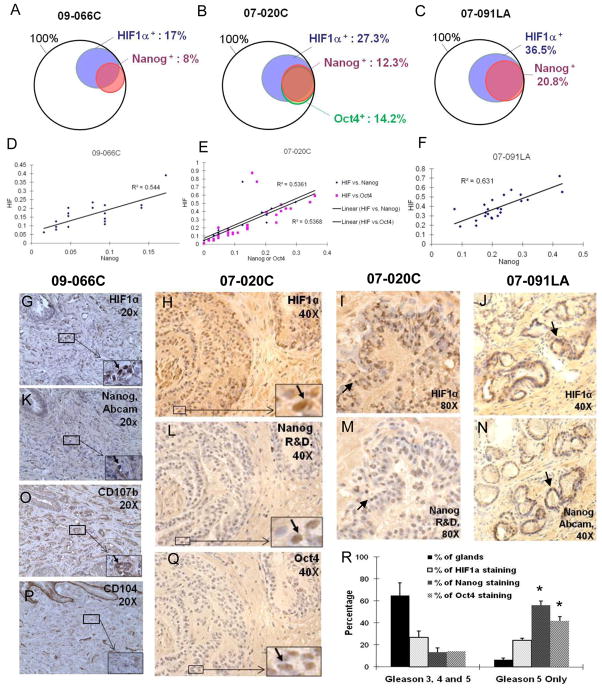
Figure 4. Overlap between HIF, NANOG and OCT4 staining in prostate tumors.
(A–C) Venn diagram representations of the staining analysis of specimen 09066C (A), 07-020C (B) and 07-091LA (C). (D–F) Quantification of HIF and NANOG, and HIF and OCT4 colocalization by correlation analysis in specimen 09-066C (D), 07-020C (E) and 07-091LA (F). Each dot represents 200 to 500 cells. The y-axis shows the ratio of HIF positive staining, and the x-axis the ratio of NANOG or OCT4 positive staining. (G–N) Representative images of HIF1α (G–J) and NANOG (K–N) immunohistochemistry staining on serial sections of specimens 09-066C, 07-020C and 07-091LA. Antibody used for NANOG staining is indicated. High magnification images are presented in the lower right corner. (O–P) CD107b (a marker for a tumor region; O) and CD104 (a marker for normal region; P) staining on serial section of the specimen 09-066C. (Q) Representative images of OCT4 immunohistochemistry staining on serial sections of specimen 07-020C. (R) NANOG and OCT4 expressions are enriched in Gleason 5 only glands, compared to the levels seen in overall glands.
Hypoxia induces stem cell miRNAs in cancer cell lines
We also detected a convergence of miRNA signatures between hypoxic cancer cell lines and undifferentiated hESC lines. Several of the hESC-enriched miRNAs (22, 29), including miR-302a-b, miR-106a-b, miR-92, miR-372, miR-19b, miR-130a, miR-30e-5p and miR-195, were found significantly up-regulated in the cancer cell lines grown in low oxygen (Fig. 1F; Suppl. Table IV), indicating that the cancer lines move towards a stem cell-like miRNA profile when exposed to hypoxia. The hypoxia-induced up-regulation of miR-302b and miR-106a in ME180 cells was validated by further qPCR analysis (Suppl. Fig. 4). We tested whether miR-302 promoter was responsive to HIF using non-degradable (nd) HIF1α and ndHIF2α (23) and a reporter-construct of the miR-302 promoter linked to luciferase. Luciferase activity was up-regulated in the presence of ndHIF1α and ndHIF2α, showing that the miR-302 promoter was responsive to ndHIF expression (Fig. 1G).
As reported previously, miR-210 was also consistently induced in response to hypoxia (Fig. 1F; 19–20). miR-210 has been shown to increase MYC activity by inhibiting MNT, the antagonist of MYC (24) and to regulate metabolism by suppressing the iron-sulfur cluster assembly protein (ISCU) (19). Therefore, HIF dependent miR-210 up-regulation might also contribute to the stem-like phenotypes in these hypoxic cancer cells.
Induction of a hESC-like expression signature is HIF dependent
To test whether the observed Oct4 upregulation was due to HIF expression, we over-expressed ndHIFs in U251 and ME180 cell lines with the Oct4-GFP construct (27; Fig. 2C). These engineered cells express GFP under the control of a 3.9-kb promoter region of OCT4 containing 3 HIF binding sites (7). Expression and activity of the ndHIF1α and ndHIF2α were first validated with Western blot analysis and HIF-binding-repeat (6xHBR)-YFP sensor constructs (Fig. 2A–B). Upon transfection of U251-Oct4-GFP and ME180-Oct4-GFP with the ndHIF constructs, GFP expression was significantly increased showing that ndHIF was able to activate the OCT4 promoter in U251 and ME180 cancer cell lines (Fig. 2C; Supp. Fig. 5A–H). Endogenous hESC marker expression was also significantly increased due to ndHIF expression (Suppl. Fig. 5I).
To further test the HIF pathway function in hESC marker expression, we used 786-O and RCC4 renal carcinoma cell lines lacking VHL activity and therefore expressing constitutively active HIF (23). In normoxia both 786 and RCC4 cells lacking VHL function showed a significant increase in the expression of endogenous hESC markers compared to the corresponding lines with active VHL (Fig. 2D; Suppl. Fig. 6A–E), Using Oct4-GFP-sensor we showed that VHL deficient cells expressed significantly higher levels of GFP compared to those with restored VHL function, suggesting activation of the OCT4 promoter by stabilized HIFs due to the lack of VHL (30× increase; Fig. 2E). A 24-fold increase in GFP-positive cells was observed in 786+VHL cells after overexpression of ndHIFs (Fig. 2E), showing that these cells can respond to stabilized HIF. These data show that the up-regulation of hESC markers in renal carcinoma lines correlated with the lack of VHL activity.
We further tested HIF dependency for hESC marker up-regulation by transfecting ME180 and HCT116 cells with siRNAs directed against HIF1α, HIF2α (EPAS1), HIFβ (ARNT) and luciferase (control). The efficacy and specificity of the siRNAs were confirmed by qPCR or microarray analysis (Suppl. Fig. 7). Hypoxia-induced expression of most hESC markers was dependent on HIFβ, HIF1α and HIF2α in ME180 and HCT116 cells (Fig. 2F–G). No increase in cell death was observed in these experiments, possibly due to the short incubation period in hypoxia after HIF knock-down (Suppl. Fig. 7B). Moreover, knock-down of HIF1α in RCC4 cells up-regulated genes non-related to apoptosis, such as TAF9B, AREG and SGK3. These same genes were down-regulated by hypoxia treatment in RCC4+VHL cells (Suppl. Fig. 7D). Thus the observed reduction of stem cell markers was a result of HIF knock-down and not of defects in viability.
Collectively, these data were consistent with the conclusion that the hypoxia induced hESC mRNA and miRNA signatures in cancer cells are HIF dependent.
Oct4, Sox2, Nanog, Lin28 and HIF overexpression in A549 lung cancer cell line
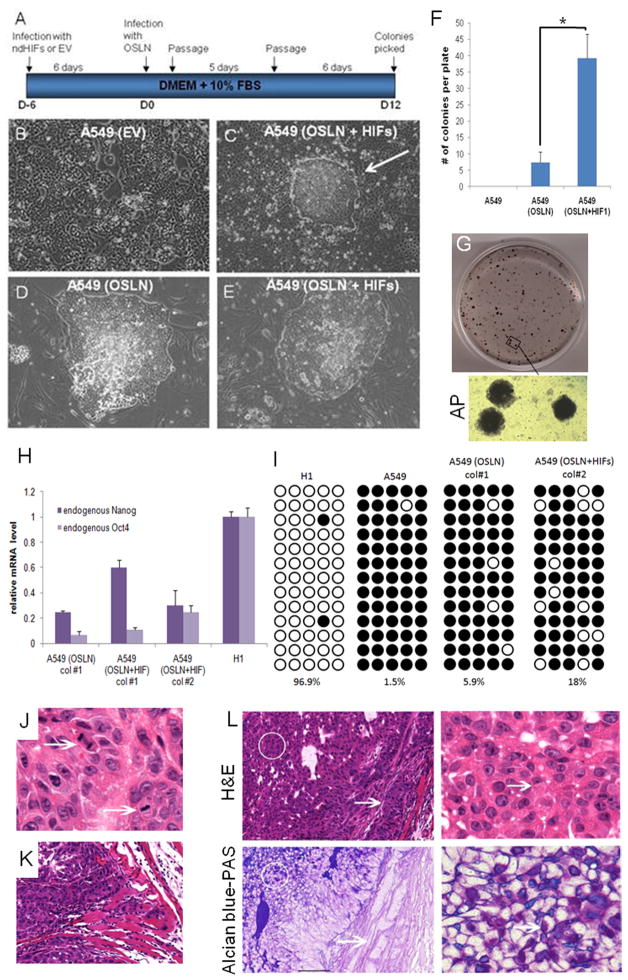
Since hypoxia results in a HIF-dependent up-regulation of stem cell markers that are shown to be sufficient to induce a pluripotent state (iPSC; 26, 33; Fig. 1–2), we tested whether these factors in our hands can induce iPSC colonies in cancer cells and whether such cells would have tumorigenic capacity. We introduced Oct4, Sox2, Nanog and Lin28 with or without ndHIFs to human lung adenocarcinoma epithelial cells (A549; Fig. 3A). Interestingly, iPSC-like colonies were observed more rapidly in the A549 cell line in these conditions than in MRC5, a normal, primary lung fibroblast line (Fig. 3A–E; A549 10days, MRC5 20days). Furthermore, introduction of ndHIF increased the efficiency of the iPS-like cell induction in the A549 cancer cell line (7 fold increase; Fig. 3F). The iPSC-like colonies generated from A549 were positive for alkaline phosphatase (AP) staining (Fig. 3G). The endogenous NANOG and OCT4 levels in the A549 iPS-like cells were lower than in normal H1 hESCs (Fig. 3H) and the promoter of OCT4 was only partially unmethylated (Fig. 3I), suggesting that the cells, while de-differentiated, were not fully re-programmed.
Figure 3. Overexpression of OCT4, SOX2, NANOG, LIN28 and HIFs in lung cancer cells.
(A) Experimental flowchart depicting the generation of iPS-like cells from A549 lung adenocarcinoma cells. (B–C) Bright field images of A549 cells infected with a pBABE empty vector control (EV, B) and of a colony obtained 12 days after infection of A549 with Oct4/Sox2, Lin28/Nanog (OSLN), ndHIF1α and ndHIF2α (C). (D–E) Bright field images of picked colonies amplified on feeders obtained from A549 infected with OSLN (D) and with OSLN+ndHIFs (E). (F) HIF1 overepression enhances the induction of iPSC-like colonies. Colonies were counted on day 20 after lentiviral transduction. (G) Alkaline phosphatase staining of A549(OSLN+HIFs) colonies grown on feeders. (H) RT-qPCR analysis of endogenous NANOG and OCT4 mRNA expression in A549(OSLN) and A549(OSLN+HIFs) clones compared to H1 cells. (I) Methylation analysis of OCT4 promoter region. White circles represent unmethylated; black circles methylated cytosine-phosphate-guanisine (CpG). Numbers indicate the percentage of unmethylated CpG. (J–K) A549(OSLN+HIFs) tumors present malignant characteristics. Representative H&E-stained section of the xenograft A549(OSLN+HIFs) showing high mitotic index (J) and local invasion (K). (L) Tumors generated by injection of A549(OSLN+HIFs) cells present regions of differentiation. Paraffin embedded sections were subjected to H&E (upper panels) or Alcian blue-PAS (lower panels) staining. Bar right panels equals 100 μm.
We hypothesized that partially re-programmed cancer cell colonies might model the process that takes place in hypoxic regions of primary tumors and thereby show high tumor aggressiveness. We challenged this hypothesis by assessing the in vivo tumorigenic capacity of these partially re-programmed A549 iPSC-like colonies by injecting them into the femoral muscle of immuno-compromised mice. Xenograft tumors obtained were large and palpable 10 days after injection of A549(OSNL+HIFs) iPSC-like colonies, revealing a very fast growth. No obvious tumor growth could be observed at 10 days in mice injected with A549 cells or A549 (HIF), while in two weeks palpations begun to reveal a small growth in mice injected with A549(HIF). A Hematoxylin and eosin staining on sections of A549 iPSC-like colony induced tumors revealed highly aggressive, malignant solid tumors with regions of central differentiation (Suppl. Fig. 8), high mitotic index (Fig. 3J), local invasion (Fig. 3K), necrosis and nuclear atypia (Suppl. Fig. 8). The small and cuboidal cells with cytoplasmic droplets observed in the central portions of the xenograft were positive for both acidic and neutral mucins (magenta and blue AB-PAS staining, Fig. 3L), attesting of the presence of some differentiated goblet cells. In accordance to partial de-differentiation, the AP-positive A549 iPSC-like colonies generated aggressive, invasive tumors, not typical teratomas (Suppl. Fig. 8).
These data show that expression of hESC markers in A549 cancer cells partially de-differentiates the cells generating highly aggressive tumors.
An overlap between HIF, NANOG and OCT4 staining in prostate tumors
We investigated the frequency of co-localization of HIF1α, NANOG and OCT4 proteins in serial sections of primary prostate tumors in patient samples. Skin, seminomas and hESCs were used as positive controls to validate the antibodies (Suppl. Fig. 9 and data not shown). In the prostate cancer specimen 09-066C (Gleason 4+3) 16.5% of tumor areas were positive for HIF1α and 7.7% were positive for NANOG (Fig. 4A, G, K; Suppl. Fig. 10A). A significant overlap between HIF1α and NANOG was observed: 69% of NANOG positive cells were also HIF1α positive (P<0.05), while 32% of the HIF1α positive cells were also NANOG positive (Fig. 4A). In addition to morphology, previously identified markers, CD107b, CD10 and CD104 were used to distinguish between tumor and benign glands (Fig. 4O, P, Suppl. Fig. 10A; 30). In specimen 07-020C, Gleason 4+3, the overlap between HIF1α and NANOG was also highly significant (91% of NANOG positive cells were also HIF1α positive) (Fig. 4B, H-I, L-M; Suppl. Fig. 10B). Furthermore, the two stem cell markers, NANOG and OCT4 were observed in the same prostate tumor regions (87% of NANOG positive cells were also OCT4 positive; Fig. 4B, L, Q). In the specimen 07–091LA, Gleason 3+4, out of the 2039 prostate glands counted 36.5% were positive for HIF1α, while 20.8% were positive for NANOG (Fig. 4C, J, N). Highly significant overlap between NANOG and HIF1α-stained glands was observed with 99% of the NANOG-positive glands also positive for HIF1α (P<0.01) while 57% HIF1α positive glands were also positive for NANOG (Fig. 4C). A detailed analysis revealed a high correlation in the number and location of positive cells in the overlapping regions for HIF, NANOG and OCT4 staining (Fig. 4D–F). We performed a goodness of fit test of a linear regression between the percentage of NANOG or OCT4 positive and that of HIF1α positive cells and obtained an R square value ranging from 0.55–0.65. The results suggest that NANOG staining correlates with HIF1α and OCT4 staining (R square >0.5), although the correlation is weakened by a few outliers and/or variation seen in different micro-areas of the tissue sections (Fig. 4D–F). These results are in accordance with the mRNA expression data (Fig. 1A–C), showing that a subset of hESC mRNAs are expressed in hypoxic cancer cells and that the protein expression of members of the hESC key regulators, NANOG and OCT4 correlates with HIF expression in primary prostate tumors. These data, however also point out that not every hypoxic cancer cell can up-regulate NANOG and OCT4, suggesting that other factors including the history of the cell and the level of HIF may play a role.
To assess the correlation of HIF1α, NANOG and OCT4 expression with clinicopathologic outcome of prostate cancer, we sampled more patients for our analysis. Since all specimens we analyzed were from recent occurrences of the disease (< 3 years) and the follow-up clinical characterizations were not available yet, we relied on Gleason score (measures the diversion from the normal gland structure), the single most important prognostic factor in prostate cancer, for indication of disease outcome (31). When we analyzed Gleason 5 glands only (most unstructured glands), we observe highly enriched expressions of NANOG and OCT4 (56.2% and 41.7%, respectively, Fig. 4R). These values are significantly higher than the observed frequencies for NANOG and OCT4 when all diseased glands are included to the analysis (Gleason 3, 4 and 5 together; Fig. 4R). We also show that the co-expression of HIF1α and NANOG in Gleason 5 only glands (20.4%) significantly increased as compared to the levels in overall diseased glands (12.3%).
Taken together, the significantly higher frequency of expressions of NANOG and OCT4 in Gleason 5 glands suggest the importance of the stem cell specific factors in the prostate cancer progression, and the increased co-expression of HIF1α and Nanog support our hypothesis that HIF1α plays a role in the cancer progression by acquiring stem cell factors expression.
Up-regulation of hESC markers in primary glioma cells and neurospheres formed in hypoxia
Our mRNA profiling of cancer cell lines also revealed an up-regulation of known cancer stem cell markers in hypoxia (Suppl. Table V). In hypoxic glioma cell line U251 we observe an up-regulation of CD133 (PROM1), a glioma stem cell (GSC) enriched marker that was previously shown to be induced at the cell surface by hypoxia in glioblastoma cells (35, 36) (Suppl. Table V-A). To test whether similar correlations between hypoxia, GSC and hESC markers were observed in primary tumors, we isolated and analyzed glioma cells from primary tumors from multiple patients. We generated cultures enriched for or depleted of the glioma stem-like subpopulation (25). Enrichment of the glioma stem cell (GSC) population was validated by several functional assays, including fluorescence-activated cell sorting (FACS) for cell surface markers such as CD133 (37). Following separation of the stem and non-stem populations, we subjected the cells to hypoxia and analyzed OCT4, NANOG, SOX2, OLIG2, CD133, cMYC, BMI-1 and MUSASHI by qPCR. Of these genes, in addition to CD133, we found that OCT4, NANOG, and cMYC were consistently up-regulated by hypoxia in populations obtained from several patient donors (Suppl. Fig. 12A). As shown previously, many primary glioma sections were found to express nuclear HIF1α (4). Some of these tumor regions also showed nuclear NANOG and OCT4 expression (Fig. 5A, Suppl. Fig. 11). We therefore tested potential functional correlation between hypoxia, neurosphere formation and iPSC inducer expression.
Figure 5. Hypoxia induces stem cell markers and enhances neurosphere formation in gliomas.
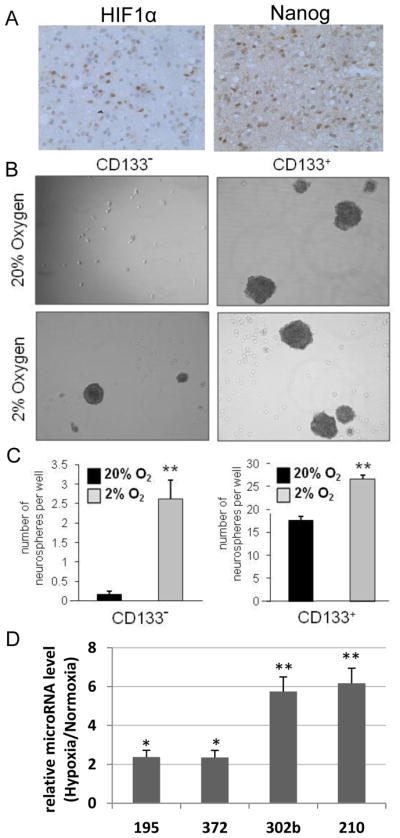
(A) Adjacent serial sections from a GBM patient tissue sample demonstrates nuclear staining of NANOG and OCT4 in a tumoral region with HIF1α immunoreactivity. Magnifications are 40X. Lower magnifications of the same region are presented in Suppl. Fig.11. (B–C) Hypoxia enhances neurosphere formation. Following collection from T4121 patient specimen, glioma stem and non-stem cells were plated in a 24 well plate at a density of 10 cells/well and cultured in either 2% O2 or 20% O2. After 12 days of treatment, representative phase contrast images were taken at 20X magnification (B). The number of neurospheres per well is presented (C). **, P < 0.01 with ANOVA.. (D) Hypoxia up-regulates hESC enriched microRNA in neurospheres generated from the non-stem population of primary gliomas. Following 12 days of exposure to normoxia or hypoxia (2% O2), the 24 wells of neurospheres derived from non-stem primary glioma cells were pooled for each condition, and the expression of hESC-enriched miRNAs was quantified by RT-qPCR.
To test whether hypoxia induced neurosphere formation capacity correlates with hESC marker expression, we first subjected glioma stem and non-stem cell populations to normoxia vs. hypoxia to test their ability to form spheroids, a functional assay linked to self-renewal (25; Fig. 5B–C, Suppl. Fig. 12B). As described previously, only CD133+ GSCs were able to form neurospheres in this media in normoxia (Fig. 5B). In contrast, we found that culture at 2% O2 supported neurosphere formation in both the stem and non-stem cell populations. Specifically, non-stem cells cultured at 2% O2 formed larger neurospheres at a higher frequency (16-fold increase in hypoxia vs. normoxia, P<0.01) (Fig. 5B–C, Suppl. Fig. 12C). This was also seen in the stem population (1.5-fold increase, P<0.01) (Fig. 5B–C). We analyzed on molecular level whether the hypoxia induced increase in neurosphere formation capacity correlates with hESC marker induction. Interestingly, many of the hESC markers were up-regulated in hypoxia induced, non stem (NS) derived neurospheres (2–6 fold increase; Fig. 5D, Suppl. Fig. 12F). In particularly, some of the key hESC enriched microRNAs were upregulated in hypoxia induced neurospheres (Fig. 5D). These data show a correlation between hypoxia induced neurosphere formation and hESC marker expression.
Hypoxia did not significantly change the growth rate of glioma non-stem cells under these experimental conditions (Suppl. Fig. 12D–E), showing that observed increase in hESC markers and neurosphere number in hypoxia are not due to increased cell growth.
DISCUSSION
We analyzed expression profiles of cancer cell lines in hypoxia and show that 11 cancer cell lines (from prostate, brain, kidney, cervix, lung, colon, liver and breast tumors) grown under hypoxic conditions share an overlapping gene expression signature with 9 hESC lines. Among the hESC enriched genes that are up-regulated in hypoxic cancer cell lines are the genes that are sufficient to induce re-programming; OCT4, NANOG, SOX2, KLF4 and cMYC. Moreover, HIF in combination of iPSC inducers, OCT4, SOX2, NANOG and LIN28 can induce efficiently partially re-programmed A549 iPS–like cells that can generate highly aggressive tumors when injected to mouse muscle. Most of the hESC enriched miRNAs are also up-regulated in hypoxic cancer cells. Further, we showed that hESC marker up-regulations in cancer cell lines were HIF dependent. We therefore analyzed the frequency of HIF and hESC marker co-localization in primary tumors and found a significant correlation between NANOG, OCT4 and HIF positive regions in primary prostate cancer. Further, the frequency of NANOG and OCT4 expression increased significantly in higher grade tumors. Many of the hypoxia-induced genes in cancer cells are previously identified markers for cancer stem cells. We therefore analyzed the correlation between hESC marker expression and stem cell character in hypoxic gliomas using a neurosphere assay as a functional read out assay, linked to self-renewal. Both, an increase of hESC markers and neurosphere formation were observed in hypoxic glioma cells, indicating a hypoxia induced correlation between stemness and neurosphere forming ability. Our data implicate HIF in acquisition of the dynamic state of stemness in pathological cases.
The overlap of gene expression signatures of hypoxic cancer cells and hESCs is enriched in primary tumors
Our large scale profiling experiments revealed a common set in 624 genes that were expressed at higher levels in hypoxic relative to normoxic cancer cell lines and in undifferentiated relative to differentiated hESC lines. Comparison to other hESC enriched gene-sets revealed a significant overlap with the identified candidate group (Suppl. Fig. 1). Importantly, a set of 31 genes that showed an overlap between all 3 hESC profiling sets and hypoxic cancer cell lines are targets for HIF, OCT4, SOX2 and/or NANOG and are enriched in Wilms tumors that show hESC epigenetic pattern (34). Previous analyses have shown that many prostate tumors express HIF (38, 39). Furthermore, NANOG was shown to be important for prostate tumor development (40). Our present work shows a significant overlap between HIF, NANOG and Oct4 expression in primary prostate tumors. Further, increase in Nanog and Oct4 expression and in Prostate Cancer Gleason score show significant positive correlation. These data support the hypothesis that one of the roles of stabilized HIF transcription factor is to activate hESC marker expression in primary tumors.
Hypoxia induces iPSC genes
We find that HIF can induce iPSC gene expression in cancer cells. Recent results have shown that hypoxia is advantageous for iPSC induction (41; JM, BS, CW, HRB, unpublished), however the mechanism is unknown. We propose that hypoxia is important in iPSC induction due to HIF transcription factor action. At least one of the iPSC inducers, OCT4 is shown to be a direct target of HIF transcription factors (7). Further studies are required to understand whether the other known iPSC inducer genes, Nanog, Sox2, Myc and Klf4 are direct targets of HIF. While the promoters of other hESC genes, including miR-302, have potential HIF-binding sites, it is not yet clear whether they are directly regulated by HIF or whether their expression is dependent on OCT4. It will also be interesting to reveal in the future whether other known HIF targets could play a role in iPSC induction.
HIF induces a hESC signature that correlates with tumor potency
We show that up-regulation of hESC signature correlates with increased neurosphere frequency in primary glioma cells (this study; 25). Increased neurosphere growth in hypoxia, even from single cells suggests that hypoxia will increase the stem-like cell population in gliomas. More experiments are required to define whether the stem-like cell group selectively expands or whether de novo de-differentiation takes place in the cancer cell population. However, the fact that a significant increase (16-fold) in neurosphere formation among the non-stem cell population is observed (Fig. 5C) suggests that the HIF pathway might have the capacity to initiate de-differentiation by up-regulating inducers of iPSC formation.
Previous work with iPSC inducers (26, 33) has shown that reversal of differentiation to recapitulate the stem cell state appears to be inherent within each cell type under the proper conditions. Therefore, it is perhaps not surprising that cancer cell can also revert to a cell with precursor character in response to stem cell marker expression (42, 43). However it raises the question whether hypoxic regions of tumors have activated HIF that can up-regulate stem cell marker expression on a level that could lead to induction of more potent tumor cells. Previous findings do suggest that this might be the case (14–16, 25). Furthermore, in order to mimic what could occur in vivo in hypoxic regions, we over-expressed iPSC inducers and HIFs in A549 lung cancer and picked colonies early in the re-programming process. We obtained partially de-differentiated iPSC-like colonies expressing lower level of endogenous NANOG and OCT4 than hESCs. Postulating that these partially reprogrammed cells might mimic the HIF induced state in tumors, we analyzed their tumor capacity in mice. Dramatically, the partially re-programmed A549 colonies generated highly aggressive tumors that grew into large size in 10 days, while the control A549 line showed no tumor growth in such short time period. These partially re-programmed A549 colonies further mimicked aggressive lung tumors in that they did not produce teratomas but could generate some differentiated endothelial cells. An article published while our work was under consideration supports our findings by showing that OCT4 and NANOG overexpression induce cancer stem cell-like properties in lung adenocarcinoma (LAC) (44). They also showed that OCT4 and NANOG are coexpressed in high grade LAC and are associated with poor survival rate of patients.
It is critical to determine which of the iPSC inducer genes have potential to increase tumor potency. Answering this question has been initiated recently by showing that NANOG function is critical in tumor development (40, 45). It will be important to further analyze the capacity of OCT4, KLF4 and miR-302 in this process (17, 18). However, not all stem cell inducers are up-regulated by hypoxia. One such exception is let-7 regulator, lin-28. Although lin-28 has been shown to promote transformation and to be associated with advanced human malignancies (46), this gene is not up-regulated in hypoxia in any of the 11 cancer cell lines used in our study (however, we cannot rule out the possibility that hypoxia affects Lin-28 protein levels). These data suggest that up-regulation of HIF pathway targets is important for stemness and thereby tumor potency, however, other factors also contribute to the process.
In summary, our data support the hypothesis that HIF targets are critical for stemness in malignant cells. Hypoxia is also beneficial for self-renewal of hESCs (1). One common denominator in these cell types, ESCs and cancer cells is low mitochondrial activity (Warburg effect in cancers and low mitochondrial activity in stem cells; 47, 48). Future studies will reveal whether HIF dependent metabolic pattern is critical for stemness.
Supplementary Material
1
2
3
4
5
6
7
8
9
Acknowledgments
We thank members of the Ruohola-Baker lab for helpful discussions. We thank the Rosetta Gene Expression Laboratory for processing microarray experiments. We thank Mei Deng (CHDD Cellular Morphology Core) and Dr. Xiaotun Zhang (Department of Urology, UW) for help with the tumor specimens. We thank Emily Liebsekind, Ausra Bendoraite, Daryl Humes and Stephanie Einsele for technical help. We thank Dr. Kaelin for providing pVHL-deficient 786-0 and RCC4 renal carcinoma cells transfected either with an empty vector or wild-type VHL, and Dr. Vogelstein for HCT116 Dicer hypomorph cell line. We thank Dr. Cui for providing the Oct4-GFP construct. This work was supported by the Brain Tumor Society, Goldhirsh Foundation and NIH grants NS047409, NS054276, CA112958, and CA116659 for JR, the French Ministry of National Education/Ecole Normale Supérieure for AH, the Robert Chang Scholarship of Sichuan University-U. Washington exchange program for AJW, the Regione Autonoma della Sardegna for CMAP, Jaconnette L. Tietze Young Scientist Award for BS and R01GM083867-01 NIGMS for HRB, 1P01GM081619-01 NIGMS and the Institute for Stem Cell and Regenerative Medicine (ISCRM) at the University of Washington for CW, CAB and HRB.
References
- 1.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–61. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–72. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–43. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 4.Mabjeesh NJ, Amir S. Hypoxia-inducible factor (HIF) in human tumorigenesis. Histol Histopathol. 2007;22:559–72. doi: 10.14670/HH-22.559. [DOI] [PubMed] [Google Scholar]
- 5.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 6.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–70. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 9.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70:2675–85. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- 11.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–93. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 12.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–5. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–93. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–40. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008;68:6533–40. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Dai J. Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells. 2010;28:885–93. doi: 10.1002/stem.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SY, Loscalzo J. MicroRNA-210: A unique and pleiotropic hypoxamir. Cell Cycle. 2010:9. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, et al. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol. 2010;3:109–13. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ware CB, Nelson AM, Blau CA. A comparison of NIH-approved human ESC lines. Stem Cells. 2006;24:2677–84. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- 22.Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, Tan Y, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–50. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Q, Bartz S, Mao M, Li L, Kaelin WG., Jr The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol. 2007;27:2092–102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–68. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Gerrard L, Zhao D, Clark AJ, Cui W. Stably transfected human embryonic stem cell clones express OCT4-specific green fluorescent protein and maintain self-renewal and pluripotency. Stem Cells. 2005;23:124–33. doi: 10.1634/stemcells.2004-0102. [DOI] [PubMed] [Google Scholar]
- 28.Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, Melen GJ, Sanchez L, Ligero G, et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–19. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, Seal S, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–41. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu AY, Roudier MP, True LD. Heterogeneity in primary and metastatic prostate cancer as defined by cell surface CD profile. Am J Pathol. 2004;165:1543–56. doi: 10.1016/S0002-9440(10)63412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein JI. An update of the Gleason grading system. J Urol. 2010;183(2):433–40.31. doi: 10.1016/j.juro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 32.Elias MC, Tozer KR, Silber JR, Mikheeva S, Deng M, Morrison RS, et al. TWIST is expressed in human gliomas and promotes invasion. Neoplasia. 2005;7:824–37. doi: 10.1593/neo.04352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 34.Aiden AP, Rivera MN, Rheinbay E, Ku M, Coffman EJ, Truong TT, et al. Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell. 2010;6(6):591–602. doi: 10.1016/j.stem.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidel S, Garvalov BK, Wirta V, von Stechow L, Schanzer A, Meletis K, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133:983–95. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 36.Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28:3949–59. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 37.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 38.Kimbro KS, Simons JW. Hypoxia-inducible factor-1 in human breast and prostate cancer. Endocr Relat Cancer. 2006;13:739–49. doi: 10.1677/erc.1.00728. [DOI] [PubMed] [Google Scholar]
- 39.Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, et al. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell. 2010;17:319–32. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeter CR, Badeaux M, Choy G, Chandra D, Patrawala L, Liu C, et al. Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells. 2009;27:993–1005. doi: 10.1002/stem.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–41. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115(20):4039–42. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci. 2010;107(1):40–5. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70(24):10433–44. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 45.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–37. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St John JC, Amaral A, Bowles E, Oliveira JF, Lloyd R, Freitas M, et al. The analysis of mitochondria and mitochondrial DNA in human embryonic stem cells. Methods Mol Biol. 2006;331:347–74. doi: 10.1385/1-59745-046-4:347. [DOI] [PubMed] [Google Scholar]
- 48.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7
8
9