Salmonella maintains the integrity of its intracellular vacuole through the action of SifA (original) (raw)
Abstract
A method based on the Competitive Index was used to identify Salmonella typhimurium virulence gene interactions during systemic infections of mice. Analysis of mixed infections involving single and double mutant strains showed that OmpR, the type III secretion system of Salmonella pathogenicity island 2 (SPI-2) and SifA [required for the formation in epithelial cells of lysosomal glycoprotein (lgp)-containing structures, termed Sifs] are all involved in the same virulence function. sifA gene expression was induced after Salmonella entry into host cells and was dependent on the SPI-2 regulator ssrA. A _sifA_– mutant strain had a replication defect in macrophages, similar to that of SPI-2 and _ompR_– mutant strains. Whereas wild-type and SPI-2 mutant strains reside in vacuoles that progressively acquire lgps and the vacuolar ATPase, the majority of _sifA_– bacteria lost their vacuolar membrane and were released into the host cell cytosol. We propose that the wild-type strain, through the action of SPI-2 effectors (including SpiC), diverts the _Salmonella_-containing vacuole from the endocytic pathway, and subsequent recruitment and maintenance of vacuolar ATPase/lgp-containing membranes that enclose replicating bacteria is mediated by translocation of SifA.
Keywords: Competitive Index/SPI-2/trafficking/type III secretion/vacuole
Introduction
Systemic infection of mice by Salmonella typhimurium is a useful and intensively studied model of typhoid fever. Following oral inoculation, bacteria can survive the acid pH of the stomach, adhere to and invade cells of the intestinal epithelium, acquire nutrients, survive in the blood, replicate within macrophages and induce cytotoxicity (Finlay, 1994; Richters-Dahlfors et al., 1997). Numerous genes that contribute to S.typhimurium virulence have been identified in recent years (Groisman and Ochman, 1997). Although the biochemical functions of some of these genes have been elucidated, little is known about their regulation in vivo and how they interact during the infectious process. Many virulence genes are clustered together on ‘pathogenicity islands’ (PAIs), which appear to have been acquired by horizontal transfer from unknown sources. SPI-1 and SPI-2 are two Salmonella PAIs that encode structurally similar but functionally distinct type III secretion systems (TTSSs) which translocate virulence proteins from bacterial to host cells during the infectious cycle (Hueck, 1998). The SPI-1 encoded TTSS, called Inv/Spa, plays an important role in invasion of epithelial cells (Galán and Curtiss, 1989; Galán, 1996). Most of the genes associated with Inv/Spa are encoded within SPI-1 at 63 centisomes (cs) on the chromosome (Mills et al., 1995), but at least two of the secreted effector proteins are encoded elsewhere: the sopB gene is present on the SPI-5 pathogenicity island (Wood et al., 1998) and SopE is encoded by a temperate bacteriophage (Hardt et al., 1998).
SPI-2, located at 30 cs, encodes the second type III secretion system (Ochman et al., 1996; Shea et al., 1996). The SsrA/B two-component regulatory system of SPI-2 is required for SPI-2 gene expression (Valdivia and Falkow, 1997; Cirillo et al., 1998; Deiwick et al., 1999). Recent work has shown that in cultured host cells, transcription of ssrA is modulated by OmpR, a two-component regulatory system protein which responds to changes in osmolarity, pH and temperature (Lee et al., 2000).
The SPI-2 secretion system plays a crucial role in systemic growth of Salmonella in its host (Hensel et al., 1995; Shea et al., 1996) and is required for bacterial proliferation in macrophages (Ochman et al., 1996; Cirillo et al., 1998; Hensel et al., 1998). Salmonella typhimurium replicates intracellularly within a vacuole that diverts from the normal phagocytic pathway (Méresse et al., 1999b). The _Salmonella_-containing vacuole (SCV) rapidly loses early endocytic markers such as EEA1 and the transferrin receptor (Steele-Mortimer et al., 1999) and acquires some lysosomal membrane glycoproteins (lgps), but does not interact extensively with either the early endocytic pathway or mature lysosomes (Garcia del Portillo and Finlay, 1995; Rathman et al., 1997). After uptake into macrophages, the SCV undergoes acidification to a pH between 4.0 and 5.0 (Rathman et al., 1996). SPI-2 mediated secretion can be induced in vitro by acidic conditions, suggesting that pH could be a physiological signal for SPI-2 secretion (Beuzón et al., 1999; Lee et al., 2000). Only one SPI-2 gene (spiC/ssaB) has been demonstrated to encode an effector protein. SpiC is reported to inhibit fusion of SCVs with lysosomes, homotypic fusion of endosomes and transferrin recycling (Uchiya et al., 1999). Two leucine-rich repeat proteins, SspH-1 and SspH-2, which are encoded outside SPI-2, have also been identified as targets of this secretion system (Miao et al., 1999), although their functions remain unknown. This raises the possibility that, as for the Inv/Spa system, other effectors may be encoded elsewhere on the chromosome.
To identify virulence genes that interact during infection, we have developed a method based on the Competitive Index (C.I.), in which single and double mutant strains are analysed by mixed infections of mice. In this paper we describe the application of this approach to SPI-2, ompR and sifA of S.typhimurium. C.I. analysis indicates that these three loci interact during systemic infection. The sifA gene is required for the formation in epithelial cells of lgp-containing tubular membrane structures termed Sifs (Garcia del Portillo et al., 1993b; Stein et al., 1996). We show that expression of sifA is strongly induced after Salmonella enters host cells and this induction is dependent on ssrA. Although Sifs are not detectable in infected macrophages, SifA function is important in these cells because a _sifA_– mutant strain has a strong replication defect in macrophages. Several hours after uptake, the majority of _sifA_– mutant bacteria lose their vacuoles and are found in the host cell cytosol. We conclude that SifA has an important role in the maintenance of the vacuolar membrane surrounding wild-type bacterial cells.
Results
Analysis of virulence gene interactions in vivo
The method we have developed for identifying virulence gene interactions relies on the additive effect of mutations in virulence genes with different functions. Because Salmonella virulence is multifactorial, combination of mutations in genes with different functions results in strains with increased attenuation (Baumler et al., 1997; Shea et al., 1999).
The C.I. is a sensitive measure of the relative degree of virulence attenuation of a particular mutant in mixed infection with the wild-type strain. It is defined as the ratio of the mutant strain to the wild-type in the output divided by the ratio of the two strains in the input (Freter et al., 1981; Taylor et al., 1987).
To establish if mutations in two different virulence genes have additive effects on virulence attenuation, we determined C.I. values for double mutant versus single mutant strains. In the case of two genes that contribute equally to the same virulence function (by encoding essential components of a macromolecular structure, for example), strains carrying mutations in either single gene would be expected to have the same level of attenuation as a strain carrying both mutations. Therefore, the C.I. of either single mutant versus the double mutant should be 1.0 (Figure 1, upper panel). In the case of two genes (a and b) with independent functions, the virulence attenuation of a double mutant strain will reflect the additive effect of both mutations (Figure 1, lower panel). The C.I. of a double mutant strain versus an _a_– strain will be equivalent to the C.I. of a _b_– strain versus the wild-type strain. Similarly, the C.I. of a double mutant strain versus a _b_– strain will be equivalent to the C.I. of an _a_– strain versus the wild-type strain, and all comparisons will have values of <1.0.
Fig. 1. Theoretical representation of Competitive Index (C.I.) analysis. C.I. is the c.f.u. ratio of double and single strains recovered from the infected animal divided by the c.f.u. ratio of double and single strains in the inoculum. Panels represent different degrees of functional relationship between two hypothetical genes (a and b) and the C.I. predicted for each case. In this case:
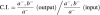
We have demonstrated previously that the C.I. of a _purD_– mutant strain of S.typhimurium (a purine auxotroph) versus the wild-type strain is not statistically different from the C.I. of a _purD_–, _ssaV_– double mutant strain versus an _ssaV_– single mutant (Shea et al., 1999), and this result was confirmed as a control for this study (Table I). The ssaV gene is thought to encode an inner membrane component of the SPI-2 secreton (Hensel et al., 1997), and is required for the secretion of SseB, which is probably a translocon component (Beuzón et al., 1999). When subjected to the analysis indicated in Figure 1, the C.I.s (Table I) confirm that purD and ssaV have totally unrelated functions, whereas sseB and ssaV are equally necessary for SPI-2 function.
Table I. Competitive Index analysis of S.typhimurium mutants.
| Mixed infectiona | C.I.b |
|---|---|
| Controls | |
| wt versus _ssaV_– | 0.006 |
| wt versus _purD_– | 0.0005 |
| _ssaV_– versus _ssaV_–, _purD_– | 0.0038d |
| _purD_– versus _ssaV_–, _purD_– | 0.019d |
| wt versus _ssaV_–, _purD_– | 0.00006 |
| wt versus _sseB_– | 0.016 |
| _ssaV_– versus _ssaV_–, _sseB_– | 1.2c |
| _sseB_– versus _ssaV_–, _sseB_– | 0.82c |
| wt versus _ssaV_–, _sseB_– | 0.014 |
| ompR/envZ | |
| wt versus _ompR_– | 0.008 |
| _ssaV_– versus _ssaV_–, _ompR_– | 0.823c |
| _ompR_– versus _ssaV_–, _ompR_– | 0.21c |
| wt versus _ssaV_–, _ompR_– | 0.010 |
| sifA | |
| wt versus _sifA_– | 0.012 |
| sifA_– p_sifA versus _sifA_– | 0.074d |
| _ssaV_– versus _ssaV_–, _sifA_– | 1.34c |
| _sifA_– versus _ssaV_–, _sifA_– | 1.17c |
| wt versus _ssaV_–, _sifA_– | 0.0019 |
SPI-2 and ompR interact in vivo
The OmpR/EnvZ two-component system responds to a variety of physicochemical changes in the environment (Heyde and Portalier, 1987; Thomas and Booth, 1992). Strains carrying mutations in ompR/envZ are highly attenuated in systemic infection of mice but mutations in genes previously known to be regulated by OmpR could not account for its virulence defect (Dorman et al., 1989; Chatfield et al., 1991). However, recently it has been shown that ssrA transcription is regulated by OmpR in infected host cells (Lee et al., 2000). Furthermore, we have found that a mutation in ompR reduces SseB levels during bacterial growth in host cells and certain laboratory media (results not shown). It was therefore of interest to determine if ompR interacts with the SPI-2 TTSS genes in vivo.
The C.I. of an _ompR_– mutant strain versus the wild-type strain following intraperitoneal (i.p.) inoculation confirmed that its level of attenuation was similar to that of a typical SPI-2 mutant strain (Table I). To investigate possible interactions between ompR and SPI-2 genes in vivo, a double mutant strain was constructed carrying mutations in ompR and ssaV. The C.I. of this strain versus either single mutant strain was not significantly different from 1.0. This result implies that the virulence functions of the two genes are related, and the in vitro regulation of SPI-2 genes by ompR (Lee et al., 2000) also occurs in vivo.
SPI-2 and sifA interact in vivo
The sifA gene is located within the potABCD operon on the Salmonella chromosome, and was probably acquired by horizontal transfer from an unknown source (Stein et al., 1996). Strains carrying mutations in sifA, ompR or envZ are all defective for the formation of Sifs (Garcia-del Portillo et al., 1993b; Stein et al., 1996; Mills et al., 1998). _sifA_– mutant strains are reported to be attenuated in virulence following oral inoculation of mice, although they do not have a replication defect in epithelial cells (Stein et al., 1996). In competition experiments, the virulence of a strain carrying a transposon insertion in sifA was found to be as attenuated as an _sseB_– mutant following i.p. inoculation of mice, and this defect was complemented by introduction of the wild-type sifA allele on a plasmid (Table I). A _sifA_– mutation was therefore introduced to the _ssaV_– mutant strain and single and double mutant strains were analysed by C.I. The double mutant strain was no more attenuated than either single mutant (Table I). Therefore, the proteins encoded by these genes appear to be involved in the same important virulence function of S.typhimurium during systemic infection.
SPI-2 mutant strains fail to induce Sifs
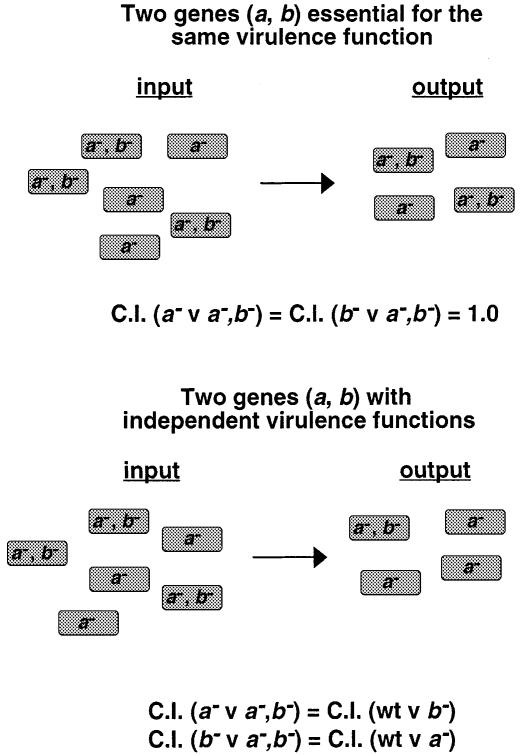
The genetic evidence linking SPI-2 and sifA (Table I) led us to ask if SPI-2 mutant strains could induce the formation of Sifs in epithelial cells. Salmonella typhimurium strains carrying a plasmid constitutively expressing green fluorescent protein (GFP) were used to infect HeLa cells, and these were subsequently stained with an anti-LAMP-1 antibody to reveal Sifs (Garcia-del Portillo et al., 1993b) (Figure 2). At 7 h post-invasion, >70% of cells infected with wild-type bacteria contained Sifs, whereas Sifs were detected in 1% or less of cells infected with strains carrying mutations in either ssaV, ssrA or ompR. Approximately 1% of cells infected with either _sseB_– or _sifA_– mutant strains contained Sifs, and the introduction of plasmids carrying functional genes to the respective mutant strains restored Sif formation to ∼40% of infected cells. Similar results to those shown in Figure 2 were obtained when infected cells were examined 16 h after bacterial invasion, which shows that the onset of Sif formation is not simply delayed in SPI-2 mutants. It has been reported that Salmonella strains with non-auxotrophic mutations affecting replication inside host cells are incapable of inducing Sif formation (Garcia del Portillo et al., 1993a). Therefore, the intracellular replication defect of SPI-2 and _ompR_– mutant strains (Figure 4; Lee et al., 2000) could account for the lack of Sifs. A _phoP_– mutant strain was used to investigate this possibility. phoP/Q exerts regulatory control over at least 40 virulence genes of Salmonella (Miller and Mekalanos, 1990), is required for bacterial replication in macrophages, but functions independently of SPI-2 in host cells (Valdivia and Falkow, 1997; results not shown). The _phoP_– strain was nevertheless capable of inducing Sifs in ∼40% of infected cells (Figure 2G). Therefore, we conclude that the failure of SPI-2 and _ompR_– mutants to induce Sifs is not simply a consequence of their replication defect, but is specifically connected with the function of these genes.
Fig. 2. Sif formation by S.typhimurium mutant strains in HeLa cells. Confocal immunofluorescence analysis of Sif formation in cells infected with: 12023 (A and B), _sseB_– (C), sseB_– p_sseB (D), _sifA_– (E) and sifA_–p_sifA (F) strains. Cells were fixed at 7 h post-infection and stained with mouse anti-LAMP-1 and TRSC-conjugated donkey anti-mouse antibodies. In (A–E) the bacterial strains carried a plasmid expressing gfp constitutively. In (F), bacterial cells were detected by goat anti-Salmonella and FITC-conjugated donkey anti-goat antibodies. _sseB_– and _sifA_– mutant strains fail to induce Sifs (C and E). This defect can be complemented by the introduction of plasmids carrying the respective wild-type alleles (D and F). Bars correspond to 4 µm; the scale in (C–F) is equivalent to that of (A). (G) represents typical results from one of three experiments where cells (n = 50) infected by each S.typhimurium strain were evaluated for Sif formation at 7 h post-invasion. Values are given as percentage of infected cells containing Sifs.
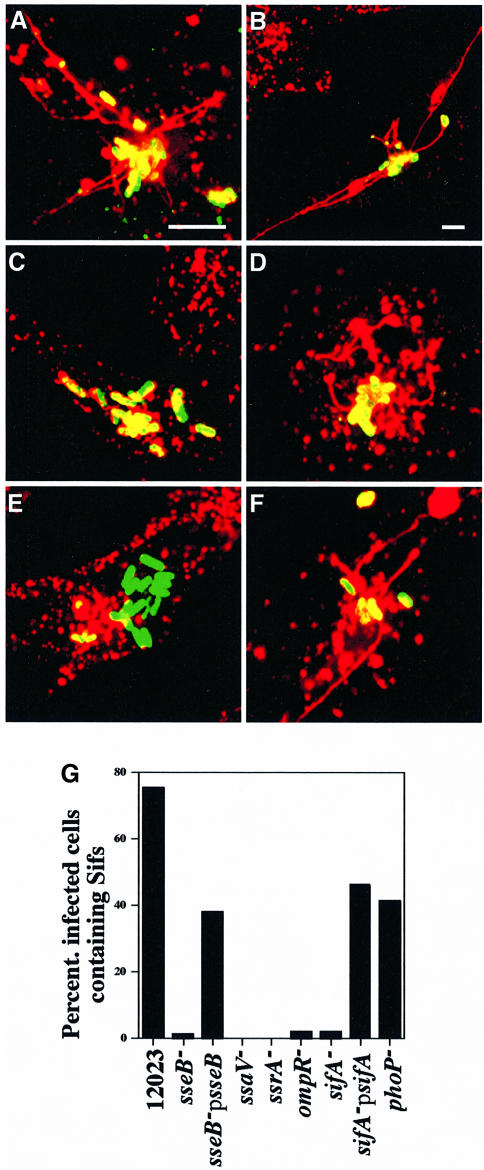
Fig. 4. A _sifA_– mutant strain is defective for replication within RAW macrophages. Opsonized bacteria were taken up by phagocytosis and at 2 and 16 h host cells were either lysed and cultured for enumeration of intracellular bacteria (gentamicin-protected), or fixed and examined by phase-contrast and confocal fluorescence microscopy. (A, B, C and D) represent typical cells infected with GFP-expressing 12023, _ssaV_–, _sifA_– and _ompR_–, respectively. In (E), the values shown represent the fold increase calculated as a ratio of the intracellular bacteria between 2 and 16 h post-uptake. Each strain was infected in triplicate and the standard errors from the means are shown. The results shown are representative of three independent experiments. Scale bar represents 4 µm.
sifA expression requires the SPI-2 regulator ssrA
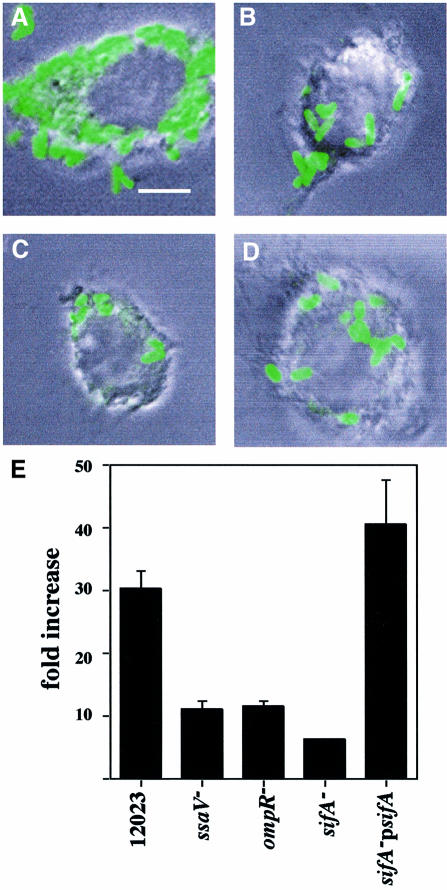
SPI-2 gene expression is induced following uptake of bacteria into host cells (Cirillo et al., 1998). The expression of some genes can be detected after 1 h (Uchiya et al., 1999), and maximal expression is achieved after 6 h (Cirillo et al., 1998). This expression is dependent on the SPI-2 two-component regulatory system SsrA/B (Cirillo et al., 1998; Hensel et al., 1998). In view of the phenotypic similarities between SPI-2 and _sifA_– mutant strains, it was of interest to know if their patterns of gene expression are similar. To analyse this, a plasmid was constructed carrying a sifA::gfp reporter fusion, transferred into different S.typhimurium strains, and expression of the fusion protein was examined during bacterial growth in HeLa cells. Wild-type bacterial cells expressed sifA::gfp only when they were intracellular (Figure 3, top panel). Approximately 2% of bacterial cells expressed sifA::gfp as early as 1 h post-invasion. The percentage of bacterial cells expressing the fusion increased gradually thereafter, with ∼90% of cells positive for GFP by 6 h post-invasion (results not shown). These kinetics are similar to those reported for SPI-2 (ssaH::gfp) expression (Cirillo et al., 1998). No sifA::gfp expression was detected in the _ssrA_– mutant strain (Figure 3, middle panel). In _ompR_– mutant strains, the number of cells expressing sifA::gfp was ∼50% that of wild-type cells, and the overall level of expression was lower than that seen in wild-type cells, consistent with the effect of an _ompR_– mutation on SPI-2 ssaH gene expression (Lee et al., 2000). As expected, the level of sifA::gfp expression in a _phoP_– mutant strain was comparable to that of the wild-type strain (Figure 3).
Fig. 3. sifA::gfp expression by S.typhimurium mutant strains in infected HeLa cells. Confocal immunofluorescence analysis in cells 6 h after invasion with different strains carrying a plasmid-borne sifA::gfp fusion (pID812). Salmonella typhimurium was detected with goat anti-Salmonella, and TRSC-conjugated donkey anti-goat antibodies. The upper panel shows three extracellular bacteria not expressing GFP, and several GFP-positive intracellular bacteria. SifA is expressed within HeLa cells in an _ssrA_-dependent manner (middle panel). SifA expression is not affected by a _phoP_– mutation but was only detected in ∼50% of bacterial cells carrying the _ompR_– mutation. Bars correspond to 4 µm.
A sifA mutant strain has a replication defect in macrophages
As most SPI-2 mutants have a replication defect inside host cells, and _sifA_– mutants are attenuated in the systemic phase of infection, a _sifA_– mutant strain was tested for replication in macrophage-like RAW cells. The _sifA_– mutant strain was found to have a replication defect comparable to that of a strain carrying a mutation in ssaV (Figure 4). This result was surprising, since _sifA_– strains are reported to be proficient for replication in HeLa cells (Stein et al., 1996). The introduction of the plasmid carrying a functional sifA gene to the _sifA_– strain restored replication to wild-type levels (Figure 4E). This demonstrates that the replication defect is not due to a polar effect of the integrated transposon on the downstream potC gene, or an unrelated mutation. In these assays, the _ompR_– mutant strain also displayed a replication defect, in agreement with a recent report (Lee et al., 2000).
Association of LAMP-1 with SCVs in RAW macrophages
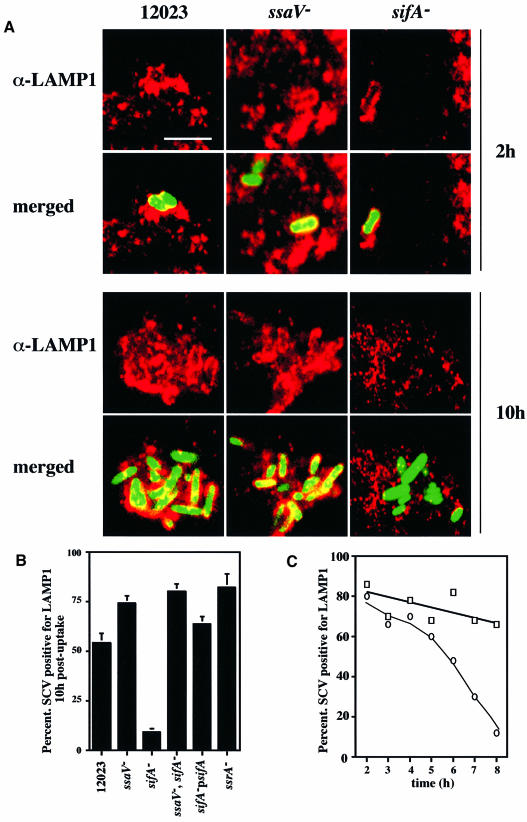
Since sifA has an important role in intra-macrophage replication of S.typhimurium we investigated the distribution of LAMP-1 in RAW macrophages infected with different S.typhimurium strains. At different time points following bacterial uptake, cells were fixed and stained with an antibody against LAMP-1, then examined by confocal immunofluorescence microscopy. No structures resembling Sifs were observed at any time point in macrophages infected with wild-type bacteria. At 2 h post-uptake, LAMP-1 was associated with ∼80% of wild-type, _ssaV_– or _sifA_– strains (Figure 5A, upper panel; Figure 5C). However, at 10 h post-uptake, LAMP-1 was present on >50% of vacuoles containing wild-type, and >70% of vacuoles containing _ssaV_– bacteria, whereas <10% of the _sifA_– bacteria were associated with this marker (Figure 5A, lower panel; Figure 5B). A time-course experiment showed that the decline in LAMP-1 association with the _sifA_– strain began ∼5 h post-uptake (Figure 5C). We conclude from these results that the maintenance of LAMP-1 on vacuoles containing wild-type bacteria is dependent on the sifA gene.
Fig. 5. The lysosomal membrane protein LAMP-1 is not associated with _sifA_– mutant bacteria 10 h post-uptake in RAW macrophages. Confocal microscopic analysis was carried out on macrophages infected with wild-type (12023), _ssaV_– or _sifA_– strains constitutively expressing GFP, at 2 h [upper panels (A)] or 10 h [lower panels (A)] post-uptake. LAMP-1 was detected using a rabbit polyclonal and TRSC-conjugated donkey anti-rabbit secondary antibodies. Scale bar corresponds to 4 µm. The percentage of bacteria (n = 50) co-localizing with LAMP-1 was determined for these and two other bacterial strains 10 h after uptake. The percentage of bacteria (n = 50) co-localizing with LAMP-1 at different time points was determined for 12023 (open squares) and _sifA_– mutant (open circles) strains (C). Results shown in (B) are the means ± SE of three independent experiments. For the sifA_–, p_sifA strain, bacteria were detected with goat anti-Salmonella and FITC-conjugated donkey anti-goat antibodies.
Trafficking of S.typhimurium strains in epithelial cells
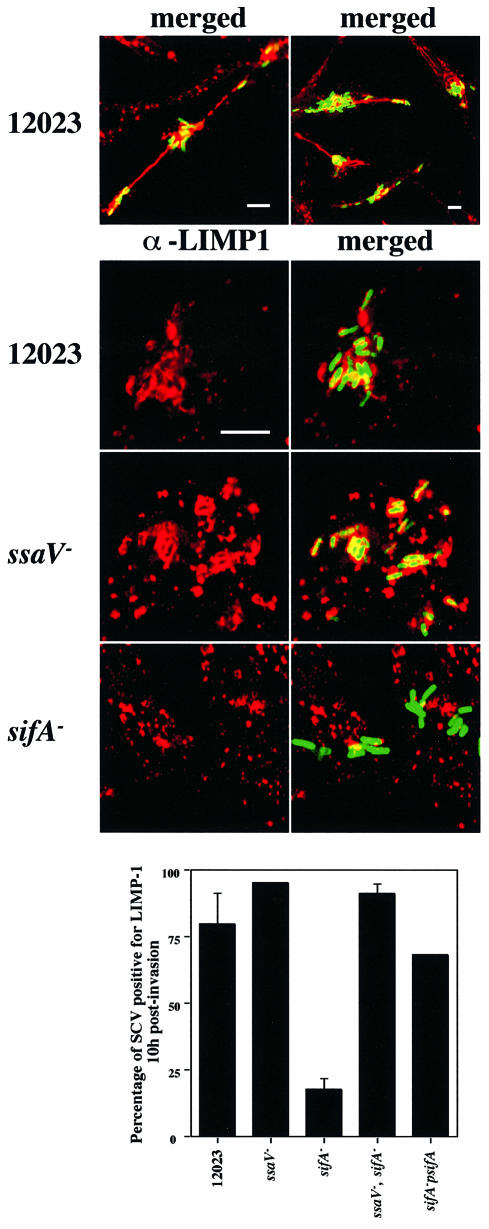
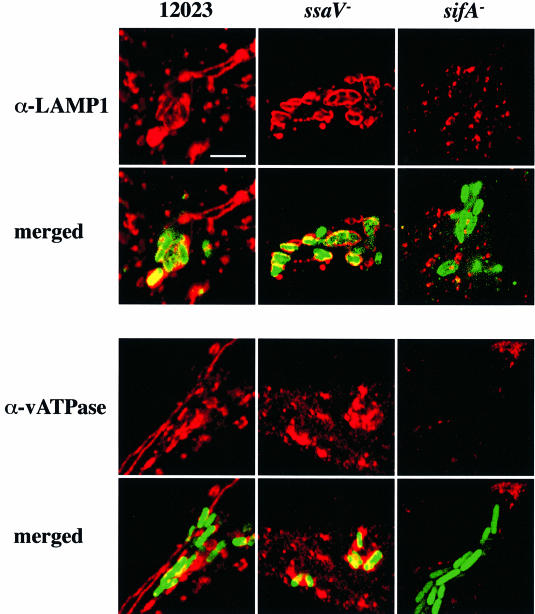
To determine if the apparent absence of lgp in the vicinity of the _sifA_– mutant cells was restricted to LAMP-1 and to macrophages, we examined the localization of another lgp, LIMP-1, also known as LAMP-3 or CD63 antigen, in infected epithelial cells. We found that LIMP-1 is a constituent of Sifs in cells infected with wild-type bacteria (Figure 6, upper panel). Furthermore, whereas LIMP-1 was present in >75% of vacuoles containing wild-type bacteria and >90% of vacuoles containing _ssaV_– bacteria, <25% of _sifA_– cells were associated with this marker (Figure 6). Similar results were obtained in HeLa cells using antibodies against LAMP-1 (Figure 7, upper panel) and LAMP-2 (not shown). LIMP-1, LAMP-1 and LAMP-2 share similar lysosomal targeting sequences, and are likely to follow the same trafficking pathway (Hunziker and Geuze, 1996). We therefore examined the acquisition of the vacuolar ATPase (vATPase), which is responsible for the acidification of endocytic vacuoles (Sato and Toyama, 1994), since its origin, function and targeting process are different to those of these lgps (Hunziker and Geuze, 1996). Interestingly, vATPase was also found to be a component of Sifs (Figure 7, lower panel), was present on vacuoles containing wild-type and _ssaV_– bacteria, but was only detected at a low frequency in association with _sifA_– bacteria (Figure 7, lower panel).
Fig. 6. LIMP-1 is a component of Sifs and is not associated with _sifA_– mutant bacteria 10 h after invasion of HeLa cells. Confocal microscopic analysis was carried out on cells infected with wild-type (12023), _ssaV_– or _sifA_– strains constitutively expressing GFP. LIMP-1 was detected using a mouse mAb and TRSC-conjugated donkey anti-mouse secondary antibodies. Low magnification images show that LIMP-1 is a component of Sifs in cells infected with wild-type bacteria (upper panel). Higher magnification images show LIMP-1-containing SCVs for wild-type and _ssaV_– mutant bacteria, but weak or no association of LIMP-1 with _sifA_– mutant cells. Scale bars correspond to 4 µm. The percentage of bacteria (n = 50) co-localizing with LIMP-1 was determined for these and two other bacterial strains. Results shown are the means ± SE of three independent experiments. For the sifA_–, p_sifA strain, bacteria were detected with goat anti-Salmonella followed by FITC-conjugated donkey anti-goat antibodies.
Fig. 7. LAMP-1 and vATPase are not associated with _sifA_– mutant bacteria 10 h post-invasion in HeLa cells. Confocal microscopic analysis was carried out on cells infected with wild-type (12023), _ssaV_– or _sifA_– strains constitutively expressing GFP. LAMP-1 and vATPase were detected using mouse monoclonal antibodies, followed by TRSC-conjugated donkey anti-mouse secondary antibodies. Scale bar corresponds to 4 µm.
Maintenance of the vacuolar membrane enclosing wild-type S.typhimurium is dependent on sifA
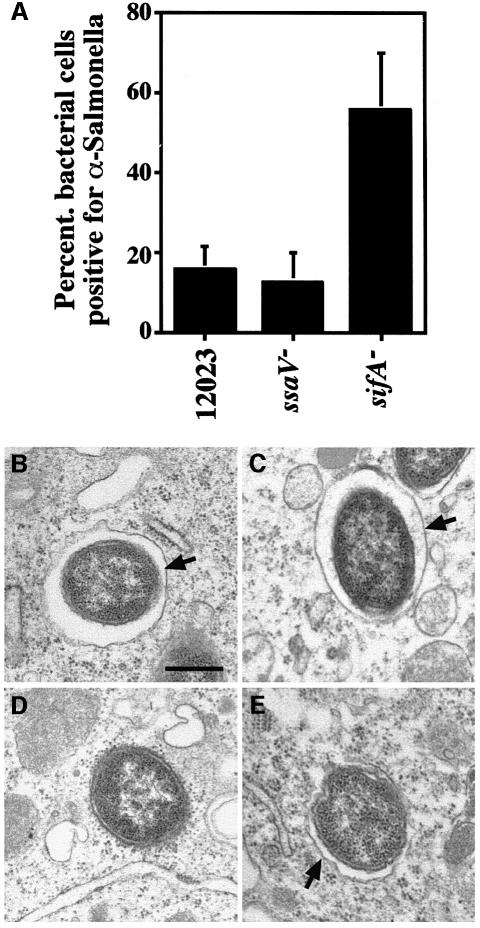
The lack of association of lgps and vATPase with _sifA_– mutant bacteria suggested that either vacuoles containing _sifA_– bacteria are non-fusogenic, as is the case for vacuoles containing some other intracellular pathogens, such as Legionella spp. (Sinai and Joiner, 1997), or that these bacteria are no longer enclosed by a membrane. The presence of a vacuolar membrane around _sifA_– bacteria was investigated by testing the accessibility of intracellular S.typhimurium strains to anti-LPS antibody in RAW cells treated with the pore-forming toxin streptolysin-O from Streptococcus pyogenes. The rationale of this experiment is that, after plasma membrane permeabilization, bacteria present in the macrophage cytosol would be accessible to the antibody added to the extracellular medium whereas bacteria protected by a vacuole would not. Infected cells were treated with streptolysin-O, fixed and stained with antibodies in the absence of detergent. At 8 h post-uptake most wild-type and _ssaV_– bacteria were protected from the anti-LPS antibody. In contrast, >55% of _sifA_– bacteria were decorated with the anti-LPS antibody, indicating their presence in the cytosol (Figure 8A). Examination of intracellular bacteria under the electron microscope confirmed the presence of cytosolic _sifA_– bacteria (Figure 8D), whereas wild-type, _ssaV_– and sifA_–, p_sifA cells were always associated with a vacuolar membrane (Figure 8B, C and E). These results show that maintenance of a lgp/vATPase-containing membrane around wild-type bacteria requires the sifA gene.
Fig. 8. Streptolysin-O permeabilization and electron microscopy of host cells infected with different bacterial strains. (A) RAW cells were infected with different strains expressing GFP for 8 h, then permeabilized with streptolysin-O, and incubated with anti-LPS antibody. The percentage of bacteria staining positive for the anti-LPS antibody was determined. Results shown are the means ± SE of three independent experiments. In each experiment, between 100 and 150 bacteria were scored for each strain. Electron microscopy analysis of RAW macrophages infected with either wild-type (B), _ssaV_– (C), _sifA_– (D) or sifA_–, p_sifA (E) strains, 10 h post-uptake. Infected cells were fixed and processed for transmission electron microscopy. Arrows in (B, C and E) point to vacuolar membranes, which are absent in (D). Scale bar corresponds to 0.5 µm.
SifA is similar to secreted proteins from Salmonella
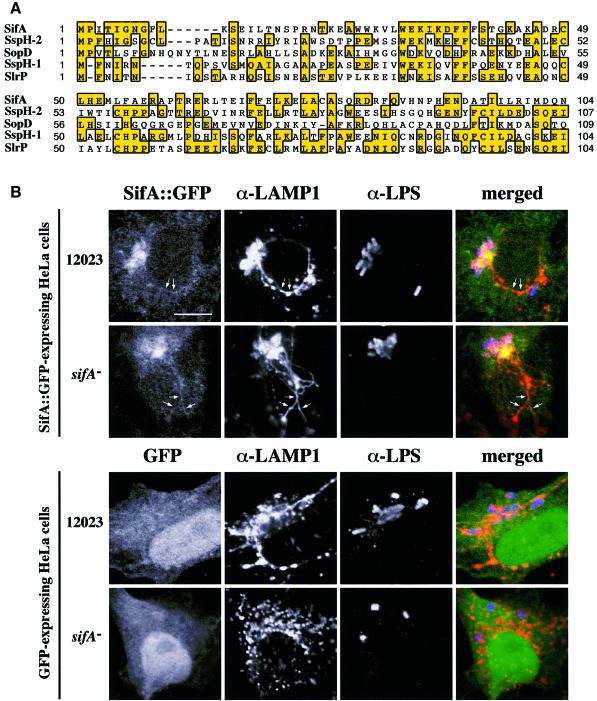
The results described above show that the maintenance of the lgp/vATPase-containing membrane of the SCV is dependent on SifA, which in turn requires the SPI-2 TTSS regulator, ssrA. The C.I. analysis also suggests that sifA and ssaV are functionally related (Table I). One obvious explanation of these results is that SifA is a SPI-2 effector protein. BLAST and FASTA searches using the predicted amino acid sequence of sifA revealed that it has weak but significant similarity at the N-terminal region to three Salmonella proteins: SspH-2, SspH-1 and SlrP (Figure 9A). SspH-1 and SspH-2 are preferentially translocated into the host cell cytosol by the SPI-1 and SPI-2 TTSS, respectively (Miao et al., 1999), but their function is unknown. The slrP gene is present on a pathogenicity island and, like SspH-1 and SspH-2, shares similarity to other proteins secreted by TTSSs (Tsolis et al., 1999). However, unlike all these proteins, SifA does not contain leucine-rich repeats. Its region of similarity is confined to approximately the first 100 amino acids of SspH-2, SspH-1 and SlrP: over this region, SifA is most similar (30% identical) to SspH-2, and has lower similarity to SlrP and SspH-1 (Figure 9A). The N-terminal regions of SspH-2 and SspH-1 are sufficient for their secretion and translocation (Miao et al., 1999), suggesting that SifA may be a secreted protein.
Fig. 9. (A) Multiple alignment of the N-terminal region of SifA with other Salmonella proteins. The alignment was generated by ClustalW (http://www2.ebi.ac.uk/clustalw/) and Multalin (http://www.toulouse.inra.fr/multalin.html) and edited by hand. (B) Formation of Sifs is restored in _sifA_– mutant-infected cells expressing SifA::GFP. HeLa cells transiently expressing SifA::GFP (upper panels) or GFP as a control (lower panels) were infected with wild-type (12023) or _sifA_– strains, and fixed with paraformaldehyde at 8 h post-infection. LAMP-1 and LPS were detected using TRSC- and Cy5-conjugated donkey anti-rabbit and anti-mouse secondary antibodies, respectively. Three-colour confocal microscopic analysis was performed. Scale bar represents 10 µm. Expression of SifA::GFP in HeLa cells restores the presence of a LAMP-1-enriched vacuole around _sifA_– bacteria. SifA–GFP is localized on SCVs and Sifs (arrows in the top panels).
As standard BLAST or FASTA database searches are not always sufficient to detect subtle sequence relationships, we used a multiple sequence alignment of the N-terminal regions of SifA, SspH-1, SspH-2 and SlrP (from Trp31 to Glu106 of the SifA amino acid sequence) to build a profile that was then used to search the non-redundant sequence database (NRDB). This method allows more sensitive database searches to be carried out, by highlighting the critical residues within a set of aligned sequences. The search using this profile revealed similarity to a portion of only one protein: the N-terminal end of Salmonella dublin SopD, with an E value of 0.0559 (indicating that the similarity is likely to be significant) (Figure 9A). SopD is translocated into HeLa cells via the Inv/Spa TTSS, its N-terminal region being sufficient to direct translocation (Jones et al., 1998).
Infection of HeLa cells expressing SifA with a sifA– mutant strain restores Sif formation
To investigate whether SifA is secreted into the host cell cytosol, three SifA–adenylate cyclase (CyaA) fusion proteins were constructed. The CyaA reporter system has been used previously to demonstrate translocation of SspH-1 and SspH-2 fusion proteins via the SPI-2 secretion system (Miao et al., 1999). The fusions contained either the N-terminal 53, 113 or 217 amino acids of SifA, which is 336 amino acids in length. The predicted sizes of the different fusion proteins were verified by immunoblotting using an anti-CyaA antibody, and their CyaA activities were demonstrated by functional complementation of an _Escherichia coli cyaA_– mutant strain (Glaser et al., 1988). These constructs were transferred into wild-type, _ssaV_– or _sifA_– strains. These were then used to infect RAW macrophages, which were assayed for CyaA activity at different time points (Miao et al., 1999). No accumulation of cAMP (reflecting CyaA activity) was detected at 6 or 8 h post-uptake for any of the strains, despite their efficient uptake by macrophages (results not shown). This suggests that either SifA is itself not translocated into the host cell cytosol, or that following translocation, its conformation or interaction with a receptor prevents determination of CyaA activity of the fusion protein.
In view of the strong circumstantial evidence favouring an effector function for SifA, HeLa cells were transiently transfected with plasmids encoding either SifA::GFP, or the GFP as a control, then infected with different S.typhimurium strains. Confocal imaging showed that the GFP was localized both in the cytoplasm and the nucleus. In contrast, the SifA::GFP fusion protein was associated mainly with membranes, vacuoles containing wild-type or _sifA_– strains, and Sifs. In addition, formation of Sifs was restored in SifA::GFP expressing cells infected with the _sifA_– mutant strain (Figure 9B). Collectively these results suggest that SifA is translocated on to the cytosolic face of the vacuolar membrane where it mediates membrane fusion events.
Maintenance of the vacuolar membrane enclosing SPI-2 mutant strains is not dependent on sifA
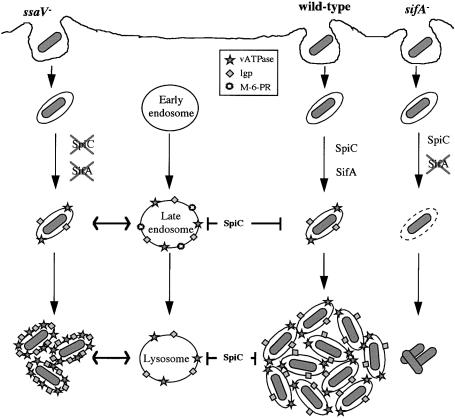
We have established that sifA expression requires the SPI-2 regulator ssrA, and that SifA is required for the maintenance of the vacuolar membrane enclosing wild-type bacteria. If, as suggested by our results, SifA is secreted by the SPI-2 TTSS, an _ssaV_– mutant strain [which lacks a functional secreton (Beuzón et al., 1999)] would be defective in SifA function and might also be expected to be released into the host cell cytosol. However, _ssaV_– mutant bacteria were found within vacuoles in macrophages (Figure 8C). If SifA is not secreted by the SPI-2 TTSS, SifA could account for the maintenance of a vacuolar membrane surrounding _ssaV_– bacteria. In this case, bacterial cells carrying both _sifA_– and _ssaV_– mutations would be released into the cytosol. However, vacuoles containing these double mutant bacteria were associated with LAMP-1 in macrophages, and with LIMP-1, LAMP-1, LAMP-2 and vATPase in HeLa cells (Figures 5 and 6 and results not shown). Furthermore, since sifA expression is ssrA dependent (Figure 3), an _ssrA_– strain would also be expected to be found in the host cell cytosol. Like _ssaV_– and _ssaV_–, _sifA_– double mutant bacteria, _ssrA_– cells were associated with LAMP-1 in macrophages (Figure 5). Therefore, sifA is not required for maintenance of the vacuolar membrane enclosing _ssaV_– or _ssrA_– bacteria, and the recruitment of lgp/vATPase-containing membrane to these cells must reflect a different process to that observed with the wild-type strain. The simplest explanation for these results is that the SPI-2 TTSS translocates other effectors which prevent the SCV from interacting with the endocytic pathway, prior to the action of SifA. An _ssaV_– strain cannot secrete any effectors, and is enclosed by a vacuole by interactions with the endocytic pathway (Figure 10).
Fig. 10. Model showing the role of the SPI-2 secretion system in the maturation of the SCV.
Discussion
In this paper we describe a general method for identifying virulence gene interactions in vivo based on the C.I. The technique involves direct comparisons between single and double mutant strains rather than combinations of different single mutants and the wild-type strain. This approach allows the relative attenuation of two mutations to be assessed directly in the same animal, avoiding host-to-host variation.
In the present work, we have used C.I. analysis to show that ompR, SPI-2 genes and sifA interact during systemic infection of mice. A functional relationship between these loci was suggested by the finding that _ompR_– mutants fail to induce Sif formation in epithelial cells (Mills et al., 1998), and the recent demonstration that OmpR regulates ssrA expression during infection of cultured macrophages (Lee et al., 2000). Our C.I. analysis indicates that in vivo, the attenuated virulence of an _ompR_– mutant can be attributed entirely to its interaction with SPI-2 genes, and vice versa. This was surprising in view of the finding that a mutation in ompR delayed but did not eliminate induction of a SPI-2 ssaH gene in cultured macrophages (Lee et al., 2000). It is possible that OmpR exerts tighter control over SPI-2 function in vivo than in cultured cells; alternatively, the partial effect of an _ompR_– mutation over SPI-2 gene expression may be sufficient to reduce expression below a functionally effective threshold. OmpR regulation of SPI-2 gene expression is therefore analagous to that of PhoP/Q over SPI-1 and SPI-3 genes (Pegues et al., 1995; Blanc-Potard and Groisman, 1997): both regulators are present in the non-pathogenic relative E.coli, but have extended their regulatory range in Salmonella to encompass virulence genes on PAIs.
In infected epithelial cells, sifA induces the formation of dramatic tubular membrane structures (Sifs), that are enriched in lysosomal membrane glycoproteins. However, the physiological relevance of these structures is not clear, despite the fact that the sifA gene is known to be important for virulence in mice (Stein et al., 1996). The C.I. tests confirm the importance of the sifA gene in S.typhimurium virulence, and suggest that sifA and SPI-2 functions are intimately connected in vivo. Since attenuated SPI-2 mutant strains fail to induce Sifs, and sifA expression is dependent on ssrA, this also explains the observation that _ompR_– mutants fail to make Sifs (Mills et al., 1998). The systemic in vivo attenuation of the _sifA_– mutant strain can be explained by its intramacrophage replication defect, which is similar to that of SPI-2 mutants. Consistent with this, the onset of sifA gene expression in cultured cells corresponds to that of SPI-2 genes (Cirillo et al., 1998).
Several lines of evidence suggest that SifA is a SPI-2 effector protein. First, the C.I. analysis implies that both loci are involved in the same function. Secondly, although sifA is regulated by ssrA, it is not present in SPI-2, which contains all the genes known to be required for secretion and translocation of SPI-2 effectors. Thirdly, sifA has a significantly lower G+C content than the average for the S.typhimurium chromosome, is flanked by repeat sequences, and is a virulence determinant specific to Salmonella, all of which suggest that it was acquired by horizontal transfer (Stein et al., 1996). A gene for an effector protein of the Inv/Spa TTSS of Salmonella was also acquired independently by horizontal transfer, which has led to the suggestion that this may allow bacterial pathogens to fine-tune their TTSSs (Hardt et al., 1998). Fourthly, SifA has weak but significant similarity to other Salmonella secreted proteins, and this is restricted to the regions reported to be sufficient for secretion and translocation. Finally, the recruitment of the SifA::GFP fusion protein to the SCV and on Sifs when expressed in host cells, and its ability to rescue Sif formation in cells infected with the _sifA_– mutant strain suggests that it is normally translocated from wild-type bacteria to the cytosolic face of the vacuole. The localization of SifA to host cell membranes is consistent with the hypothesis that it mediates membrane fusion events (see below). If SifA binds to a receptor in the vacuolar membrane this might also explain the failure to demonstrate translocation using SifA::Cya fusions, since this could interfere with CyaA activity of this fusion protein.
Our results show that the maintenance of a lgp/vATPase- containing membrane around wild-type but not SPI-2 mutant strains of S.typhimurium requires a functional sifA gene. We present a model (Figure 10) to account for the intracellular phenotypes of different mutant strains. The vacuole containing wild-type bacteria, through the action of the SPI-2 TTSS effector SpiC (Uchiya et al., 1999) and possibly other SPI-2 effectors, does not interact with late endosomes and lysosomes, and SifA actively recruits and maintains the lgp/vATPase-containing membrane. It is known that acquisition of lgps by the SCV in epithelial cells occurs by recruitment of pre-existing lgp-containing vesicles in a rab7-dependent manner, without direct interaction with lysosomes (Méresse et al., 1999a). Through the action of unknown SPI-2 effectors, wild-type Salmonella also inhibits trafficking of the NADPH oxidase to the SCV (Vázquez-Torres et al., 2000). These processes create an environment conducive to bacterial replication. Strains containing mutations in ssaV or ssrA do not have a functional SPI-2 secretion system, and the vacuoles acquire lgps and vATPase through interactions with late endosomes and lysosomes. A strain carrying a mutation in sifA retains the ability to translocate SpiC and other effectors, which prevent interactions with late stages of the endocytic pathway. However, because this strain cannot recruit or maintain lgp/vATPase-containing membrane through the action of SifA, the vacuole cannot be sustained, and the bacterial cell is released into the cytoplasm. For intracellular pathogens to replicate but remain within a membrane-bound vacuole, there must be a progressive net increase in the surface area of the vacuolar membrane. SifA is therefore likely to control directed membrane fusion events, and the lack of SifA would lead to insufficient membrane acquisition to enclose the replicating bacterial cells, which would become exposed to the cytosol.
The intra-macrophage replication defect displayed by the _sifA_– mutant strain is comparable to the phenotype of S.dublin or E.coli K-12 strains carrying the hly gene of Listeria monocytogenes. These bacteria can also escape to the macrophage cytosol by secreting listeriolysin, but they fail to replicate (Gentschev et al., 1995). This suggests that, unlike other intracellular pathogens such as Listeria which can replicate in the macrophage cytosol (de Chastellier and Berche, 1994), Salmonella cannot exploit this environment for growth. To our knowledge, SifA is the first example of a bacterial protein responsible for the maintenance of a membrane-bound vacuole. Specificity of vesicular trafficking in the eukaryotic cell is conferred by the integral membrane vSNARE and tSNARE proteins (Rothman and Wieland, 1996), and is modulated by rab GTPases (Schimmoller et al., 1998). In view of the involvement of the rab7 GTPase in the biogenesis of SCVs (Méresse et al., 1999a) and its presence on Sifs (S.Méresse and J.-P.Gorvel, unpublished observations) it is possible that SifA acts as an activator of this membrane fusion machinery, thereby ensuring a continuous flow of membrane towards the SCV.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table II. H3P6 carries a mini-Tn_5_ transposon insertion that was mapped between nucleotides 44 and 139 of the sifA open reading frame (ORF; 1012 nucleotides in length) and was identified but not further characterized in the original STM screen of S.typhimurium (Hensel et al., 1995). The mutation was transduced by phage P22 into strain 12023 prior to use in this study. Double mutant strains were also constructed by P22-mediated transductions, carried out as described previously (Davis et al., 1980). Bacteria were grown in Luria–Bertani (LB) medium supplemented with ampicillin (50 µg/ml), kanamycin (50 µg/ml), tetracycline (25 µg/ml) or chloramphenicol (50 µg/ml for plasmid-containing strains, 10 µg/ml for chromosomal integrants) as appropriate. CJD359 and CS105 were kindly provided by C.Dorman and S.Miller, respectively.
Table II. Strains used in this study.
| Name | Description | Source or reference |
|---|---|---|
| 12023s | wild-type | NTCC |
| P3F4 | ssrA::mTn_5_ in 12023s | Hensel et al. (1995) |
| P3H6 | sifA::mTn_5_ in 12023s | this study |
| P4E3 | purD::mTn_5_ in 12023s | Hensel et al. (1995) |
| CS105 | phoP-102::Tn_10d_Cm in 12023s | Miller et al. (1989) |
| CJD359 | ompR-1009::Tn_10_ in SL1344 | Dorman et al. (1989) |
| HH102 | ΔsseB::aphT in 12023s | Hensel et al. (1998) |
| HH109 | ssaV::aphT in 12023s | Deiwick et al. (1999) |
| HH110 | _ssaV::_Cmr in 12023s | Shea et al. (1999) |
| HH111 | ssaV::Cmr, purD::mTn_5_ in 12023s | Shea et al. (1999) |
| HH123 | ssaV::Cmr, sseB::aphT in 12023s | this study |
| HH172 | ssaV::Cmr, sifA::mTn_5_ in 12023s | this study |
| HH182 | ompR-1009::Tn_10_ in 12023s | this study |
| HH187 | ssaV::aphT, ompR-1009::Tn_10_ in 12023s | this study |
Plasmids
Plasmid p_sseB_, used to complement the Δ_sseB_ mutant strain, has been described previously (Hensel et al., 1998). Plasmid pFPV25.1, carrying gfpmut3A under the control of a constitutive promoter, was introduced into bacterial strains for fluorescence visualization where indicated (Valdivia and Falkow, 1997). pID812 is a derivative of pFPV25, a vector carrying promoterless gfpmut3A (Valdivia and Falkow, 1997) containing a translational fusion of a fragment of sifA to the gfp gene. A fragment including 160 bp of the sifA promoter region and the sequence encoding the N-terminal 270 amino acids of SifA was amplified by PCR from 12023s genomic DNA using SIFA1 (5′-GGATCCGACAGTAATGCGTTTATACGCGAAGCTCTC-3′) and SIFA2 (5′-CATATGGGTCTTATCCGCTAGTAAAACCTCTTTTAC-3′) primers. The 1 kb PCR product, containing terminal _Bam_HI and Nde_I sites, was digested and ligated into pFPV25, generating pID812. p_sifA is a derivative of pACYC184 (Chang and Cohen, 1978) carrying the sifA gene under the control of a constitutive promoter. A DNA fragment including the complete ORF of sifA and its ribosomal binding site (rbs) was amplified by PCR from 12023s genomic DNA using SIFN-B (5′-GGATCCTTACTCCAGTATAAGTGAG-3′) and SIFAT2 (5′-CTCGAGGTGACGTCTGAGAAAG-3′) primers. The ∼1 kb PCR product, containing terminal _Bam_HI and _Xho_I sites, was digested and ligated into pACYC184 Eco_RV–_Sal_I sites inside the Tetr gene generating p_sifA. All the constructions were verified by DNA sequencing.
Mouse mixed infections
Female BALB/c mice (20–25 g) were used for all infection studies and were inoculated i.p. with a 0.2 ml volume of bacterial cells suspended in physiological saline. To prepare the inocula bacteria were grown overnight at 37°C in LB broth with shaking (150 r.p.m.) and then used to inoculate fresh medium (1:100) and grown under the same conditions for 2–3 h until an OD550 of 0.35–0.6 was reached. Cultures were then diluted in physiological saline to a concentration of 2.5 × 105 bacteria per strain and mixed before the infection. The c.f.u. were enumerated by plating a dilution series of the inoculum, using antibiotics to distinguish between the strains. To confirm that the presence of the antibiotic did not result in lower bacterial recovery, strains carrying antibiotic resistance were plated on LB and LB with the appropriate antibiotic. Mice were killed at 48 h post-inoculation by carbon dioxide inhalation. The spleens were removed and bacteria recovered and enumerated after plating a dilution series on to LB agar and LB agar with the appropriate antibiotics (Shea et al., 1999).
Statistical analysis
Each C.I. value is the mean of three independent mice infections. Student’s _t_-test was used to analyse the C.I.s of single mutant strains versus double mutant strains (i.e. _a_– versus _a_–_b_–) with two null hypotheses: mean C.I. is significantly different from 1.0 and mean C.I. is significantly different from the C.I. of the single mutant versus the wild-type strain (wt versus b). Probabilities (p) of ≤0.05 were considered significant.
Sequence analysis
Sequence database similarity searches were carried out by Blast2 (http://www.ncbi.nlm.nih.gov/BLAST/), WU-Blast2 and Fasta3 (http://www2.ebi.ac.uk/blast2/ and /fasta3/) (Pearson and Lipman, 1988; Altschul et al., 1997). All the searches used derived amino acid sequences.
The multiple alignment shown in Figure 9A was generated by ClustalW (http://www2.ebi.ac.uk/clustalw/) and Multalin (http://www.toulouse.inra.fr/multalin.html) (Corpet, 1988; Thompson et al., 1994). The sequences included can be found under the SWALL accession numbers Q56061 (SifA), Q9XDN9 (SspH-1), AAF00615 (SspH-2) and Q9XCV2 (SlrP). A ClustalW multiple alignment of the region of interest was used to generate a profile by ProfileWeight. This profile was then used to search the NRDB using the program Profilesearch. Both programs are available at the Bioccelerator Web server (http://shag.embl-heidelberg.de:8000/Bic/).
Antibodies and reagents
The mouse monoclonal antibodies (mAbs) anti-LAMP-1 H3A4 and anti-CD63 H5C6 developed by J.T.August and J.E.K.Hildreth were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa (Department of Biological Siences, Iowa, IA), and were used at a dilution of 1:2000. Anti-LAMP-2 mouse mAb CD3 was obtained from Dr Minoru Fukuda (The Burnam Institute, La Jolla Cancer Research Foundation, La Jolla, CA), and was used at a dilution of 1:1000, and anti-vATPase mouse mAb OSW2 was obtained from Dr Satoshi B.Sato (Kyoto University, Kyoto, Japan) and was used at a dilution of 1:2000. Anti-LAMP-1 rabbit polyclonal antibody 156 against the 11-residue cytoplasmic domain of LAMP-1 has been described previously (Steele-Mortimer et al., 1999), and was used at a dilution of 1:1000. Anti-Salmonella goat polyclonal antibody CSA-1 was purchased from Kirkegaard and Perry Laboratories (Gaithersburg, MD) and was used at a dilution of 1:400. Anti-Salmonella LPS mouse mAb 1E6 was purchased from Biodesign International (Kennebunk, ME) and was used at a dilution of 1:10 000. Texas Red sulfonyl chloride (TRSC)-, cyanine 5 (Cy5)- and fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse, anti-rabbit and anti-goat antibodies were purchased from Jackson Immunoresearch Laboratories, Inc. (West Grove, PA), and used at a dilution of 1:400.
Cell culture
RAW 264.7 cells were obtained from ECACC (ECACC 91062702). HeLa (clone HtTA1) cells were kindly provided by Dr H.Bujard (Heidelberg, Germany). Cells were grown in Dulbecco’s modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 2 mM glutamine at 37°C in 5% CO2.
Bacterial infection of HeLa cells
HeLa cells were seeded on to glass coverslips (12 mm diameter) in 24-well plates at a density of 5 × 104 cells per well, 24 h before infection. Bacteria were incubated for 16 h at 37°C with shaking, diluted 1:33 in fresh LB broth and incubated in the same conditions for 3.5 h. The cultures were diluted in Earle’s buffered salt solution (EBSS) pH 7.4 and added to the HeLa cells at a multiplicity of infection (m.o.i.) of ∼100:1. The infection was allowed to proceed for 15 min at 37°C in 5% CO2. The monolayers were washed once with DMEM containing FCS and 100 µg/ml gentamicin and incubated in this medium for 1 h, after which the gentamicin concentration was decreased to 16 µg/ml.
Bacterial infection of macrophages and survival assays
Macrophages were seeded at a density of 4 × 105 cells per well in 24-well tissue culture plates, 24 h before use. Bacteria were cultured at 37°C with shaking until they reached an OD600 of 2.0. The cultures were diluted to an OD600 of 1.0 and opsonized in DMEM containing FCS and 10% normal mouse serum for 20 min. Bacteria were added to the monolayers at an m.o.i. ∼100:1, centrifuged at 170 g for 5 min at room temperature and incubated for 25 min at 37°C in 5% CO2. The macrophages were washed once with DMEM containing FCS and 100 µg/ml gentamicin and incubated in this medium for 1 h. The medium was replaced with DMEM containing FCS and 16 µg/ml gentamicin for the remainder of the experiment. For enumeration of intracellular bacteria, macrophages were washed three times with phosphate-buffered saline (PBS), lysed with 0.1% Triton X-100 for 10 min and a dilution series was plated on to LB agar.
Immunofluorescence and electron microscopy
For immunofluorescence, cell monolayers were fixed in 3.7% paraformaldehyde in PBS pH 7.4, for 15 min at room temperature and washed three times in PBS. Antibodies were diluted in 10% horse serum, 1% bovine serum albumin (BSA), 0.1% saponin in PBS. Coverslips were washed twice in PBS containing 0.1% saponin, incubated for 30 min with primary antibodies, washed twice with 0.1% saponin in PBS and incubated for 30 min with secondary antibodies. Coverslips were washed twice in 0.1% saponin in PBS, once in PBS and once in H2O, and mounted on Mowiol. Samples were analysed using a fluorescence microscope (BX50; Olympus Optical Co., Ltd) or a confocal laser scanning microscope (LSM510, Zeiss).
For transmission electron microscopy, cell suspensions were fixed in 3% glutaraldehyde prepared in 0.1 M cacodylate buffer pH 7.3. Fixation was for 1–2 h at room temperature, after which the cells were washed in fresh buffer before post-fixing in 1% osmium tetroxide in the same buffer. The cells were encased in agar (Ryder and MacKenzie, 1981), dehydrated through a graded series of alcohols and embedded in Araldite epoxy resin. Ultrathin sections were cut on a diamond knife and stained in alcoholic uranyl acetate and lead citrate before examination in a transmission electron microscope operated at 75 kV.
Streptolysin-O permeabilization of RAW cells
Streptolysin-O was obtained from Dr S.Bhakdi (Mainz, Germany). RAW cells were grown on coverslips, infected for 8 h with different strains, then rapidly washed with ice-cold ICT buffer (50 mM HEPES–KOH pH 7.1, 4 mM MgCl2, 10 mM EGTA, 8.4 mM CaCl2, 78 mM KCl, 1 mM DTT, 1 mg/ml BSA). They were then incubated for 5 min at 4°C with 2 µg/ml streptolysin-O in ICT buffer, extensively washed, incubated for 5 min at 37°C in the same buffer and immediately fixed in 3.7% paraformaldehyde. Coverslips were treated for immunofluorescence by double labelling as described above but in the absence of detergent. Mouse anti-Salmonella LPS was used to detect bacteria present in the cytosol. Antibody raised against the LAMP-1 cytoplasmic domain was used as control for plasma membrane permeabilization.
Expression of SifA::GFP in HeLa cells
The full sifA ORF was amplified from S.typhimurium genomic DNA by PCR using the following primers: 5′-AAAAAAGAATTCCACCATGCCGATTACTATAGGGAATGG-3′ and 5′-AAAAAACCCGGGCTAAAAAACAACATAAACAGCCGC-3′. The PCR product was subcloned into the unique _Eco_RI and _Xma_I sites of pEGF-C1 vector (Clontech Laboratories) into the same reading frame as GFP, generating psifA::gfp. Vectors encoding GFP or SifA::GFP were used to transfect HeLa cells with FuGene 6 (Boehringer Mannheim) following the manufacturer’s instructions. Cells were further incubated for 24–48 h and infected with bacterial strains as described. The expressed SifA::GFP fusion protein was checked by western blot analysis using an anti-GFP mouse mAb JL-8 (Clontech) to be of the expected size (583 amino acid residues, 66 kDa).
Acknowledgments
Acknowledgements
We thank Agnès Ullmann for providing an antibody against adenylate cyclase, Antoine Danchin for E.coli strain TP610pVUC1, and Christoph Tang and Herb Arst for valuable suggestions. We are very grateful to Colin Gleeson for practical assistance. This project was supported by grants from the MRC (UK) to D.W.H., and from CNRS and INSERM to S.M. C.B. was supported by an EMBO Fellowship.
References
- Altschul S., Madden,T., Schaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped blast and PSI-blast: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler A., Tsolis,R., Valentine,P., Ficht,T. and Heffron,F. (1997) Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect. Immun., 65, 2254–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzón C., Banks,G., Deiwick,J., Hensel,M. and Holden,D.W. (1999) pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol., 33, 806–816. [DOI] [PubMed] [Google Scholar]
- Blanc-Potard A. and Groisman,E. (1997) The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J., 16, 5376–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.C. and Cohen,S.N. (1978) Construction and characterisation of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol., 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield S., Dorman,C., Hayward,C. and Dougan,G. (1991) Role of ompR dependent genes in Salmonella typhimurium virulence: mutants deficient in both OmpC and OmpF are attenuated in vivo. Infect. Immun., 59, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo D., Valdivia,R., Monack,D. and Falkow,S. (1998) Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol., 30, 175–188. [DOI] [PubMed] [Google Scholar]
- Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.H., Botstein,D. and Roth,J.R. (1980) Advanced Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- de Chastellier C. and Berche,P. (1994) Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun., 62, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiwick J., Nikolaus,T., Erdogan,S. and Hensel,M. (1999) Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol., 31, 1759–1773. [DOI] [PubMed] [Google Scholar]
- Dorman C., Chatfield,S., Higgins,C., Hayward,C. and Dougan,G. (1989) Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect. Immun., 57, 2136–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B.B. (1994) Molecular and cellular mechanisms of Salmonella pathogenesis Curr. Top. Microbiol. Immunol., 192, 163–185. [DOI] [PubMed] [Google Scholar]
- Freter R., O’Brien,P. and Macsai,M. (1981) Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect. Immun., 34, 234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J.E. (1996) Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol., 20, 263–271. [DOI] [PubMed] [Google Scholar]
- Galán J.E. and Curtiss,R.,III (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl Acad. Sci. USA, 86, 6383–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F. and Finlay,B.B. (1995) Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose-6-phosphate receptors. J. Cell Biol., 129, 81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F., Zwick,M.B., Leung,K.Y. and Finlay,B.B. (1993a) Intracellular replication of Salmonella within epithelial cells is associated with filamentous structures containing lysosomal membrane glycoproteins. Infect. Agents Dis., 2, 227–231. [PubMed] [Google Scholar]
- Garcia-del Portillo F., Zwick,M.B., Leung,K.Y. and Finlay,B.B. (1993b) Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl Acad. Sci. USA, 90, 10544–10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentschev I., Sokolovic,Z., Mollenkopf,H.J., Hess,J., Kaufmann,S.H., Kuhn,M., Krohne,G.F. and Goebel,W. (1995) Salmonella strain secreting active listeriolysin changes its intracellular localization. Infect. Immun., 63, 4202–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P., Ladant,D., Sezer,O., Pichot,F., Ullmann,A. and Danchin,A. (1988) The calmodulin-sensitive adenylate cyclase of Bordetella pertussis: cloning and expression in Escherichia coli. Mol. Microbiol., 2, 19–30. [PubMed] [Google Scholar]
- Groisman E. and Ochman,H. (1997) How Salmonella became a pathogen. Trends Microbiol., 5, 343–349. [DOI] [PubMed] [Google Scholar]
- Hardt W.-D., Urlaub,H. and Galán,J.E. (1998) A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl Acad. Sci. USA, 95, 2574–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M., Shea,J.E., Gleeson,C., Jones,M.D., Dalton,E. and Holden,D.W. (1995) Simultaneous identification of bacterial virulence genes by negative selection. Science, 269, 400–403. [DOI] [PubMed] [Google Scholar]
- Hensel M., Shea,J.E., Raupach,B., Monack,D., Falkow,S., Gleeson,C. and Holden,D.W. (1997) Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol., 24, 155–167. [DOI] [PubMed] [Google Scholar]
- Hensel M. et al. (1998) Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol., 30, 163–174. [DOI] [PubMed] [Google Scholar]
- Heyde M. and Portalier,R. (1987) Regulation of major outer membrane porin proteins of Escherichia coli K 12 by pH. Mol. Gen. Genet., 208, 511–517. [DOI] [PubMed] [Google Scholar]
- Hueck C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev., 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker W. and Geuze,H.J. (1996) Intracellular trafficking of lysosomal membrane proteins. BioEssays, 18, 379–389. [DOI] [PubMed] [Google Scholar]
- Jones M.A., Wood,M.W., Mullan,P.B., Watson,P.R., Wallis,T.S. and Galyov,E.E. (1998) Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect. Immun., 66, 5799–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A., Detweiler,C. and Falkow,S. (2000) OmpR regulates the two-component system SsrA–SsrB in Salmonella pathogenicity island 2. J. Bacteriol., 182, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méresse S., Steele-Mortimer,O., Finlay,B.B. and Gorvel,J.P. (1999a) The rab7 GTPase controls the maturation of _Salmonella typhimurium_-containing vacuoles in HeLa cells. EMBO J., 18, 4394–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méresse S., Steele-Mortimer,O., Moreno,E., Desjardins,M., Finlay,B.B. and Gorvel,J.P (1999b) Controlling the maturation of pathogen-containing vacuoles: a matter of life and death. Nature Cell Biol., 1, 183–188. [DOI] [PubMed] [Google Scholar]
- Miao E., Scherer,C., Tsolis,R., Kingsley,R., Adams,L., Baumler,A. and Miller,S. (1999) Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol., 34, 850–864. [DOI] [PubMed] [Google Scholar]
- Miller S.I. and Mekalanos,J.J. (1990) Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol., 172, 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.I., Kukral,A.M. and Mekalanos,J.J. (1989) A two-component regulatory system (phoP/phoQ) controls Salmonella typhimurium virulence. Proc. Natl Acad. Sci. USA, 86, 5054–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D.M., Bajaj,V. and Lee,C.A. (1995) A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol., 15, 749–759. [DOI] [PubMed] [Google Scholar]
- Mills S., Ruschkowski,S., Stein,M. and Finlay,B. (1998) Trafficking of porin-deficient Salmonella typhimurium mutants inside HeLa cells: ompR and envZ mutants are defective for the formation of _Salmonella_-induced filaments. Infect. Immun., 66, 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Soncini,F.C., Solomon,F. and Groisman,E.A. (1996) Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl Acad. Sci. USA, 93, 7800–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. and Lipman,D. (1988) Improved tools for biological sequence analysis. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegues D.A., Hantman,M.J., Behlau,I. and Miller,S.I. (1995) PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol., 17, 169–181. [DOI] [PubMed] [Google Scholar]
- Rathman M., Sjaastad,M.D. and Falkow,S. (1996) Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun., 64, 2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathman M., Barker,L.P. and Falkow,S. (1997) The unique trafficking pattern of _Salmonella typhimurium_-containing phagosomes in murine macrophages is independent of the mechanism of bacterial entry. Infect. Immun., 65, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Dahlfors A., Buchan,A. and Finlay,B. (1997) Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med., 186, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.E. and Wieland,F.T. (1996) Protein sorting by transport vesicles. Science, 272, 227–234. [DOI] [PubMed] [Google Scholar]
- Ryder T.A. and MacKenzie,M.L. (1981) The routine preparation of seminal fluid specimens for transmission electron microscopy. J. Clin. Pathol., 34, 1006–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S.B. and Toyama,S. (1994) Interference with the endosomal acidification by a monoclonal antibody directed toward the 116 (100)-kD subunit of the vacuolar type proton pump. J. Cell Biol., 127, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmoller F., Simon,I. and Pfeffer,S.R. (1998) Rab GTPases, directors of vesicle docking. J. Biol. Chem., 273, 22161–22164. [DOI] [PubMed] [Google Scholar]
- Shea J.E., Hensel,M., Gleeson,C. and Holden,D.W. (1996) Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl Acad. Sci. USA, 93, 2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea J., Beuzón,C., Gleeson,C., Mundy,R. and Holden,D.W. (1999) Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect. Immun., 67, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai A.P. and Joiner,K.A. (1997) Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu. Rev. Microbiol., 51, 415–462. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O., Méresse,S., Gorvel,J.P. and Finlay,B.B. (1999) Biogenesis of _Salmonella typhimurium_-containing vacuoles in epithelial cells involves interactions with the early endocytic pathways. Cell Microbiol., 1, 33–49. [DOI] [PubMed] [Google Scholar]
- Stein M., Leung,K., Zwick,M., Garcia-del Portillo,F. and Finlay,B. (1996) Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol., 20, 151–164. [DOI] [PubMed] [Google Scholar]
- Taylor R., Miller,V., Furlong,D. and Mekalanos,J. (1987) Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl Acad. Sci. USA, 84, 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A.D. and Booth,I.R. (1992) The regulation of expression of the porin gene ompC by acid pH. J. Gen. Microbiol., 138, 1829–1835. [DOI] [PubMed] [Google Scholar]
- Thompson J., Higgins,D. and Gibson,T. (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis R.M., Townsend,S.M., Miao,E.A., Miller,S.I., Ficht,T.A., Adams,L.G. and Baumler,A.J. (1999) Identification of a putative Salmonella enterica serotype typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun., 67, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiya K., Barbieri,M., Funato,K., Shah,A., Stahl,P. and Groisman,A. (1999) A Salmonella virulence protein that inhibits cellular trafficking. EMBO J., 18, 3924–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R. and Falkow,S. (1997) Fluorescence-based isolation of bacterial genes expressed within host cells. Science, 277, 2007–2011. [DOI] [PubMed] [Google Scholar]
- Vázquez-Torres A., Xu,Y., Jones-Carson,J., Holden,D.W., Lucia,S.M., Dinauer,M.C., Mastroeni,P. and Fang,F.C. (2000) Salmonella Pathogenicity Island 2-dependent evasion of the phagocyte NADPH oxidase. Science, 287, 1655–1658. [DOI] [PubMed] [Google Scholar]
- Wood M., Jones,M., Watson,P., Hedges,S., Wallis,T. and Galyov,E. (1998) Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol., 29, 883–891. [DOI] [PubMed] [Google Scholar]