Diversity of the Human Skin Microbiome Early in Life (original) (raw)
Abstract
Within days after birth, rapid surface colonization of infant skin coincides with significant functional changes. Gradual maturation of skin function, structure, and composition continues throughout the first years of life. Recent reports have revealed topographical and temporal variations in the adult skin microbiome. Here we address the question of how the human skin microbiome develops early in life. We show that the composition of cutaneous microbial communities evolves over the first year of life, showing increasing diversity with age. Although early colonization is dominated by Staphylococci, their significant decline contributes to increased population evenness by the end of the first year. Similar to what has been shown in adults, the composition of infant skin microflora appears to be site specific. In contrast to adults, we find that Firmicutes predominate on infant skin. Timely and proper establishment of healthy skin microbiome during this early period might have a pivotal role in denying access to potentially infectious microbes and could affect microbiome composition and stability extending into adulthood. Bacterial communities contribute to the establishment of cutaneous homeostasis and modulate inflammatory responses. Early microbial colonization is therefore expected to critically affect the development of the skin immune function.
Introduction
The importance of the human skin microbiome in skin health and in the overall well-being of the individual has recently been started to be appreciated (Gao et al., 2007; Costello et al., 2009; Grice et al., 2009; Peterson et al., 2009). Most of this work has focused on adult skin, and little attention has been given so far to skin microbiology during the first weeks to years of life (Leyden, 1982; Dominguez-Bello et al., 2010). The time of birth marks a significant change for the skin of the newborn as it undergoes transitions from an aqueous and mostly sterile environment of the womb to a gaseous one with constant microbial interaction. During the first days after birth, rapid surface colonization coincides with significant skin barrier function changes. Reduction in transepidermal water loss, skin pH, and sebaceous activity, and increase in water content (Chiou and Blume-Peytavi, 2004) are some of the changes implicated in creating an environment conducive to the colonization of some bacterial species and prohibitive to others. Changes in skin barrier and water-handling functions together with high rates of surface expansion and epidermal cell proliferation continue through the first months to years of life (Nikolovski et al., 2008; Stamatas et al., 2010). During this time of development, infant skin has been shown to differ from adult skin in structure, function, and biochemical composition (Stamatas, 2010). Recent reports describe the topographical and temporal differences in the adult skin microbiome (Gao et al., 2007; Costello et al., 2009; Grice et al., 2009; Peterson et al., 2009) and point to its relative stability over time for the same individual (Costello et al., 2009; Grice et al., 2009).
Here we try to answer whether the infant skin microbiome is distinct from that of adult, given the particular differences in skin structure and function. Further, we examine whether the infant skin microbiome evolves along with the structural and functional development of infant skin and whether it varies across body sites.
Results
We first assessed the overall bacterial richness and diversity of infant skin (Supplementary Table S1 online and Supplementary data online). When the data from three skin sites were combined (arm, buttock, and forehead), the number of operational taxonomic units (OTUs) of bacteria was not found to significantly change with age. However, within each age group, differences were detected in the number of OTU across the three skin sites. Although there were no differences in OTU across body sites in the 1- to 3-month-olds, we identified fewer OTU on the arm relative to the buttock and forehead in the 4- to 6-month-olds, and to the buttock in the 7- to 12-month-olds. Although the number of genera in an area does not significantly change over the first year, the relative abundance of the community (evenness) significantly increases with age. An indication of microbiome stabilization in infants may be demonstrated by this increased diversity evenness in the microbial populations on the skin over the first year of age.
Taxonomic composition analysis revealed, in decreasing order, Bacilli, Clostridia, and Actinobacteria as the most abundant classes within all infant skin samples. Differences for the relative amounts were seen as a function of age and body site (Supplementary Table S2 online). In contrast to adult skin where Proteobacteria, Actinobacteria, and Firmicutes dominate in that order (Gao et al., 2007; Grice et al., 2008, 2009; Costello et al., 2009), infants are colonized predominantly by Firmicutes, followed in abundance by Actinobacteria, Proteobacteria, and Bacteriodetes. The difference in the colonization of adults versus infants is more likely due to the state of development, in terms of structure and composition of infant skin, which may represent a distinct environment for microbial colonization.
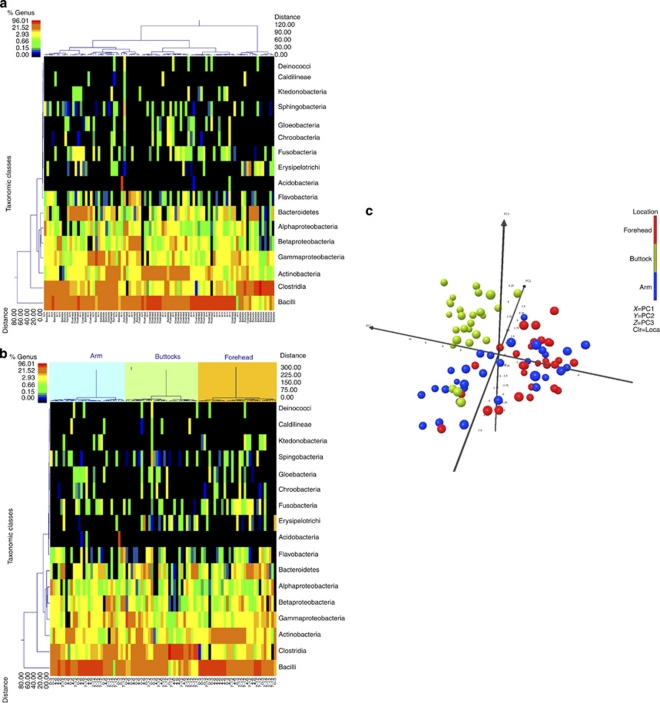
The class information was also evaluated through dual hierarchical clustering in which the top 17 classes were assessed without (Figure 1a) and with (Figure 1b) body site groups as clustering parameters. The dendrograms indicate that the buttock site has higher levels of Clostridia (phylum Firmicutes), whereas the arm and forehead samples show heightened levels of the Bacilli class of Firmicutes. Samples taken from the forehead have higher levels of the Actinobacteria class as compared with the other two groups, demonstrating similarity with adult sebaceous sites (Grice et al., 2009).
Figure 1.
Hierarchical clustering of infant skin samples taken from arms, foreheads, and buttock (a) taken together and (b) analyzed by sampled site. The most predominant 17 classes were chosen to be used in hierarchical clustering to assess the relationships between samples. (c) UNIFRAC-based principal component analysis demonstrates clustering according to body site. Each point corresponds to samples from a single location for each study participant. The percent variability for each principal component shown is PC1: 42.97%, PC2: 13.09%, and PC3: 8.29%. Clr, cluster; Loca, location.
The ability of samples to separate by skin site was assessed by the UNIFRAC-based principal component analysis (PCA, Figure 1c). The data indicate some cluster overlapping within the groups, suggesting common bacteria on the arm, buttock, and forehead. However, the samples taken from the buttock are highly divergent from samples taken from the arm and forehead. There is noticeable overlap between the clusters of the arm and the forehead samples, suggesting similarities between these skin environments.
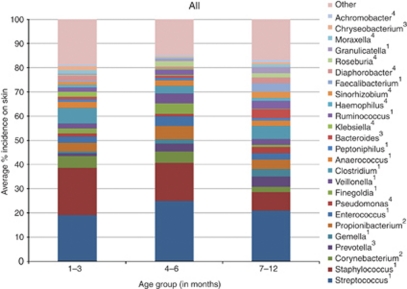
Analysis of the major microbial populations at the genus level as a function of age (Figure 2) shows that in the two youngest infant groups (1–3 and 4–6 months), there is a predominance of Streptococci and Staphylococci, which account for up to 40% of the total skin microbiome, with over 23 other genera making up the remaining population. Thus, 25 genera with predominance >1% make up on average 80–85% of the total population. As the infant ages, the abundance of the low predominance genera (<10%) increases, whereas levels of Staphylococcus, and Streptococcus statistically decrease, suggesting an expansion in diversity and development of the human skin microbiome.
Figure 2.
Relative abundance of the most predominant genera in the various age groups, averaged over all body sites. Superscripts indicate phylum: 1, Firmicutes; 2, Actinobacteria; 3, Bacteriodetes; 4, Proteobacteria.
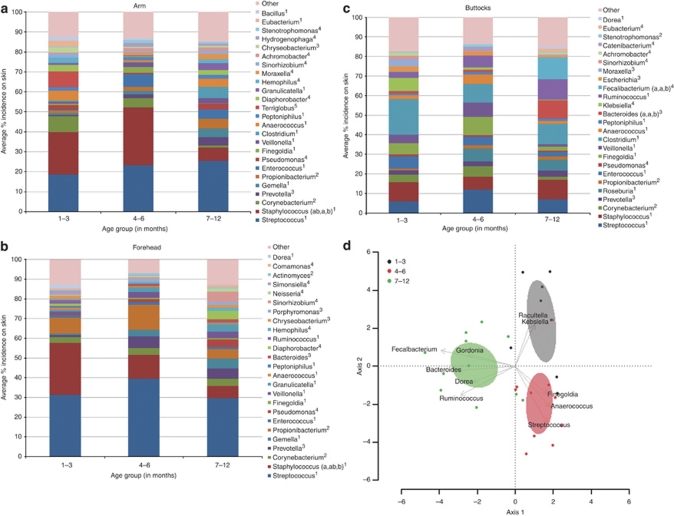
In further support of the idea of a still-developing microbiome, our data showed that the bacterial genera found on the arm, forehead, and buttock changes as a function of age (Figure 3 and Supplementary Table S3 online). Hierarchical clustering at the genus level demonstrates the evolution of skin microflora with age at the sampled sites (Supplementary Figure S1 online). Although the total number of bacteria genera remained the same with age, distance-based redundancy analysis at the genus level showed that the population evenness significantly increased in all sampled sites. On the arm and forehead, the abundance of Staphylococci significantly changes with age (Figure 3a and b, and Supplementary Table S3 online), which may explain in part the changes in population evenness. The population structure significantly changed with age for the buttock site (Figure 3c and d).
Figure 3.
Relative abundance of the most predominant genera on the arm (a), forehead (b), and buttock (c) in the various infant age groups. The predominant genera seen were Streptococcus, Staphylococcus, and Corynebacterium in the arm samples and Streptococcus, Staphylococcus, and Propionibacterium in the forehead samples. Along with Streptococcus and Staphylococcus, Clostridium, Runinococcus, and Finegoldia were also predominately seen in samples taken from the buttocks. Statistical analysis was performed across age groupings and significance indicated by the notation a, b, where a is significantly different than b. Superscripts indicate phylum: 1, Firmicutes; 2, Actinobacteria; 3, Bacteriodetes; 4, Proteobacteria; 5, Acidobacteria. (d) Biplot of the distance based redundancy analysis of microbial communities on the buttocks. Coordinates for the species arrows are correlations with each axis, and only the 10 most important (based on the magnitude of correlations) genera are shown. The 95% confidence ellipses for the age groups also are shown.
Discussion
Skin microbiome is known to depend on the local “micro-environment” of the skin site. In adults, there are differences in the bacterial population composition, diversity, and evenness between sites that are relatively more sebaceous, moist, or dry (Grice et al., 2009). Infant skin water-handling processes undergo a maturation process over the first year of life (Nikolovski et al., 2008). It has been shown, for example, that stratum corneum of infant is relatively better hydrated than adult. It is, therefore, expected that the microbiome of infant skin would resemble closer that of the relatively more moist skin sites in adult. Indeed, the overall relatively higher abundance of Staphylococcus in infant compared with adult skin agrees with the data reported for moist adult skin sites (Grice et al., 2009). Moreover, adult sebaceous sites were less diverse, less even, and less rich than other sites (Grice et al., 2009). We find that this is in agreement with the data of our youngest group (Supplementary Table S1 online) when sebaceous activity may still be present, because of influence of maternal hormones, but reduces drastically beyond the first weeks of life (Henderson et al., 2000).
Immediately after birth, bacterial communities on infant skin have been reported to be undifferentiated across body sites (Dominguez-Bello et al., 2010). Our results indicate that site-specific evolution of such communities appears to begin within the first 3 months of life. In contrast to what has been found in adult skin, namely that the microbiome is established and maintained over time (Gao et al., 2007; Grice et al., 2009), microflora differences between body sites in infants, coupled with the measured increase in population evenness with age, indicate that the infant skin microbiome is unstable. This instability may present the opportunity for aberrant skin development if the normal establishment of commensal microbiota goes awry.
Despite limited sebum production in infants (Agache et al., 1980), Propionibacteria were detected in the top five most predominant genera in infant forearm and forehead samples. Although the study participants did not have any skin conditions, it is interesting to note that the observed increase in Propionibacteria in the forehead samples at ages 4–6 months correlates with the period in which infantile acne begins to appear (Hello et al., 2008). We have found that the buttock area is unique in its microflora. Notably, we detected the genus Finegoldia predominantly on the buttock, which is a newly described, protease-elaborating commensal bacterium that because of its abundance on the buttock skin could have a role in the development of irritation in the diaper area (Goto et al., 2008). Early colonization of Clostridium and other gut-derived and/or anaerobic bacteria, particularly on the youngest infants and coupled with a significant increase in Bacteroides with age, is likely driven by the proximity to the gastrointestinal tract and the fact that there is a diaper covering the area that may change the oxygen potential, pH, and water-holding capacity of the skin (Visscher et al., 2000). This hypothesis is further supported by the fact that in agreement with published data of infant stool microbiota (Palmer et al., 2007), we detected early aerobic colonizers (Staphylococcus, Streptococcus, and Enterococcus) and other variable or transient taxa such as Prevotella, Veillonella, and _Clostridia_on at the buttock site. This observation, and the less likely competition from other skin microbiota during the early development of the buttock skin, could explain the microbial populations specific to this site.
It has been recently reported that the skin microbiome of newborns within 24 hours after birth correlates strongly with the mode of delivery; the microbiome of infants delivered vaginally is close to the one of mother's vagina and the one of infants delivered by Cesarean section relates to that of the mother's skin (Dominguez-Bello et al., 2010). Our data did not show any significant classification distinction with regard to mode of delivery, whether the samples were taken from infants of all ages tested or from the youngest group tested (1–3 months). This observation leads us to believe that the newborn skin microbiome is dynamic enough that initial differences (within 24 hours of delivery as described in the Dominquez-Bello et al., 2010) arising from mode of delivery disappear within a month of life.
Infant skin is known to be sensitive and prone to inflammatory conditions such as eczema and diaper dermatitis, and infections such as candidiasis. Of particular importance is the recent increase in the incidence of infantile atopic dermatitis (Mancini et al., 2008). Beyond their role in pathogenic infection, bacterial communities contribute to cutaneous homeostasis by directly affecting inflammatory responses (Lai et al., 2009). For example, Staphylococcal species have been found to modulate inflammatory responses through specific lipoteichoic acid mediators and are involved in homeostatic control of skin inflammation (Lai et al., 2009). Erythema toxicum neonatorum, a common rash of healthy newborn infants, particularly associated with Staphylococcus spp., triggers the local skin immune system and a systemic acute phase response in the first day after birth (Marchini et al., 2005). Interestingly, Staphylococcus may have a role in promoting early immune response and their subsequent decline may be an orchestrated response that the skin and/or the bacteria themselves conduct to facilitate microbiome and immunity development. Infant adaptive immunity continues to develop during early life (Holt and Jones, 2000), and the evolution of our skin microbiome may induce maturation of the immune system by providing the epitopes to train the adaptive immune system (Marchini et al., 2005) or even a direct form of defense (Cogen et al., 2008, 2010). Proteases secreted by certain species that colonize the skin can also affect its barrier function (Hirasawa et al., 2010) and it remains to be seen whether the skin microbiome has a role in educating keratinocytes for the formation of an adequate skin barrier during the time of functional development with obvious consequences to skin immunity. Interestingly, it has been proposed that immune pathways in the skin, as in the gut, are linked to the development of allergy (Spergel and Paller, 2003; Callard and Harper, 2007) or asthma (Benn et al., 2002).
Although microbial colonization of the human skin begins immediately after birth (Dominguez-Bello et al., 2010), we demonstrate here that it is not fully established within the first few weeks or even months of life, but rather evolves over the first year and likely beyond. Timely and proper establishment of a healthy skin microbiome (commensal genera) has a pivotal role in denying access to transient, harmful, and potentially infectious microbes. Thus, this early period could affect long-term microbiome stability. Microbial colonization of infant skin is expected to critically affect the development of the skin immune function and perhaps the maturation of other skin barrier functions, as well as the development of the systemic immune system.
Materials and Methods
Sample collection
A total of 31 healthy infants ranging in age from 3 to 52 weeks were recruited for this study. The infants were equally distributed among three age groups (1–3, 4–6, and 7–12 months of life). These groups were selected based on previous data on the maturation process of infant skin properties (Nikolovski et al., 2008), with the addition of a younger group (1–3 months). In addition, samples were taken from five mothers to serve as internal adult microbiome controls. There was a relatively equal distribution of male and female infants. All infants and mothers were Caucasian and reside in New Jersey. Swabs were taken between December 2008 and January 2009 from three skin sites: lower volar forearm, forehead, and buttock (center of the gluteal area). Infants had no presence of preexisting or dormant dermatological skin conditions including diaper dermatitis. The study was performed with an independent Institutional Review Board approval and following the Declaration of Helsinki Principles. A parent or legal guardian signed a written informed consent before the start of the study.
In order to normalize the routines of all subjects, mothers were instructed to bathe themselves and their infants with the same mild baby cleanser (Johnson's Head-to-Toe Baby Wash, Johnson & Johnson Consumer Products, Skillman, NJ) the evening before their scheduled appointment. No product usage (including application of topical products) was allowed after the bath, the night before and during the day of their scheduled appointment. Infants who had been on antibiotics, steroidal medication, or used medicated soaps within the past 30 days of the study were not included.
Skin flora was sampled using a swab technique (Paulino et al., 2006). A 2 × 2 cm area of the skin was sampled by swabbing the skin with a cotton pledget that was soaked in 0.15 NaCl and 0.1% Tween 20. The head of each swab was aseptically cut from the handle and placed into a sterile microcentrifuge tube and frozen at −20 °C, until shipment to sequencing facility.
DNA extraction
Swab samples from 31 infant and 5 adult subjects were taken out of the −80 °C freezer. Cotton tips of the swabs were shaved off using sterile surgical scalpels. The samples were placed into a TissueLyser (Qiagen, Valencia, CA) with a mix of 600 μl RLT buffer, a 5-mm sterile stainless steel bead, and 500 μl of glass disruption beads. The samples were processed for 5 minutes at 30 Hz. A subsample of 100 μl of lysate was then transferred into a 1.7 ml microcentrifuge tube containing a mix of 250 μl 100% ethanol and 350 μl of RLT buffer with β-mercaptoethanol. Standard procedures for the Qiagen DNA Extraction kit (Qiagen) were then followed after the mix was transferred into a spin column. A volume of 500 μl of buffers AW1 and AW2 were used for washing. Finally, the DNA was eluted with 100 μl of buffer AE and stored at −80 °C.
bTEFAP
We utilized the bacterial tag-encoded FLX-titanium amplicon pyrosequencing approach (bTEFAP; pronounced beta-FAP) as described previously (Wolcott et al., 2009; Bailey et al., 2010; Finegold et al., 2010; Gontcharova et al., 2010; Pitta et al., 2010), to evaluate the bacterial populations from infant skin swabs. Briefly, all DNA samples were diluted to 100 ng ml−1 and a 100 ng aliquot of each DNA sample was used for a 50 ml step 1 PCR reaction. The 16S universal Eubacterial primers 530F (59-GTGCCAGCMGCNGCGG) and 1100R (59-GGGTTNCGNTCGTTG) were used for amplifying the 600 bp region of 16S rRNA genes. HotStarTaq Plus Master Mix Kit (Qiagen) was used for PCR. A step 2 PCR was performed for 454-amplicon sequencing under the same condition by using designed special fusion primers with different tag sequences. After secondary PCR, all amplicon products from different samples were mixed in equal volumes, and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, Beverly, MA). In preparation for FLX sequencing (Roche, Nutley, NJ), the DNA fragment sizes and concentrations were accurately measured using DNA chips under a Bio-Rad Experion Automated Electrophoresis Station (Bio-Rad Laboratories, Hercules, CA) and a TBS-380 Fluorometer (Turner Biosystems, Sunnyvale, CA). A 9.6 E+06 sample of double-stranded DNA molecules per ml with an average size of 625 bp were combined with 9.6 million DNA capture beads, and then amplified by emulsion PCR. After bead recovery and bead enrichment, the bead-attached DNA fragments were denatured with NaOH, and sequencing primers were annealed. A two-region 454-sequencing run was performed on a 70675 GS PicoTiterPlate using the Genome Sequencer FLX System (Roche). Twenty tags were used on region of the PicoTiterPlate. All FLX procedures were performed using Genome Sequencer FLX System manufacturer's instructions (Roche). All molecular procedures and bioinformatics were performed at Research and Testing Laboratory, Lubbock, TX.
bTEFAP sequence processing pipeline
Methods of analysis for pyrosequencing data used herein have been described previously (Wolcott et al., 2009; Bailey et al., 2010; Finegold et al., 2010; Gontcharova et al., 2010; Pitta et al., 2010). Briefly, sequences, which were <250 base pairs after phred20-based quality trimming were not considered. Sequences were analyzed by a script optimized for high-throughput data to identify potential chimeras in the sequence files and all definite chimeras were depleted as described previously (Gontcharova et al., 2010). Bacterial species were identified within the resulting FASTA files for each sample using BLASTn comparison and alignments to curated ribosomal databases derived from and with taxonomic designations, based upon the National Center for Biotechnology Information. Comparison of hit IDs to the NCBI taxonomy database was used to create taxonomic compilations.
Diversity and richness analyses were performed as described previously (Dowd et al., 2008) from raw reads of phred20 quality that were trimmed to 350 bp. Recently it has been suggested that OTU predictions derived from 454 pyrosequencing using methods such as MOTHUR (Schloss et al., 2009) provides overestimations of diversity (Quince et al., 2009). Particularly at the 1% divergence, estimators, such as rarefaction, ACE, and Chao1, may create overestimation of OTU's when using pyrosequencing data. Here, in order to reduce such overestimation errors, we used the 3 and 5% divergence to classify OTUs for species and genus levels, correspondingly, and in addition each sample was subject to the same analyses.
Statistics
Microbial diversity analyses (Finkel and Kolter, 1999; Hong et al., 2006; Sogin et al., 2006; Ptacnik et al., 2008) were performed by clustering sequence tags into groups of defined sequence variation as previously described (Schloss and Handelsman, 2005). Diversity measurements (rarefaction, Shannon index, ACE, and Chao1) were analyzed using a blocked analysis of variance. All analyses were conducted using R programming language (R Development Core Team, 2009).
Distance-based redundancy analysis (Legendre and Anderson, 1999), as implemented in the R package “vegan” (Oksanen et al., 2010), was used to test for differences in microbial communities among subjects belonging to one of three age groupings. This method uses a permutation test for the effects of constraints (in this case, age group) on a community ordination. This ordination is based on inter-sample (i.e., inter-subject) distances. Distances among subjects with respect to microbial communities were examined using abundances that were scaled so that each species was given equal weight. Bray–Curtis distances were used to characterize differences among subjects.
Principal component analysis
To assess the separability of the samples, PCA was implemented. It is widely used for dimensionality reduction to aid in visualization of high-dimensional data. These data are visualized by plotting the samples on primary eigenvectors defined by the principal components. Samples more similar to each other should appear closer together according to the respective axis reflecting the variation among all samples. This technique is useful in displaying clusters existing within data. Two sets of variable PCA analyses were performed on the data. In the first approach, we used methods as described previously (Suchodolski et al., 2009; Pitta et al., 2010) to formulate the environment and NEXUS tree files for a distance matrix, based on the UNIFRAC (Lozupone et al., 2006) formula. This sequence distance matrix-based PCA was evaluated using R and visualized using Xlstat (Addinsoft USA, New York, NY). In the second approach, the variables (features) were the relative bacterial composition (percentage of each taxonomic unit) at a given taxonomic level with PCA and visualization performed using Xlstat.
Clustering
To analyze the relationships and clustering between groups of samples, dual hierarchal dendrogram was calculated, based on bacterial composition information at the class and genus taxonomic levels. The analysis was performed using the NCSS using weighted pair clustering, based upon Manhattan distance measurements.
Acknowledgments
We thank the volunteers who participated in this study as well as Diana Friscia, Catherine Mack, and Mark Green for assistance with sample collection. We thank Prof. Richard Gallo for his thoughtful review and comments. We also thank Katharine Martin and Dr Nikiforos Kollias for thier comments on the the study and manuscript.
Glossary
NCBI
National Center for Biotechnology Information
OTU
operational taxonomic unit
PCA
principal component analysis
This study was sponsored in full by Johnson & Johnson Consumer Companies.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
Supplementary Information
References
- Agache P, Blanc D, Barrand C, et al. Sebum levels during the first year of life. Br J Dermatol. 1980;103:643–649. doi: 10.1111/j.1365-2133.1980.tb01686.x. [DOI] [PubMed] [Google Scholar]
- Bailey MT, Dowd SE, Parry NM, et al. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacterrodentium. Infect Immun. 2010;78:1509–1519. doi: 10.1128/IAI.00862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn CS, Thorsen P, Jensen JS, et al. Maternal vaginal microflora during pregnancy and the risk of asthma hospitalization and use of antiasthma medication in early childhood. J Allergy Clin Immunol. 2002;110:72–77. doi: 10.1067/mai.2002.125833. [DOI] [PubMed] [Google Scholar]
- Callard RE, Harper JI. The skin barrier, atopic dermatitis and allergy: a role for Langerhans cells. Trends Immunol. 2007;28:294–298. doi: 10.1016/j.it.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Chiou YB, Blume-Peytavi U. Stratum corneum maturation. A review of neonatal skin function. Skin Pharmacol Physiol. 2004;17:57–66. doi: 10.1159/000076015. [DOI] [PubMed] [Google Scholar]
- Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence. Br J Dermatol. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen AL, Yamasaki K, Sanchez KM, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Callaway TR, Wolcott RD, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Dowd SE, Gontcharova V, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, et al. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontcharova V, Youn E, Sun Y, et al. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol J. 2010;4:8–19. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Yamashita A, Hirakawa H, et al. Complete genome sequence of Finegoldia magna, an anaerobic opportunistic pathogen. DNA Res. 2008;15:39–47. doi: 10.1093/dnares/dsm030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hello M, Prey S, Leaute-Labreze C, et al. Infantile acne: a retrospective study of 16 cases. Pediatr Dermatol. 2008;25:434–438. doi: 10.1111/j.1525-1470.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- Henderson CA, Taylor J, Cunliffe WJ. Sebum excretion rates in mothers and neonates. Br J Dermatol. 2000;142:110–111. doi: 10.1046/j.1365-2133.2000.03249.x. [DOI] [PubMed] [Google Scholar]
- Hirasawa Y, Takai T, Nakamura T, et al. Staphylococcusaureus extracellular protease causes epidermal barrier dysfunction. J Invest Dermatol. 2010;130:614–617. doi: 10.1038/jid.2009.257. [DOI] [PubMed] [Google Scholar]
- Holt PG, Jones CA. The development of the immune system during pregnancy and early life. Allergy. 2000;55:688–697. doi: 10.1034/j.1398-9995.2000.00118.x. [DOI] [PubMed] [Google Scholar]
- Hong SH, Bunge J, Jeon SO, et al. Predicting microbial species richness. Proc Natl Acad Sci USA. 2006;103:117–122. doi: 10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Di Nardo A, Nakatsuji T, et al. Commensal bacteria regulate toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Anderson MJ. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr. 1999;69:1–24. [Google Scholar]
- Leyden JJ.1982Bacteriology of infant skin Neonatal Skin: Structure and Function(Maibach H, Boisits E, eds),New York: Marcel Dekker; 167–181. [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini AJ, Kaulback K, Chamlin SL. The socioeconomic impact of atopic dermatitis in the United States: a systematic review. Pediatr Dermatol. 2008;25:1–6. doi: 10.1111/j.1525-1470.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- Marchini G, Nelson A, Edner J, et al. Erythema toxicum neonatorum is an innate immune response to commensal microbes penetrated into the skin of the newborn infant. Pediatr Res. 2005;58:613–616. doi: 10.1203/01.pdr.0000176836.27156.32. [DOI] [PubMed] [Google Scholar]
- Nikolovski J, Stamatas GN, Kollias N, et al. Barrier function and water-holding and transport properties of infant stratum corneum are different from adult and continue to develop through the first year of life. J Invest Dermatol. 2008;128:1728–1736. doi: 10.1038/sj.jid.5701239. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. 2010Vegan: community ecology package. R package version 1.17-2Accessed at 〈http://cran.r-project.org/web/packages/vegan/〉
- Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino LC, Tseng CH, Strober BE, et al. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta DW, Pinchak E, Dowd SE, et al. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb Ecol. 2010;59:511–522. doi: 10.1007/s00248-009-9609-6. [DOI] [PubMed] [Google Scholar]
- Ptacnik R, Solimini AG, Andersen T, et al. Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc Natl Acad Sci USA. 2008;105:5134–5138. doi: 10.1073/pnas.0708328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol. 2003;112:S118–S127. doi: 10.1016/j.jaci.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Stamatas GN.2010The structural and functional development of skin during the first year of life: investigations using non-invasive methods Textbook of Aging Skin(Farage MA, Miller KW, Maibach HI, eds),Berlin: Springer-Verlag; 715–724. [Google Scholar]
- Stamatas GN, Nikolovski J, Luedtke MA, et al. Infant skin microstructure assessed in vivo differs from adult skin in organization and at the cellular level. Pediatr Dermatol. 2010;27:125–131. doi: 10.1111/j.1525-1470.2009.00973.x. [DOI] [PubMed] [Google Scholar]
- Suchodolski JS, Dowd SE, Westermarck E, et al. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol. 2009;9:210. doi: 10.1186/1471-2180-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher MO, Chatterjee R, Munson KA, et al. Changes in diapered and nondiapered infant skin over the first month of life. Pediatr Dermatol. 2000;17:45–51. doi: 10.1046/j.1525-1470.2000.01711.x. [DOI] [PubMed] [Google Scholar]
- Wolcott RD, Gontcharova V, Sun Y, et al. Bacterial diversity in surgical site infections: not just aerobic cocci any more. J Wound Care. 2009;18:317–323. doi: 10.12968/jowc.2009.18.8.43630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information