Interleukin-1exerts distinct actions on different cell types of the brain in vitro (original) (raw)
Abstract
Interleukin-1 (IL-1) is a critical neuroinflammatory mediator in the central nervous system (CNS). In this study, we investigated the effect of IL-1 on inducing inflammation-related gene expression in three astrocyte, two microglial, and one brain endothelial cell line. Interleukin-1 beta (IL-1β) is found to be produced by the two microglial cell lines constitutively, but these cells do not respond to IL-1β stimulation. The three astrocyte cell lines responded to IL-1β stimulation by expressing MCP-1, CXCL-1, and VCAM-1, but different subtypes of astrocytes exhibited different expression profiles after IL-1β stimulation. The brain endothelial cells showed strongest response to IL-1β by producing MCP-1, CXCL-1, VCAM-1, ICAM-1, IL-6, and COX-2 mRNA. The induction of endothelial COX-2 mRNA is shown to be mediated by p38 MAPK pathway, whereas the induction of other genes is mediated by the NF-κB pathway. These results demonstrate that IL-1 exerts distinct cell type-specific action in CNS cells and suggest that IL-1-mediated neuroinflammation is the result of the summation of multiple responses from different cell types in the CNS to IL-1.
Keywords: astrocyte, microglia, endothelial cells, signal transduction pathways, gene expression
Introduction
Interleukin-1 (IL-1) is involved in multiple functions in the central nervous system (CNS). In neuroimmune communication, IL-1 induces fever, increased slow wave sleep, increased hypothalamic-pituitary-adrenal (HPA) activation, and sickness behaviors.1 During brain injury, IL-1 modulates neural inflammation to exacerbate2 or reduce secondary tissue damage, depending on specific conditions.3 In Alzheimer’s disease, IL-1 was implicated as a critical inflammatory cytokine that could contribute to the progression of this disease.4 Current literature suggests that IL-1 is involved in numerous physiological and pathological processes in the CNS.
The diverse activities of IL-1 in the CNS have been associated with the different cell types that IL-1 might act on. For example, IL-1-induced sickness behavior has been associated with IL-1’s action on brain endothelial cells;5 IL-1-mediated changes in brain tissue injury and recovery have been associated with IL-1’s action on astrocytes;6 and IL-1-induced changes in brain cytokine levels have been associated with IL-1’s action on microglia.7 Whether IL-1 induces distinct effects from these different CNS cell types has not been clarified.
In vivo, when IL-1 is injected directly into the cerebral ventricles, many different cell types are activated, including neurons, astrocytes, microglia, and endothelial cells.8 Activation of these cell types causes discrete patterns of gene expression in different cell types: eg, c-fos in paraventricular nucleus (PVN) neurons, glial fibrillary acidic protein (GFAP) in astrocytes, IL-1 in microglia, and COX-2 in endothelial cells. It is not clear, however, which cells are activated by the direct action of IL-1 on the receptors of IL-1 on the cell membrane and which cells are activated, not by IL-1 directly, but indirectly by other signaling molecules induced by IL-1 from a different cell type.
Previous studies have attempted to isolate the direct action of IL-1 on a specific CNS cell type by examining the effects of IL-1, in vitro, in primary cultures.9–11 These primary cultures were typically generated from neonatal brains; the results generated from these cultures may not reflect IL-1’s effect on adult CNS cells.12 More importantly, cells in the primary culture are generally not pure, rendering complete exclusion of an observed effect from contaminating cell types difficult.
In this study, we tested patterns of selected gene expression in several CNS cell lines representing different cell types of the CNS. In addition, we examined the signal transduction pathways mediating IL-1-induced effects in these cell types.
Materials and methods
Cell culture
The murine astrocyte cell lines (C8-D1A, C8-D30, and C8-S), murine brain endothelial cell line (bEnd.3), and murine peripheral endothelial cell line (SVEC4-10) were cultured in Dulbecco’s modified eagle medium (DMEM) (supplemented with 10% fetal bovine serum [FBS], 100 units/mL penicillin G, 100 μg/mL streptomycin). The murine microglial cell lines (EOC2 and EOC20) were cultured in the growth medium (DMEM, 70%; LADMAC [mouse bone marrow, producing colony stimulating factor-1]-conditioned media, 20%; and FBS, 10%). Cultures were maintained at 37°C with 95% humidity and 5% CO2. Cells were used at approximately 90% confluency.
Experimental manipulation
Cells were seeded in 6-well plates with 2 × 106 cells/well. Recombinant mouse interleukin-1 beta (IL-1β) (1 ng/mL, R&D Systems®, Minneapolis, MN) was used to stimulate the astrocytes and brain endothelial cells for 2, 4, or 16 hours. Culture medium (MC) was used as control. In signal transduction blockade experiments, SP600125 (JNK inhibitor II, 40 μM, Calbiochem®, EMD Chemicals Inc, Gibbstown, NJ), PD98059 (MEK1 inhibitor, 20 μM, Calbiochem®), SB203580 (p38 MAPK inhibitor, 20 μM, Calbiochem®), or BAY11-7082 (NF-κB pathway inhibitor 5 μM, Biomol®, Enzo Life Sciences, Plymouth Meeting, PA) was first dissolved in dimethyl sulfoxide (DMSO) and then added to the culture medium 1 hour before IL-1β stimulation. Cells were harvested 2 hours after IL-1β stimulation.
RNA isolation and cDNA preparation
Cells were harvested and mRNA was isolated (Qiagen RNeasy® Mini Kit, Valencia, CA) from both IL-1β-treated cells and control cells. The RNase-Free DNase Set (Qiagen, Valencia, CA) was used to digest DNA during RNA purification. To generate cDNA, 1 μg of RNA was mixed with 300 ng of random primer (Invitrogen™, Carlsbad, CA) and 1 μL of 10 mM dNTPs for each sample. The mixture was incubated at 70°C for 5 minutes and then chilled on ice. Then, the mixture was incubated with 2 μL reverse transcription buffer and 1μL M-MuLV reverse transcriptase (New England Biolabs ®, Ipswich, MA) at 37°C for 1 hour. Finally, the resulting cDNAs were heated to 95°C for 5 minutes to inactivate the enzyme and were stored at −20°C until use.
Real-time polymerase chain reaction
Polymerase chain reaction (PCR) analyses were performed to detect the expression of iNOS, Cox-2, TNF-α, IL-6, IL-1β, IL-1R1, MCP-1, CXCL-1, VCAM-1, ICAM-1, and G3PDH (as an internal control). PCR primers used in this study are listed in Table 1 (TNF-α primers were obtained from Applied Biosystem TaqMan® gene expression assay, Carlsbad, CA). The expression of IL-1 receptor, type I (IL-1R1) gene was detected by SYBR® green PCR system (Applied Biosystem, Carlsbad, CA), while the expression of all other genes was detected by the iCycler iQ real-time system (Bio-Rad, Hercules, CA). Real-time PCR results were expressed as relative levels of RNA over that of the G3PDH. The detection limit of real-time PCR was set by the threshold cycle number of a PCR, which detects a single copy of a given sequence. A sample was deemed undetectable if the threshold PCR cycle number was over the detection limit.
Table 1.
PCR primers for real-time PCR measurement of target genes
| Name | Forward | Reverse | Probe |
|---|---|---|---|
| IL-1R1 | 5′-GTCCCTGACTTCAAGAATTACC | 5′-CTCCAAGACCTCAGGCAACAG | NA |
| CXCL-1 | 5′-CTGCACCCAAACCGAAGTC | 5′-AGCTTCAGGGTCAAGGCAAG | 5′-CACTCAAGAATGGTCGCGAGGC |
| MCP-1 | 5′-TTGGCTCAGCCAGATGCA | 5′-CCTACTCATTGGGATCATCTTGC | 5′-AACGCCCCACTCACCTGCTGCTAC |
| ICAM-1 | 5′-CACAGAACTGGATCTCAG | 5′-GAGCTTTGGGATGGTAGCTG | 5′-TGGCATTGTTCTCTAATGTCTCCGAGGC |
| VCAM-1 | 5′-CGATTGCTCAAATCGGTGAC | 5′-GTCTATCTGGGTTCTCCATG | 5′-TCCATGGCCCTCACTTGCAGCA |
| COX-2 | 5′-CAGCCAGGCAGCAAATCC | 5′-TCAAATCCTGTGCTCATACATT | 5′-TGCTGTTCCAATCCATGTCAAAAC |
| IL-1β | 5′-GGCCTCAAAGGAAAGAATCTATACC | 5′-GTATTGCTTGGGATCCACACTCT | 5′-ATGAAAGACGGCACACCCACCCTG |
| IL-6 | 5′-TATGAAGTTCCTCTCTGCAAGAGA | 5′-TAGGGAAGGCCGTGGTT | 5′-CCAGCATCAGTCCCAAGAAGGCAA |
| iNOS | 5′-CAGCTGGGCTGTACAACCTT | 5′-TGAATGTGATGTTTGCTTCGG | 5′-CGGGCAGCCTGTGAGACCTTTGAT |
| G3PDH | 5′-GGCAAATTCAACGGCACAGT | 5′-GGGTCTCGCTCCTGGAAGAT | 5′-AAGGCCGAGAATGGGAAGCTTGTCATC |
| Markers | IL-1β | |||
|---|---|---|---|---|
| Astrocytes | Brain endo. | |||
| C8-D1A | C8-S | C8-D30 | bEnd.3 | |
| CXCL-1 | + + | + + | + + + | + + + + |
| MCP-1 | + + | + | + + + | + + + |
| ICAM-1 | − | − | − | + + + |
| VCAM-1 | + + | − | + + | + + + |
| COX-2 | + | − | − | + + + |
| IL-6 | − | − | − | + + + |
| IL-1β/BAY11-7082 | ||||
| CXCL-1 | + | − | + | + |
| MCP-1 | + | − | + | + |
| ICAM-1 | − | − | − | − |
| VCAM-1 | + | − | − | + |
| COX-2 | − | − | − | + + + |
| IL-6 | − | − | − | − |
| IL-1β/SB203580 | ||||
| CXCL-1 | + + | + + | + + + | + + + + |
| MCP-1 | + + | + | + + + | + + + |
| ICAM-1 | − | − | − | + + + |
| VCAM-1 | + + | − | + + | + + + |
| COX-2 | + | − | − | − |
| IL-6 | − | − | − | + + + |
ELISA
ELISA kits for MCP-1, VCAM-1, COX-2 (R&D Systems®, Minneapolis, MN, catalog number: MEJ00, MVC00, and KCB4198, respectively), and IL-6 (BD Biosciences, San Diego, CA, catalog number 555240) were used. Protein levels of MCP-1, VCAM-1 and IL-6 were measured in cell supernatant according to the manufacturer’s instructions. A measure of 2.0 × 106 C8-S, C8-D1A, C8-D30, or bEnd.3 cells were stimulated by 1 ng/mL of IL-1β for 4 hours. Culture medium was used as control. Cell supernatant samples were centrifuged to remove particulates and were aliquoted and stored at −20°C before use. Protein levels were calculated using standard curves generated from reference proteins provided in the ELISA kits.
COX-2, an intracellular protein, was measured by a cell-based ELISA. A total of 20,000 C8-S, C8-D1A, C8-D30, or bEnd.3 cells were seeded into each well of the 96-well microplate overnight. Cells were treated with 1 ng/mL of IL-1β for 4 hours or with control medium. Then, the cells were fixed in 4% formaldehyde, quenched with a wash buffer containing 0.6% H2O2 and blocked with a blocking buffer containing 10% FBS (these buffers were provided in the kit). Cellular COX-2 and GAPDH were detected by a mouse anti-COX-2 and a rabbit anti-GAPDH. The anti-COX-2 antibody was labeled by a secondary antibody conjugated with horse radish peroxidase (HRP) and visualized by a fluorescence substrate (F1, detectable at 600 nm) and the anti-GAPDH antibody was labeled by a secondary antibody conjugated with alkaline phosphatase (AP) and visualized by a different fluorescence substrate (F2, detectable at 450 nm). Control wells, with no primary antibody added, were included to establish background fluorescence. Background fluorescence was subtracted from the fluorescence in sample wells. COX-2 protein levels were calculated by dividing the COX-2-detecting fluorescence at 600 nm in each well by the GAPDH-detecting fluorescence at 450 nm in each well to obtain relative fluorescence unit (RFU).
Statistical analysis
All the data are presented as mean ± standard error of the mean (SEM). Variations in mRNA expression levels were evaluated by one-way analysis of variance (ANOVA) followed by post-hoc analysis (Tukey test). All tests were considered significant if P < 0.05.
Results
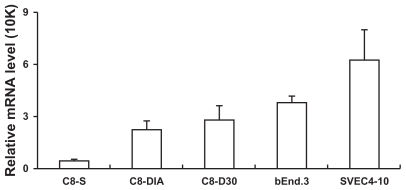
Figure 1 shows that IL-1R1 was detectable in all three astrocyte cell lines and the brain endothelial cell line; it was not detected in the two microglial cell lines (data not shown). We also detected IL-1R1 in the peripheral endothelial cell line, SVEC4-10, which is known to express IL-1R1, to verify our analysis (Figure 1). In the astrocyte lines, IL-1β induced increased expression of MCP-1, CXCL-1, and VCAM-1 (Figure 2). Increased expression of MCP-1 was found in all three astrocyte lines, with the highest induction levels occurring at 2 hours after the IL-1β stimulation and the level of induced MCP-1 declining precipitously at 4 and 16 hours post-IL-1β stimulation. The strongest induction occurred in the C8-D30 line and the weakest in the C8-S line. Similarly, CXCL-1 was induced by IL-1β in all three astrocyte lines; the strongest induction occurred again in the C8-D30 line, but the weakest induction was found in C8-D1A cells. VCAM-1 was not induced in C8-S cells, but in both C8-D1A and C8-D30 cells, with the stronger induction in C8-D1A cells. IL-1β did not induce the expression of ICAM-1, COX-2, iNOS, TNFα, IL-1β, and IL-6 in these astrocyte lines (data not shown).
Figure 1.
IL-1R1 mRNA levels in different cell lines.
Notes: Means and standard errors of the mean (SEM) derived from three separate experiments are presented. The relative IL-1R1 mRNA levels have been normalized to the G3PDH levels in the same sample.
Figure 2.
Time course of gene expression after IL-1β stimulation in the three astrocyte cell lines.
Notes: The relative mRNA levels of MCP-1, CXCL-1, and VCAM-1 (normalized to the G3PDH levels in the same sample) are presented. *Indicates significant increase of an mRNA after IL-1β stimulation over the control mRNA values (P < 0.05).
Abbreviation: MC, culture medium.
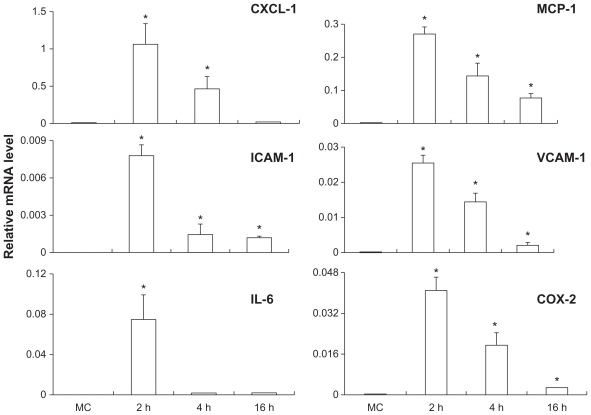
IL-1β-induced gene expression in the brain endothelial cell line is shown in Figure 3. The endothelial genes that were induced by IL-1β include MCP-1, ICAM-1, CXCL-1, VCAM-1, IL-6, and COX-2, whereas the expression of iNOS, TNFα, and IL-1β was not induced by IL-1β (data not shown). Again, peak induction levels were seen at 2 hours post-IL-1β stimulation. Whereas MCP-1, ICAM-1, CXCL-1, VCAM-1, and COX-2 mRNA expression gradually declined thereafter, the IL-6 expression abruptly returned to the basal level at 4 hours post-IL-1β stimulation.
Figure 3.
Time course of gene expression after IL-1β stimulation in the brain endothelial cell line bEnd.3.
Notes: The relative mRNA levels of MCP-1, CXCL-1, ICAM-1, VCAM-1, IL-6, and COX-2 are presented. Means and standard errors of the mean (SEM) were generated from three separate experiments. *Indicates significant increase of an mRNA level after IL-1β stimulation as compared with the level after vehicle treatment (P < 0.05).
Abbreviation: MC, culture medium.
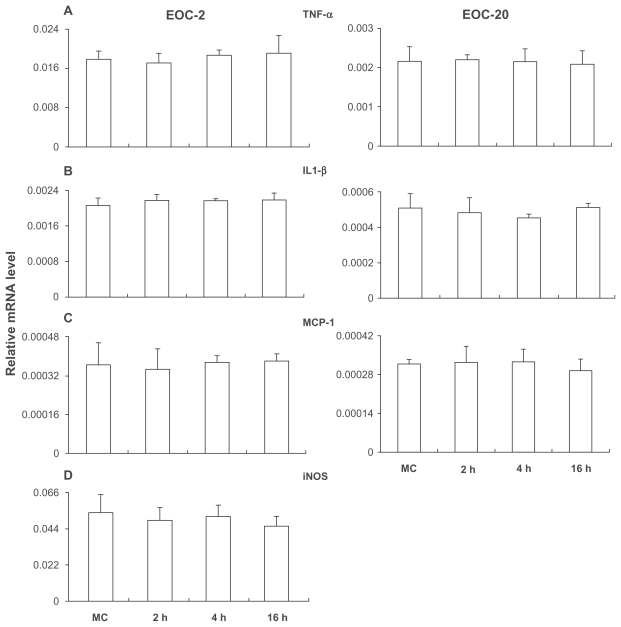
Previous studies have shown that the microglial cell is a major producer of inflammatory mediators. In addition, cells with very low expression of IL-1R1 were known to respond to IL-1β stimulation.13 Therefore, we examined IL-1β-induced gene expression in the two microglial cell lines, although IL-1R1 mRNA was not detected in either line. Figure 4 shows that TNFα, IL-1β, MCP-1, and iNOS were constitutively expressed in the EOC2 cells (Figures 4A–D). After IL-1β stimulation, the expression of these genes was not changed at 2, 4, and 16 hours post stimulation. Similarly, in EOC20 cells, TNFα, IL-1β, and MCP-1 were constitutively expressed and their expression was not altered after the IL-1β stimulation (Figures 4A–C).
Figure 4.
Gene expression in EO C2 (A–D) and EO C20 (A–C).
Notes: With vehicle treatment, TNF-α, IL-1β, MCP-1, and iNOS were expressed in EO C2 and TNF-α, IL-1β, and MCP-1 were expressed in EO C20. With IL-1β treatment, these expression patterns were not changed.
Abbreviation: MC, culture medium.
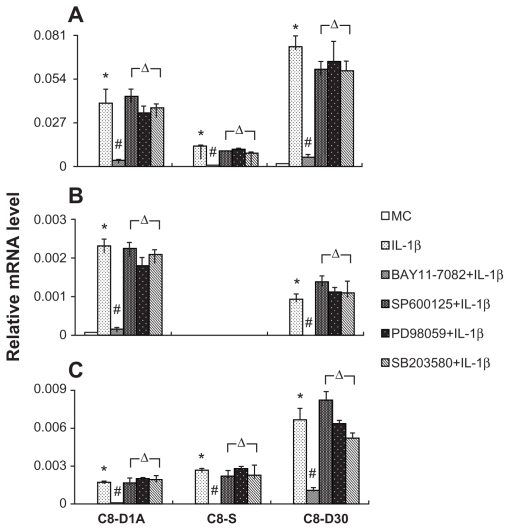
Next, we used specific inhibitors of signal transduction to determine the signaling pathways that mediate IL-1β-induced gene expression. NF-κB pathway inhibitor (BAY11-7082), JNK pathway inhibitor (JNK inhibitor II), ERK pathway inhibitor (PD98059), or p38 MAPK inhibitor (SB203580) was used. Because the highest gene expression occurred at 2 hours after IL-1β stimulation, we pretreated cells with an inhibitor for 1 hour and stimulated cells with IL-1β for 2 hours. Figure 5 shows that in astrocyte lines, the induced expression of MCP-1, VCAM-1, and CXCL-1 can all be completely inhibited by the NF-κB inhibitor, BAY11-7082. Other inhibitors did not alter IL-1β-induced gene expression in these astrocyte lines. Figure 6 shows that in the brain endothelial cell line, pretreatment of BAY11-7082 abrogated the induction of MCP-1, CXCL-1, VCAM-1, ICAM-1, and IL-6. Other inhibitors did not alter IL-1β-induced expression of these genes (data not shown). In contrast, IL-1β-induced COX-2 expression was not inhibited by BAY-11-7082, but by SB203580.
Figure 5.
NF-κB pathway inhibitor (BAY11-7082) inhibited the IL-1β-induced expression of MCP-1, CXCL-1, and VCAM-1 in astrocytes.
Notes: BAY11-7082 (5 μM) was added 1 hour before the cells received the IL-1β stimulation. *Indicates significant increase of an mRNA level after IL-1β stimulation compared with the level after vehicle treatment (P < 0.05). #Indicates significant decrease of an mRNA after BAY11-7082 + IL-1β treatment as compared with the level after IL-1β treatment alone (P < 0.05). ΔIndicates inhibitors for MEK1 (PD98059), JNK (SP600125), and p38 MAPK (SB 203580) did not alter IL-1β-induced increase of an mRNA level.
Abbreviation: MC, culture medium.
Figure 6.
NF-κB pathway inhibitor (BAY11-7082) inhibited the IL-1β-induced expression of MCP-1, CXCL-1, VCAM-1, ICAM-1, and IL-6 in bEnd.3.
Notes: p38 MAPK inhibitor (SB 203580) inhibited the IL-1β-induced expression of COX-2 in bEnd.3. BAY11-7082 (5 μM) or SB 203580 (20 μM) was added 1 hour before the cells received IL-1β stimulation. *Indicates significant increase of an mRNA level after IL-1β stimulation compared with the level after vehicle treatment (P < 0.05). #Indicates significant decrease of an mRNA expression level after BAY11-7082 or SB 203580 + IL-1β treatment compared with the level after IL-1β treatment alone (P < 0.05).
Abbreviation: MC, culture medium.
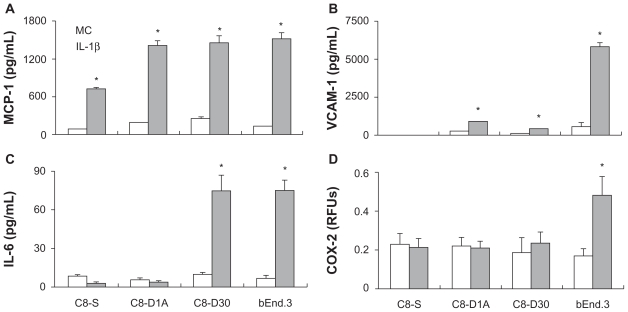
To examine whether the distinct induction patterns observed at the mRNA level are mirrored at the protein level, we measured IL-1β-induced MCP-1, VCAM-1, IL-6, and COX-2 protein levels in the three astrocyte cell lines and brain endothelial cell line (Figure 7). MCP-1 protein was induced in all four cell lines, with the lowest induction in the C8-S cells (Figure 7A). VCAM-1 was induced in C8-D1A, C8-D30, and brain endothelial bEnD.3 cells, but not in C8-S cells (Figure 7B). The strongest induction occurred in the bEnD.3 cells withVCAM-1 in bEnD.3 more than seven times higher than those in the other cells. Whereas IL-6 was not induced in C8-S and C8-D1A cells, it was dramatically induced in both C8-D30 and bEnD.3 cells (Figure 7C). COX-2 was not induced in all three astrocytes, but was induced in the brain endothelial cells (Figure 7D).
Figure 7.
MCP-1 (A), VCAM-1 (B), IL-6 (C), and COX-2 (D) protein levels in three astrocyte cell lines and in the brain endothelial cell line after cells were stimulated for 4 hours by culture medium (MC) or by IL-1β.
Notes: *Indicates significant increase of a protein level after IL-1β stimulation compared with the level after vehicle treatment (P < 0.05).
Discussion
IL-1R1 is expressed at very different levels in the tested brain endothelial, astrocyte, and microglial cell lines. The highest expression of IL-1R1 is found in brain endothelial cells (Figure 1). This is consistent with previous studies that showed brain endothelial cell as the major cell type that carries CNS IL-1R1.14–16 In addition, endothelial IL-1R1 has been shown to mediate fever, brain endothelial COX-2 expression, and CNS leukocyte infiltration induced by intracerebral IL-1β injection.5 In addition, our real-time PCR assay also detected IL-1R1 in a peripheral endothelial cell line, SVEC4-10, confirming the ability of the assay to detect known IL-1R1 expression.17
Intermediate levels of IL-1R1 mRNA were detected in the three astrocyte lines and IL-1R1 mRNA was not detected in the two microglial cell lines. In vivo microglial and astrocyte expression of IL-1R1 was not found in many studies in the normal adult brain by in situ hybridization or by immunohistochemistry.14–16 After brain injury, however, IL-1R1 expression in astrocytes and microglia near the site of injury has been reported.18 In vitro, IL-1 can directly alter the phenotype of astrocytes19 and induce the expression of proMMP9,20 S100B,9 and EP3 receptors on astrocytes;21 direct action of IL-1 on primary (postnatal) mouse microglial culture has been shown to induce the expression of IL- 17.7 Thus, IL-1R1 may be expressed in activated astrocytes and microglia or it may be expressed in these cell types during development. The astrocyte and microglial cell lines used in the present study may represent only part of the in vivo phenotype spectrum of these cell types.
The three astrocyte cell lines used in the present study represent the type 1 (C8-D1A), type 2 (C8-S), and type 3 (C8-D30) astrocytes. In culture, C8-D1A and C8-S have small somata while C8-D30 has a large somata and no processes. C8-D1A contains multiple short processes while C8-S has only two processes with one of them being very thin and long. These cells may resemble different subtypes of astrocytes in vivo. Thus far, C8-D1A has been used most frequently in the analysis of astrocyte functions in vitro.22,23 It is interesting to note that C8-S, which has the morphology of structural fibrous astrocytes in vivo, expresses the least amount of IL-1R1 (IL-1R1/G3PDH = 4.1 × 10−5 ± 2 × 10−5), whereas C8-D30, which has morphology resembling a phagocytic or reactive astrocyte in tissue,24 expresses the highest amount of IL-1R1 (IL-1R1/G3PDH = 2.8 × 10−4 ± 8.4 × 10−5).
IL-1R1 is not detected in the two microglial cell lines. These cells may resemble themicroglia in vivo that have not been activated to the state of IL-1R1 expression.
In the three astrocytes, IL-1β induced significant expression of MCP-1, a monocyte chemokine, and CXCL-1, a neutrophil chemokine, with the strongest induction seen in the C8-D30 cells (MCP-1/G3PDH = 0.069 ± 0.006; CXCL-1/G3PDH = 0.046 ± 0.004). These results suggest that the activated astrocytes may contribute to chemotaxis when they are stimulated by IL-1β. IL-1β also induced the expression of a cell adhesion molecule, VCAM-1, in C8-D30 and C8-D1A cells. Interestingly, the highest induction of VCAM is seen in the C8-D1A cells (VCAM-1/G3PDH = 0.0018 ± 0.0002), not C8-D30 cells (VCAM-1/G3PDH = 9.8 × 10−4 ± 6 × 10−4). It is possible, the type 1 astrocytes (C8-D1A) are more associated with the brain blood vessels in vivo than the type 3 astrocytes (C8-D30) and, therefore, are more involved in vascular leukocyte infiltration that requires the presence of cell adhesion molecules at the blood brain barrier.25 It is striking that no VCAM-1 expression was induced in the C8-S cells. This is consistent with the notion that these type 2 astrocytes may be similar to the fibrous astrocytes in vivo, which are mainly present in the white matter as structural glial cells.26
The broadest and the strongest induction of gene expression by IL-1β in the present study was found in the endothelial cell line. IL-1β induced the expression of both chemokines (MCP-1 and CXCL-1), both adhesion molecules (ICAM-1 and VCAM-1), a cytokine (IL-6), and an inflammatory mediator (COX-2) expression. The IL-1β-induced expression levels of MCP-1, CXCL-1, and VCAM-1 in the endothelial cells were all significantly higher than their counterparts found in any of the three astrocytes. Thus, IL-1β acting directly on brain endothelial cells may be the dominant action during neuroinflammation. This is consistent with the observation that direct injection of IL-1β into the cerebral ventricles induces significant endothelial expression of a large number of inflammation related genes.8 In addition, the induction of COX-2 and IL-6 by IL-1β is only observed in the brain endothelial cells, not in the astrocytes. Thus, astrocytes may contribute to a smaller set of gene expression during IL-1β-induced neuroinflammation.
The two microglial cell lines tested did not respond to IL-1β stimulation, consistent with the fact that IL-1R1 could not be detected on these cells in the present study. Because previous studies showed that IL-1 may exert powerful effects on cells carrying very little IL-1R1,13 we stimulated these microglial cells with IL-1β in case our method was not sensitive enough to detect minute amounts of IL-1R1. IL-1β stimulation did not induce mRNA expression in any of the genes tested. Interestingly, in EOC2 cells, TNFα, IL-1β, MCP-1, and iNOS are constitutively expressed, whereas in EOC20 cells, only TNFα, IL-1β, and MCP-1 are constitutively expressed. Therefore, EOC2 and EOC20 may represent microglial cells that are at different activation states. Previous studies showed that microglia is a major cellular source of the proinflammatory cytokines IL-127–29 and TNFα30,31 in the CNS. The present results confirm this notion, but they also show that microglia may not be the responder cells to IL-1. This is different from some peripheral monocytes in which IL-1 induces its own expression.32 Therefore, unless microglia is activated to the state of an IL-1R1-expressing cell, it is not likely to trigger self-amplifying neuroinflammation in the brain.
IL-1β-induced expression of MCP-1, CXCL-1, and VCAM-1 in the astrocytes is blocked by the inhibitor of NF-κB, not the inhibitors of ERK, JNK, and p38 MAPK, demonstrating that IL-1β induces the expression of these genes exclusively via the NF-κB pathway in astrocytes. In the brain endothelial cells, IL-1β-induced expression of MCP-1, CXCL-1, VCAM-1, ICAM-1, and IL-6 was also blocked by the inhibitor of NF-κB, not the inhibitors of other signal transduction pathways. On the other hand, IL-1β-induced expression of COX-2 is not blocked by the inhibitor of NF-κB, but by the inhibitor of p38 MAPK in the brain endothelial cells. This is surprising because IL-1β-induced COX-2 expression has been shown to be mediated by ERK, JNK, p38, and NF-κB in fibroblasts,33 and by NF-κB in epithelial cells34 in the literature. Therefore signaling pathways that mediate IL-1-induced COX-2 expression and the expression of other inflammatory genes may be segregated as a unique feature of brain endothelial cells. This has important implications in how IL-1-induced neuroinflammation and neural signaling may be separately regulated. Previously, studies have shown that in vivo COX-2 expression in brain endothelial cells is an intermediate step in transmitting peripheral IL-1 signals to the brain,5,35 and brain endothelial NF-κB activation is associated with CNS IL-1-mediated neuroinflammation.8 Interestingly, both CNS and peripheral IL-1 can induce brain endothelial COX-2 expression, but only CNS IL-1 can induce neuroinflammation that is characterized by leukocyte infiltration into the brain.8 It is intriguing to speculate that IL-1-induced neuroinflammation, via endothelial NF-κB, and IL-1-induced neural signaling, via endothelial COX-2, could be separated due to the fact that the different endothelial signal transduction pathways are operative for these two different functions.
The distinct cell type-specific induction patterns caused by IL-1β stimulation occurred at both mRNA and protein levels. Results presented in Figure 7 show IL-1β-induced protein expression of MCP-1, VCAM-1, IL-6, and COX-2 in different cell lines largely resembles the patterns found for the expression of mRNAs of these molecules. For example, VCAM-1 mRNA was induced by IL-1β in both C8-D1A and C8-D30 cells, but not in C8-S cells (Figure 5). Similarly, VCAM-1 protein was induced in the C8-D1A and C8-D30 cells, but not in C8-S cells (Figure 7). In addition, COX-2 protein was induced only in the brain endothelial cell line, not in the astrocyte cell lines. This result parallels the results of COX-2 mRNA induction patterns described in Figures 2 and 3. Among the three astrocyte cell lines, C8-S was the least responsive to IL-1β stimulation in terms of induction of MCP-1, VCAM-1, and IL-6 at both mRNA and protein levels. The only disparate induction patterns between mRNA and protein levels were found in IL-6 expression levels in C8-D30 cells. In these cells, IL-1β did not induce increased IL-6 mRNA expression (data not shown), but induced significant IL-6 protein expression (Figure 7). It is known that in certain cell types, IL-1 can induce increased production of IL-6 without increased transcription of IL-6 mRNA via a mechanism of mRNA stabilization.36 It is possible, that in C8-D30 cells, IL-1β induces increased IL-6 protein levels via this mechanism.
In summary, IL-1 acting directly on astrocytes, microglia, and endothelial cells induces cell type-specific responses. Microglial cells tested in the present study constitutively expressed IL-1, but did not respond to IL-1. The astrocyte responses to IL-1 can be differentiated by the subtype of astrocytes tested, with the reactive astrocytes exhibiting the strongest induction of neuroinflammatory genes and the structural astrocytes the weakest induction of these genes. The strongest responders to IL-1 were the brain endothelial cells, which used NF-κB-mediated pathways to express genes related to leukocyte infiltration, but p38 MAPK to express COX-2.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Quan N, Banks WA. Brain-immune communication pathways. Brain Behav Immun. 2007;21(6):727–735. doi: 10.1016/j.bbi.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Relton JK, Rothwell NJ. Interleukin-1 receptor antagonist inhibits ischaemic and excitotoxic neuronal damage in the rat. Brain Res Bull. 1992;29(2):243–246. doi: 10.1016/0361-9230(92)90033-t. [DOI] [PubMed] [Google Scholar]
- 3.Pinteaux E, Trotter P, Simi A. Cell-specific and concentration-dependent actions of interleukin-1 in acute brain inflammation. Cytokine. 2009;45(1):1–7. doi: 10.1016/j.cyto.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Griffin WS, Mrak RE. Interleukin-1 in the genesis and progression of and risk for development of neuronal degeneration in Alzheimer’s disease. J Leukoc Biol. 2002;72(2):233–238. [PMC free article] [PubMed] [Google Scholar]
- 5.Ching S, Zhang H, Belevych N, et al. Endothelial-specific knockdown of interleukin-1 (IL-1) type 1 receptor differentially alters CNS responses to IL-1 depending on its route of administration. J Neurosci. 2007;27(39):10476–10486. doi: 10.1523/JNEUROSCI.3357-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John GR, Lee SC, Song X, Rivieccio M, Brosnan CF. IL-1-regulated responses in astrocytes: relevance to injury and recovery. Glia. 2005;49(2):161–176. doi: 10.1002/glia.20109. [DOI] [PubMed] [Google Scholar]
- 7.Kawanokuchi J, Shimizu K, Nitta A, et al. Production and functions of IL-17 in microglia. J Neuroimmunol. 2008;194(1–2):54–61. doi: 10.1016/j.jneuroim.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Proescholdt MG, Chakravarty S, Foster JA, Foti SB, Briley EM, Herkenham M. Intracerebroventricular but not intravenous interleukin-1beta induces widespread vascular-mediated leukocyte infiltration and immune signal mRNA expression followed by brain-wide glial activation. Neuroscience. 2002;112(3):731–749. doi: 10.1016/s0306-4522(02)00048-9. [DOI] [PubMed] [Google Scholar]
- 9.De Souza DF, Leite MC, Quincozes-Santos A, et al. S100B secretion is stimulated by IL-1beta in glial cultures and hippocampal slices of rats: Likely involvement of MAPK pathway. J Neuroimmunol. 2009;206(1–2):52–57. doi: 10.1016/j.jneuroim.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Hinkerohe D, Smikalla D, Haghikia A, et al. Effects of cytokines on microglial phenotypes and astroglial coupling in an inflammatory coculture model. Glia. 2005;52(2):85–97. doi: 10.1002/glia.20223. [DOI] [PubMed] [Google Scholar]
- 11.Singh RJ, Mason JC, Lidington EA, et al. Cytokine stimulated vascular cell adhesion molecule-1 (VCAM-1) ectodomain release is regulated by TIMP-3. Cardiovasc Res. 2005;67(1):39–49. doi: 10.1016/j.cardiores.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Wang XF, Yin L, Hu JG, et al. Expression and localization of p80 interleukin- 1 receptor protein in the rat spinal cord. J Mol Neurosci. 2006;29(1):45–53. doi: 10.1385/JMN:29:1:45. [DOI] [PubMed] [Google Scholar]
- 13.Sims JE, Gayle MA, Slack JL, et al. Interleukin 1 signaling occurs exclusively via the type I receptor. Proc Natl Acad Sci U S A. 1993;90(13):6155–6159. doi: 10.1073/pnas.90.13.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ericsson A, Liu C, Hart RP, Sawchenko PE. Type 1 interleukin-1 receptor in the rat brain: distribution, regulation, and relationship to sites of IL-1-induced cellular activation. J Comp Neurol. 1995;361(4):681–698. doi: 10.1002/cne.903610410. [DOI] [PubMed] [Google Scholar]
- 15.Yabuuchi K, Minami M, Katsumata S, Satoh M. Localization of type I interleukin-1 receptor mRNA in the rat brain. Brain Res Mol Brain Res. 1994;27(1):27–36. doi: 10.1016/0169-328x(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 16.Konsman JP, Vigues S, Mackerlova L, Bristow A, Blomqvist A. Rat brain vascular distribution of interleukin-1 type-1 receptor immunoreactivity: relationship to patterns of inducible cyclooxygenase expression by peripheral inflammatory stimuli. J Comp Neurol. 2004;472(1):113–129. doi: 10.1002/cne.20052. [DOI] [PubMed] [Google Scholar]
- 17.Chen Q, Zhang H, Li Q, et al. Three promoters regulate tissue- and cell type-specific expression of murine interleukin-1 receptor type I. J Biol Chem. 2009;284(13):8703–8713. doi: 10.1074/jbc.M808261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman WJ. Cytokines regulate expression of the type 1 interleukin-1 receptor in rat hippocampal neurons and glia. Exp Neurol. 2001;168(1):23–31. doi: 10.1006/exnr.2000.7595. [DOI] [PubMed] [Google Scholar]
- 19.John GR, Chen L, Rivieccio MA, Melendez-Vasquez CV, Hartley A, Brosnan CF. Interleukin-1beta induces a reactive astroglial phenotype via deactivation of the Rho GTPase-Rock axis. J Neurosci. 2004;24(11):2837–2845. doi: 10.1523/JNEUROSCI.4789-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CY, Hsieh HL, Sun CC, Tseng CP, Yang CM. IL-1 beta induces proM-MP- 9 expression via c-Src-dependent PDGFR/PI3K/Akt/p300 cascade in rat brain astrocytes. J Neurochem. 2008;105(4):1499–1512. doi: 10.1111/j.1471-4159.2008.05318.x. [DOI] [PubMed] [Google Scholar]
- 21.Waschbisch A, Fiebich BL, Akundi RS, et al. Interleukin-1 beta-induced expression of the prostaglandin E-receptor subtype EP3 in U373 astrocytoma cells depends on protein kinase C and nuclear factor-kappaB. J Neurochem. 2006;96(3):680–693. doi: 10.1111/j.1471-4159.2005.03599.x. [DOI] [PubMed] [Google Scholar]
- 22.Huppmann S, Romer S, Altmann R, Obladen M, Berns M. 17beta-estradiol attenuates hyperoxia-induced apoptosis in mouse C8-D1A cell line. J Neurosci Res. 2008;86(15):3420–3426. doi: 10.1002/jnr.21777. [DOI] [PubMed] [Google Scholar]
- 23.Reeves MA, Bellinger FP, Berry MJ. The neuroprotective functions of selenoprotein M and its role in cytosolic calcium regulation. Antioxid Redox Signal. 2010;12(7):809–818. doi: 10.1089/ars.2009.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusaka H, Hirano A, Bornstein MB, Moore GR, Raine CS. Transformation of cells of astrocyte lineage into macrophage-like cells in organotypic cultures of mouse spinal cord tissue. J Neurol Sci. 1986;72(1):77–89. doi: 10.1016/0022-510x(86)90037-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhu SN, Dana MR. Expression of cell adhesion molecules on limbal and neovascular endothelium in corneal inflammatory neovascularization. Invest Ophthalmol Vis Sci. 1999;40(7):1427–1434. [PubMed] [Google Scholar]
- 26.Doering LC, Fedoroff S, Devon RM. Fibrous astrocytes and reactive astrocyte-like cells in transplants of cultured astrocyte precursor cells. Brain Res. 1983;282(2):183–189. doi: 10.1016/0165-3806(83)90096-2. [DOI] [PubMed] [Google Scholar]
- 27.Giulian D, Baker TJ, Shih LC, Lachman LB. Interleukin 1 of the central nervous system is produced by ameboid microglia. J Exp Med. 1986;164(2):594–604. doi: 10.1084/jem.164.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Dam AM, Brouns M, Louisse S, Berkenbosch F. Appearance of interleukin-1 in macrophages and in ramified microglia in the brain of endotoxin-treated rats: a pathway for the induction of non-specific symptoms of sickness? Brain Res. 1992;588(2):291–296. doi: 10.1016/0006-8993(92)91588-6. [DOI] [PubMed] [Google Scholar]
- 29.Griffin WS, Sheng JG, Gentleman SM, Graham DI, Mrak RE, Roberts GW. Microglial interleukin-1 alpha expression in human head injury: correlations with neuronal and neuritic beta-amyloid precursor protein expression. Neurosci Lett. 1994;176(2):133–136. doi: 10.1016/0304-3940(94)90066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricciardi-Castagnoli P, Pirami L, Righi M, et al. Cellular sources and effects of tumor necrosis factor-alpha on pituitary cells and in the central nervous system. Ann N Y Acad Sci. 1990;594:156–168. doi: 10.1111/j.1749-6632.1990.tb40476.x. [DOI] [PubMed] [Google Scholar]
- 31.Chao CC, Hu S, Sheng WS, Peterson PK. Tumor necrosis factor-alpha production by human fetal microglial cells: regulation by other cytokines. Dev Neurosci. 1995;17(2):97–105. doi: 10.1159/000111278. [DOI] [PubMed] [Google Scholar]
- 32.Toda Y, Tsukada J, Misago M, Kominato Y, Auron PE, Tanaka Y. Autocrine induction of the human pro-IL-1beta gene promoter by IL-1beta in monocytes. J Immunol. 2002;168(4):1984–1991. doi: 10.4049/jimmunol.168.4.1984. [DOI] [PubMed] [Google Scholar]
- 33.Ogata S, Kubota Y, Yamashiro T, et al. Signaling pathways regulating IL-1alpha-induced COX-2 expression. J Dent Res. 2007;86(2):186–191. doi: 10.1177/154405910708600215. [DOI] [PubMed] [Google Scholar]
- 34.Newton R, Kuitert LM, Bergmann M, Adcock IM, Barnes PJ. Evidence for involvement of NF-kappaB in the transcriptional control of COX-2 gene expression by IL-1beta. Biochem Biophys Res Commun. 1997;237(1):28–32. doi: 10.1006/bbrc.1997.7064. [DOI] [PubMed] [Google Scholar]
- 35.Quan N, He L, Lai W. Endothelial activation is an intermediate step for peripheral lipopolysaccharide induced activation of paraventricular nucleus. Brain Res Bull. 2003;59(6):447–452. doi: 10.1016/s0361-9230(02)00951-6. [DOI] [PubMed] [Google Scholar]
- 36.Witowski J, Jorres A, Coles GA, Williams JD, Topley N. Superinduction of IL-6 synthesis in human peritoneal mesothelial cells is related to the induction and stabilization of IL-6 mRNA. Kidney Int. 1996;50(4):1212–1223. doi: 10.1038/ki.1996.430. [DOI] [PubMed] [Google Scholar]