ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif (original) (raw)
Abstract
Dysregulated bone morphogenetic protein (BMP) signaling in endothelial cells (ECs) and pulmonary arterial smooth muscle cells (PASMCs) are implicated in human genetic disorders. Here, we generated genome-wide maps of Smad1/5 binding sites in ECs and PASMCs. Smad1/5 preferentially bound to the region outside the promoter of known genes, and the binding was associated with target gene upregulation. Cell-selective Smad1/5 binding patterns appear to be determined mostly by cell-specific differences in baseline chromatin accessibility patterns. We identified, for the first time, a Smad1/5 binding motif in mammals, and termed GC-rich Smad binding element (GC-SBE). Several sequences in the identified GC-SBE motif had relatively weak affinity for Smad binding, and were enriched in cell type-specific Smad1/5 binding regions. We also found that both GC-SBE and the canonical SBE affect binding affinity for the Smad complex. Furthermore, we characterized EC-specific Smad1/5 target genes and found that several Notch signaling pathway-related genes were induced by BMP in ECs. Among them, a Notch ligand, JAG1 was regulated directly by Smad1/5, transactivating Notch signaling in the neighboring cells. These results provide insights into the molecular mechanism of BMP signaling and the pathogenesis of vascular lesions of certain genetic disorders, including hereditary hemorrhagic telangiectasia.
INTRODUCTION
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-β (TGF-β) family, which regulate a variety of cellular processes including differentiation, proliferation, migration and cell death in a cell type-specific and context-dependent manner (1). Perturbations of BMP signaling pathways have been implicated in a diverse set of developmental disorders, tumorigenesis and diseases including ectopic ossification and cardiovascular diseases. Mutations in ENG, ACVRL1 or SMAD4 genes have been shown to cause hereditary hemorrhagic telangiectasia (HHT) (2–4), which is a multisystemic vascular disorder characterized by epistaxis, telangiectases and arteriovenous malformation (AVM). The ACVRL1 gene encodes an endothelial-specific type I receptor for TGF-β members, ALK-1, whose signals are transmitted through BMP-specific receptor-regulated Smads (BR-Smads; Smad1/5/8) (5). Recent work has indicated that haploinsufficiency of ALK-1 causes HHT (6). The ENG gene encodes Endoglin, which is an endothelial expressed co-receptor and modulates ALK-1 signaling (7). The SMAD4 gene encodes a common mediator Smad (co-Smad), which makes a heterotrimeric complex with BR-Smads and regulate transcription of specific target genes (8). Therefore, dysregulated BMP signaling through ALK-1 in endothelial cells (ECs) is implicated in the pathogenesis of HHT. Interestingly, BMP signaling activated by BMP type I receptors, other than ALK-1 in ECs, are not able to compensate for the loss of function of ALK-1. On the other hand, aberrant BMP signaling through BMP type II receptor (encoded by BMPR2), especially in pulmonary arterial smooth muscle cells (PASMCs), are implicated in the pathogenesis of pulmonary arterial hypertension (PAH) (9,10). Therefore, readout of BMP signaling depends on the strength of BMP signaling, types I and II receptors and co-receptors, and cell types.
A binding sequence for BR-Smad was originally identified in Drosophila. Kim and colleagues (11) indicated that GCCGnCGC is a consensus binding sequence for Mad (Drosophila Smad1). In mammals, similar GC-rich sequences, e.g. GCCG or GGCGCC, have been evaluated in the promoter regions of well-known BMP target genes (12,13). Recombinant protein of the DNA binding domain of mouse Smad1 (Smad1 MH1) has been shown to bind to the GGCGCC sequence in vitro (14). This GGCGCC sequence is widely accepted as a binding sequence for BR-Smads, while a binding motif for BR-Smad has not been clearly defined in mammals.
Recent advances in microarray and sequencing technologies have made it possible to analyze global gene expression profiles and genome-wide maps of protein binding sites or epigenetic marks (15). Two groups have reported genome-wide analyses of BR-Smads in mouse ES cells (mESCs) using chromatin immunoprecipitation (ChIP) coupled with promoter array (ChIP-chip) and ChIP followed by sequencing (ChIP-seq) analyses (16,17). Through profiling of the global binding sites of 13 transcription factors and 2 transcription regulators in mESCs, Chen and colleagues (16) hypothesized that Smad1 makes an enhancer complex with Sox2-Oct4 (also known as Pou5F1), which defines ES-specific binding patterns of Smads. However, it has not been clarified whether a transcription factor complex, or an enhancer complex, determines the cell type-specific binding patterns of Smads in other cell types.
Here, we performed ChIP-seq to map Smad1/5 occupancy at high resolution in two different primary human cells treated with several BMP isoforms; human umbilical vein endothelial cells (HUVECs) with BMP-9 or BMP-6 and PASMCs with BMP-4. Smad1/5 preferentially bound to the region outside the promoter of known genes, and their binding was associated with upregulation of target genes. In HUVECs, Smad1/5 binding regions overlapped with reported enhancer regions. Comparison of HUVECs and PASMCs revealed that about 20% of the binding regions were overlapped. In contrast, most of the Smad1/5 binding sites in HUVECs treated with BMP-6 overlapped with those with BMP-9, especially in the regions with higher affinity for Smads. Cell-selective Smad1/5 binding patterns appear to be determined mostly by cell-specific differences in baseline chromatin accessibility patterns. In addition, a Smad1/5 binding motif was identified and termed a GC-rich Smad Binding Element (GC-SBE). Interestingly, GGAGCC sequence was enriched in the HUVEC- or PASMC-specific Smad1/5 binding regions compared with the GGCGCC sequence. We revealed that mutations of GC-SBE affected binding of Smad complex in a cell type-specific manner. Furthermore, we characterized EC-specific Smad1/5 target genes and found that several Notch signaling pathway-related genes were induced in ECs. Among them, a Notch ligand, JAG1 was regulated directly by Smad1/5, transactivating Notch signaling in the neighboring cells. These results provide insights into the molecular mechanism of BMP signaling and the pathogenesis of vascular lesions of HHT.
MATERIALS AND METHODS
Cell culture
HEK293T, HepG2 and HeLa cells were obtained from the American Type Culture Collection (ATCC). HUVECs and PASMCs were obtained from Lonza. HMEC-1, an immortalized human dermal microvascular EC line, was obtained from Dr T. Lawley (Emory University, Atlanta, GA, USA). 293T, HepG2 and HeLa cells were maintained in Dulbecco's modified Eagle's medium (Gibco), supplemented with 10% (v/v) fetal bovine serum (FBS) (HyClone) and 1% penicillin–streptomycin (Gibco). HUVECs and HMEC-1 were cultured in EGM-2 medium (Lonza). PASMCs were cultured in SmGM-2 (Lonza).
Reagents and antibodies
Recombinant human BMP-4, BMP-6 and BMP-9 were purchased from R&D Systems. TNF-α was from PeproTech. Cycloheximide (CHX) was purchased from Sigma-Aldrich.
The following antibodies were used: anti-Flag M2 (Sigma-Aldrich), anti-α-tubulin (AC-15; Sigma-Aldrich), anti-HDAC-1 (2E10; Upstate Millipore), anti-Smad1 (Bio Matrix Research, Chiba, Japan), which recognizes both Smad1 and Smad5 for ChIP procedure, anti-Smad1/5/8 (N-18; Santa Cruz Biotechnology) for western blotting, anti-phospho-Smad1/5 (Cell Signaling Technology), anti-phospho-Smad1/5/8 (Cell Signaling) and anti-JAG1 (H-114; Santa Cruz).
ChIP
Chromatin isolation, sonication and immunoprecipitation (IP) using anti-Smad1/5 antibody were performed essentially as described (18).
ChIP-sequencing (ChIP-seq) and data analysis
High-throughput sequencing of the ChIP fragments was performed using Illumina Genome Analyzer (Illumina) following the manufacturer's protocols. One flow cell lane was used for sequencing of each pooled sample. Unfiltered 36 bp sequence reads were aligned against the human reference genome (NCBI Build 36, hg18) using ELAND (Illumina). Peaks were called using CisGenome v1.2 (19) by two-sample analysis, where input genomic DNA was used as a negative control (Supplementary Table S1). Assigning a binding site to the nearest gene within 100 kb from a peak was performed using CisGenome.
A set of random genomic control regions for 3750 Smad1/5 binding regions was generated by randomly picking up the same number of 301 bp chromosome-matched sequences. In order to calculate the frequency of transcription factor binding site (TFBS)-positive sequences, MATCH score of position-specific scoring matrix (PSSM) for each transcription factor was computed. The highest MATCH score (HMS) was assigned to each sequence, and the number of sequences with HMS greater than or equal to a threshold was counted. For obtaining background data against those of 3750 Smad1/5 binding regions, the chromosome-matched sequences were generated randomly for 1000 times. The distribution of HMSs in 1000 sets of 3750 sequences was used as background control for each PSSM. The threshold was set to the mode of the distribution of HMSs. PSSMs were obtained from JASPAR database (20). A set of non-overlapping matched genomic control sequences was generated by CisGenome. The frequency of TFBSs in these sequences (motif counts per set of sequences) was computed with the likelihood ratio ≥500 (default value of CisGenome) or 200 (for shorter motifs such as MEME4). Fifty sets of matched control sequences were used as background data. Mapping of TFBSs to the specific genomic regions were calculated by the CisGenome.
The sequences of Smad1/5 binding sites were input to the MEME (21) with several options: mod = oops, nmotifs = 5, minw = 6, maxw = 8, revcomp and other settings default. The logo plots were generated using the seqLogo package in R (http://bioconductor.org/packages/2.6/bioc/html/seqLogo.html). Enriched binding motifs were also obtained from the Cis-regulatory Element Annotation System (CEAS) website as described (http://ceas.cbi.pku.edu.cn/index.html) (22,23). Overrepresented gene ontology (GO) categories for genes associated with Smad1/5 binding regions were determined using the Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7; http://david.abcc.ncifcrf.gov) (24).
ChIP and quantitative-PCR
The real-time PCR was conducted as described (23). Primer sequences are given in Supplementary Table S2 in the Supplementary Data. The amount of immunoprecipitated DNA was calculated relative to the input.
RNA isolation, quantitative real-time reverse transcription–PCR and conventional RT–PCR
Extraction of total RNA, qRT–PCR and conventional RT–PCR were performed as described (23). Primer sequences are given in Supplementary Table S2.
Gene expression profiling
HUVECs and PASMCs were serum starved overnight and treated with or without BMP-9 (1 ng/ml), BMP-6 or BMP-4 (50 ng/ml) treatment for 2 or 24 h. Gene expression profiling was performed with a GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix) as described (18). The 8544 and 8067 genes, whose signal intensity exceeded 100 at any time point were considered to be expressed and functional in HUVECs and PASMCs, respectively. The heatmaps were produced using the heatmap.2 function from the gplots library in R (http://cran.r-project.org/web/packages/gplots/).
Histone modification data
Genome-wide histone modification map for monomethylation of histone H3 lysine 4 (H3K4me1), trimethylation of histone H3 lysine 4 (H3K4me3) and acetylation of histone H3 lysine 27 (H3K27ac) of HUVECs were produced and released from the ENCODE Project (25) and were downloaded from UCSC (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/encodeDCC/wgEncodeBroadChipSeq/).
Plasmid construction
FLAG-tagged Smad constructs were previously described (18). Each fragment of Smad1/5 binding regions was amplified from human genomic DNA by PCR, cloned into a modified pGL4.10 reporter plasmid (Promega) driven by minimal adenoviral major late promoter (MLP) (12). A point mutation was introduced by site-directed mutagenesis using PCR with specific primers. A reporter construct with six multimerized CTGGAGCC sequence (pGL4-6xGC-SBE-Luc) was constructed as follows. A fragment with one copy of the CTGGAGCC sequence was cloned into the pcDNA3.1 vector (Invitrogen). The sequences of the oligonucleotides were 5′-AGATCTTCGAACAGCTCTGGAGCCAGATGGCCTGGATCC-3′ (sense) and 5′-GGATCCAGGCCATCTGGCTCCAGAGCTGTTCGAAGATCT-3′ (antisense). This fragment was multimerized in tandem, and the fragment containing six tandem copies was subcloned into the modified pGL4-MLP plasmid. Six multimerized dimeric CBF1/Suppressor of Hairless/Lag1 (CSL) binding sites with Epstein-Barr virus (EBV) TP1 promoter sequence of the pGA981-6 (26) was transferred into pGL4.10 reporter plasmid (pGL4-12xCSL-Luc) and used for Notch reporter assay. A plasmid encoding GST-hSmad1-MH1 was constructed by PCR amplification of the MH1 domain of human Smad1 (1–143 amino acid). The fragment was subcloned into pGEX-6P-1 vector (GE Healthcare, Chalfont St Giles, UK). All constructs were DNA sequence verified.
Protein production and purification
The bacterially expressed GST fusion proteins containing residues for human Smad11–143 (GST-hSmad1-MH1) were purified with Glutathione Sepharose 4B beads (GE Healthcare) followed by cleavage with PreScission Protease (GE Healthcare) at 4°C overnight according to the recommendations of the manufacturer. The concentration of the protein was measured by BCA Protein Assay Kit (Pierce).
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSA) was conducted essentially as described previously (14) and detected with LightShift Chemiluminescent EMSA kit (Pierce). The sequences of the DNA oligos are provided in Supplementary Table S2.
Lentiviral infection and luciferase assays
Since transfection efficacy is very low in HUVECs and it is rather toxic, we adapted lentiviral expression system. pGL4 constructs were subcloned between EcoRI and XhoI sites of the lentiviral vector construct CS-CDF-CG-PRE. Recombinant lentiviral vectors were generated as reported previously (23).
Stably expressing cells were stimulated with indicated doses of BMP-9 or BMP-6, and then they were harvested and assayed for luciferase activity at 12 h after stimulation. Luciferase activities of the cell lysates were determined using the Dual-luciferase Reporter Assay System (Promega).
Transient transfection and dual-luciferase assays
Transient transfection was carried out using FuGENE 6 (Roche) for HEK293T cells, Lipofectamine 2000 (Invitrogen) for HepG2 cells and Lipofectamine LTX (Invitrogen) for HMEC-1 cells and PASMCs according to the recommendations of the manufacturer.
Cells were transiently transfected with 1 μg of the luciferase reporter constructs along with 0.05 μg of Renilla luciferase reporter vector pGL4.74[hRluc/TK] (Promega) as an internal control. For HMEC-1 and PASMCs, the medium was changed at 3 h after transfection. Cells were stimulated with 1 ng/ml BMP-9 (HMEC-1), 50 ng/ml BMP-6 (HepG2) or 30 ng/ml BMP-4 (PASMCs) at 24 h after transfection, and then they were harvested and assayed for luciferase activity at 16 h after stimulation.
RNA interference
Duplexes of small interfering RNA (siRNA) against human Smad4 (D-003902-05) were synthesized by Dharmacon (Thermo Fisher Scientific), and were transfected using Lipofectamine RNAiMAX (Invitrogen) according to the recommendations of the manufacturer. The final concentration of siRNA in the culture media was 10 nM. At 24 h after transfection, cells were serum starved overnight, treated with or without BMP-9 for 2 or 24 h and subjected to qRT–PCR.
Western blotting
Western blotting was performed essentially as described (18). Cytoplasmic and nuclear fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) according to the recommendations of the manufacturer.
Immunofluorescence microscopy
HUVECs were treated with 1 ng/ml BMP-9 for 24 h, fixed in 10% formalin for 20 min and incubated overnight at 4°C with primary antibodies (JAG1, 1:100 dilution) diluted in Blocking One solution (Nacalai Tesque, Kyoto, Japan). The cells were washed with PBST (PBS with 0.1% Triton X-100), and then incubated with secondary antibodies (Alexa Fluor 488 goat anti-rabbit IgG, Invitrogen, 1:500 dilution) for 2 h and TOTO-3 (Invitrogen) for 10 min at room temperature. Images were obtained with a Zeiss LSM 510 Meta confocal microscope (Carl Zeiss).
Transactivation (coculture) Notch assay
One day prior to transfection, HeLa cells were seeded at a density of 5.0 × 104 cells per well in 12-well plate. Cells were transiently transfected with 1 μg of the pGL4-12xCSL-Luc reporter construct along with 0.05 μg of pGL4.74[hRluc/TK] (Promega) using Lipofectamine 2000 (Invitrogen). After 16 h transfection, medium was changed to 1:1 mixture of DMEM and EGM-2 and then 1.0 × 105 HUVECs were added. After adhesion of HUVECs (about 2 h later), cells were treated with or without 5 ng/ml BMP-9 for 24 h, and subjected to luciferase assay.
Statistical analysis
The difference between experimental groups of equal variance was analyzed using Student's _t_-test with P < 0.05 being considered significant. All experiments were performed at least three times independently and similar results were obtained.
RESULTS
Genome-wide identification and characterization of Smad1/5 binding sites in HUVECs and PASMCs
We performed ChIP-seq analyses using HUVECs stimulated with BMP-9 (1 ng/ml) or BMP-6 (50 ng/ml) and PASMCs treated with BMP-4 (50 ng/ml). Doses of the ligands for HUVECs were determined based on the phosphorylation status of BR-Smads and the physiological range of the circulating ligands. BMP-9 has been identified as a major circulating ligand for ALK-1 (27). Serum concentration of BMP-9 ranges from 1 to 12 ng/ml, which is enough for full activation of ALK-1 (Supplementary Figure S1A) (28,29). Thus, it is thought to play important roles in the control of vascular quiescence. BMP-6 transduces its signal mainly through the BMP type I receptor ALK-2 (encoded by ACVR1), which is also a receptor for BMP-9 (1). Notably, BMP-6 exists in FBS at concentrations of 2–10 ng/ml (29), and BMP-6 has been reported to activate ECs (30). However, 10 ng/ml was not enough to activate Smad1/5 in HUVECs (Supplementary Figure S1A). We selected a BMP-6 concentration of 50 ng/ml for our experiments, which gave an equivalent induction of ID1 mRNA (Supplementary Figure S1B), and almost as high phospho-Smad1/5/8 level in the nuclear fraction as stimulation with 1 ng/ml BMP-9 (Supplementary Figure S1C). We also confirmed that 50 ng/ml BMP-4 was enough for full activation in PASMCs (Supplementary Figure S1D). Both HUVECs and PASMCs expressed Smad1 and Smad5 (Supplementary Figure S1E).
The anti-Smad1/5 antibody worked efficiently in IP under formalin-fixed condition (Supplementary Figure S1F and S1G). Human genomic DNA sequences that corresponded to known BMP responsive elements (BREs) in mouse Id1 (12) and mouse Hey1 (13) promoters were used as positive control regions. In HUVECs, a comparable enrichment of Smad1/5 was confirmed at the ID1 promoter after BMP-6 or BMP-9 treatment, while weak Smad1/5 binding was observed at the HEY1 promoter after BMP-6 stimulation (Supplementary Figure S1H). Since maximal Smad1/5 binding was observed at 1.5 h after stimulation with BMPs, we adopted this time of stimulation for ChIP-seq analyses, the same stimulation time that was used in similar studies of Smad2/3 (18) and Smad4 (31).
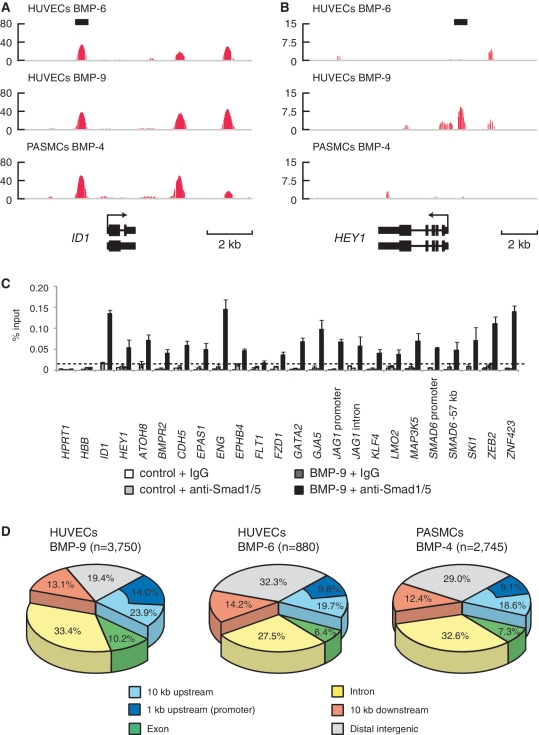
The ChIP DNA and the control input DNA were then submitted to high-throughput sequencing analyses. The enriched genomic regions were determined using CisGenome (19). Using a false discovery rate (FDR) cut off of 0.1, a total of 3750 Smad1/5 binding regions were identified in the ChIP-seq data of HUVECs treated with BMP-9, 880 in HUVECs treated with BMP-6 and 2745 in PASMCs treated with BMP-4 (Figure 1A and B; Supplementary Figure S1I). To validate the results, BMP-9-dependent Smad1/5 enrichment was confirmed by ChIP–qPCR at 20 novel Smad1/5 binding regions of variable peak intensity (Figure 1C). The ChIP-seq peaks were annotated to a total of 2179, 563 and 1,609 genes, respectively (Supplementary Table S1). Approximately 30% of the binding sites were located in the introns of known genes and 20% in the promoter regions within 10 kb upstream of known transcription start sites (TSSs) (Figure 1D). These Smad1/5 binding regions were highly conserved among multiple vertebrate species (Supplementary Figure S1J).
Figure 1.
Genome-wide identification and characterization of Smad1/5 binding sites in HUVECs. (A and B) Genomic loci of ID1 (A) and HEY1 (B) are shown together with the results of ChIP-seq (in red). The direction of transcription is shown by the arrow beginning at the TSS. Horizontal black bars represent the positions of previously reported Smad1/5/8 binding regions. (C) HUVECs were stimulated with 1 ng/ml BMP-9 for 1.5 h and subjected to ChIP assays with anti-Smad1/5 antibody or control IgG. The ChIP samples were quantified by real-time PCR with locus specific primers and normalized to input DNA. The dashed line indicates 0.01% of input. The data are the mean of triplicate values ± SD. (D) Genome-wide location summary of Smad1/5 binding regions relative to known genes in human genome (hg18). Ten kilo base pairs upstream and downstream regions are defined as ≤10 kb upstream from the TSS or ≤10 kb downstream from the transcription end site (TES) of a gene, respectively. Distal intergenic refers to all locations outside the intragenic and the 10 kb flanking regions.
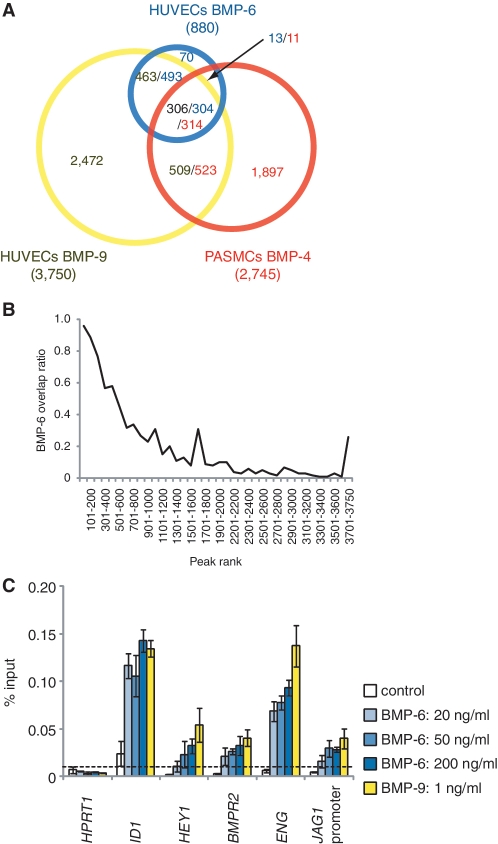
Comparison of the three ChIP-seq data revealed that ∼20% of Smad1/5 binding regions overlapped between HUVECs and PASMCs (Figure 2A), while most of the Smad1/5 binding sites in HUVECs after BMP-6 stimulation overlapped with those after BMP-9 stimulation, especially in the higher ranked peaks (Figure 2A and B). Common Smad1/5 binding sites shared with HUVECs treated with BMP-9 and those with BMP-6, including those at ID1 and ID3 loci, had comparable levels of Smad1/5 binding, suggesting that these sites had higher affinity for Smad1/5, while the BMP-9 specific sites (Figure 1A and B and Supplementary Figure S1I) had weaker affinity. In line with this hypothesis, increasing concentrations of BMP-6 dose dependently enhanced the Smad1/5 binding to the BMP-9 specific binding sites, e.g. at HEY1 and JAG1 loci (Figure 2C), whereas common Smad1/5 binding sites, e.g. at ID1 and ENG loci, had enough enrichment when stimulated with only 20 ng/ml BMP-6 (Figure 2C). These dose response data also indicated that 50 ng/ml BMP-6 was not enough to achieve Smad1/5 binding to target sites with relatively lower affinity for Smad1/5 in ECs.
Figure 2.
Comparison of Smad1/5 binding regions in HUVECs and PASMCs. (A) A Venn diagram represents the overlaps of Smad1/5 binding regions of HUVECs treated with BMP-9 (Yellow) or BMP-6 (Blue) and PASMCs treated with BMP-4 (Red). The number of binding regions of HUVECs treated with BMP-9 (Black) or BMP-6 (Blue) and that of PASMCs treated with BMP-4 (Red) are also shown. The numbers of overlapped regions are not identical, since some of the peaks are not on a one-by-one correspondence. (B) ChIP-seq peaks of HUVECs treated with BMP-9 are ranked by peak height. Fraction of peaks overlapped with the peaks of HUVECs treated with BMP-6 is calculated for every 100 peak and plotted. Overlapped peaks are enriched in the high ranked peaks. (C) HUVECs were starved overnight and stimulated with the indicated concentration of BMP-6 or BMP-9 for 1.5 h and were subjected to ChIP assays with anti-Smad1/5 antibody. The ChIP samples were quantified by real-time PCR with locus-specific primers and normalized to input DNA. The dashed line indicates 0.01% of input. The data are the mean of triplicate values ± SD.
Smad1/5 bind to enhancer regions already accessible in specific cell types
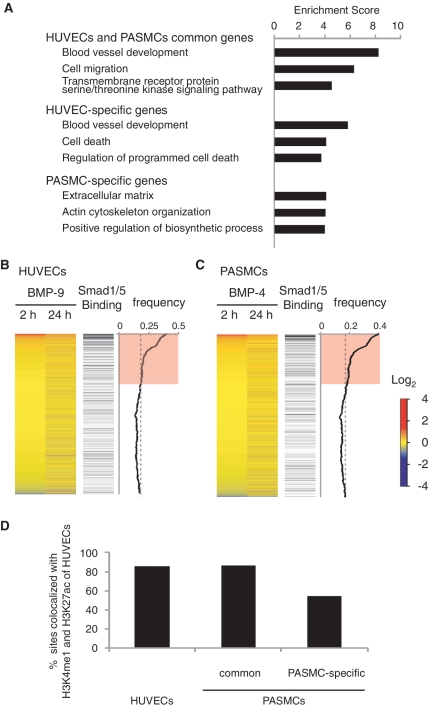
To investigate the biological functions associated with Smad1/5 binding in HUVECs and PASMCs, the significance of functional annotation clustering of the GO of the genes related to Smad1/5 binding was assessed using DAVID (24). This analysis showed that the highest enriched GO category of biological function for HUVEC-specific genes with BMP-9 stimulation was related to blood vessel development, while that for PASMC-specific genes with BMP-4 stimulation was related to extracellular matrix production (Figure 3A). Thus, Smad1/5 bind to different sets of target sites in different cell types, which may be related to the cell type-specific function.
Figure 3.
Genome-wide identification of cell type-specific BMP target genes and gene expression profiles. (A) GO enrichment analysis was performed to elucidate the biological processes and pathways associated with each gene cluster. The top three annotation clusters are shown in bar plots. The value reflects the Enrichment Score. Group names are based on interpretation of enriched GO annotations. (B and C) Gene expression profiles of HUVECs of HUVECs with BMP-9 stimulation or PASMCs with BMP-4 are illustrated by heat map. Probes are sorted by fold change relative to time 0 at early phase (2 h after stimulation) (left panel). Increased or decreased mRNA expression is represented by red or blue, respectively. Black horizontal bars represent probes of genes associated with Smad1/5 binding regions (middle panel). Moving average of the frequency of probes with Smad1/5 binding is plotted in a 1000-probe sliding window (right panel). The red-colored areas indicate the probes, whose Smad binding frequency is higher than the expected average. The dashed line indicates the expected average. (D) Frequency of the Smad1/5 binding regions co-localized with enhancer regions of HUVECs. The Smad1/5 binding regions in PASMCs are divided into two groups, those shared with HUVECs or PASMC-specific sites.
In order to identify the cell type-specific binding mechanism for Smad complex, we sought known binding motifs enriched in the Smad1/5 binding regions using the CEAS website (22). Interestingly, ETS, AP-1, AP-2 and SP-1 binding sites were enriched in the Smad1/5 binding regions in both HUVECs and PASMCs, while other motifs occurred only in a small proportion of sequences analyzed (Supplementary Table S3). We also conducted de novo motif prediction in order to find overrepresented motifs in the HUVEC- and PASMC-specific Smad1/5 binding regions using MEME (21). Obtained motifs were then compared with TRANSFAC (32) and JASPAR (20) database of known motifs, and ranked by their similarity using the TOMTOM program (33). The predicted motifs were similar to the ones identified by CEAS (Supplementary Figure S2). These results suggest that these transcription factors do not determine the cell type-specific BR-Smad binding pattern.
To evaluate the association between Smad1/5 binding and gene expression regulation, expression microarray analyses were performed at several time points (0, 2 and 24 h). We confirmed an equivalent induction of ID proteins after BMP-6 or BMP-9 stimulation in HUVECs (Supplementary Figure S3A). Combining the mapping data with gene expression profiles revealed that Smad1/5 binding regions were enriched in early upregulated genes (Figure 3B and C). Notably, in HUVECs treated with BMP-9, 108 genes were upregulated and 37 were downregulated more than 2-fold in early phase, and 70 of the 108 upregulated genes (64%) and 9 of the 37 downregulated genes (24%) were associated with Smad1/5 binding regions (Supplementary Figure S3B). We consider these 70 upregulated genes (corresponding to 170 binding sites) as putative direct target genes of ALK-1 in ECs (Supplementary Table S4). We also identified 19 putative direct target genes in PASMCs using the same criteria (Supplementary Table S4).
Smad1/5 binding regions in HUVECs were further characterized using differential histone modification marks, which were produced and released from the ENCODE Project (25). H3K4me3 is associated with promoters and H3K4me1 is preferentially associated with enhancers. H3K27ac is associated with active regulatory regions (34). As many as 3651 Smad1/5 binding peaks (97.4%) overlapped with H3K4me1 or H3K4me3 regions of HUVECs. Among them, 3201 Smad1/5 binding peaks (85.4%) overlapped with enhancer regions, characterized with both H3K4me1 and H3K27ac (Figure 3D and Supplementary Figure S3C) (34). In PASMCs, 86.5% (724/837) of common Smad1/5 binding peaks shared with HUVECs and PASMCs overlapped with enhancer regions of HUVECs characterized with both H3K4me1 and H3K27ac, while only 54.3% (1036/1908) of PASMC-specific peaks overlapped with endothelial enhancers (Figure 3D). Thus, these data also suggest that Smad1/5 preferentially bind to enhancer regions already accessible in specific cell types.
GC-SBE is a direct binding motif for Smad1/5
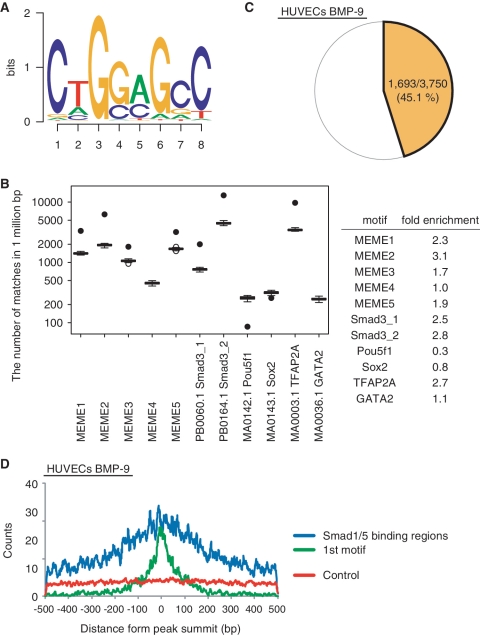
To elucidate a specific binding motif in Smad1/5 binding regions, a de novo motif prediction was performed using MEME (21). Since ChIP experiments may detect indirect Smad1/5-DNA binding through protein–protein interaction, we focused on the 170 Smad1/5 binding regions of BMP-9 target genes in HUVECs. Five overrepresented motifs were identified and designated as MEME1–5 (Figure 4A and Supplementary Figure S4A). These motifs were validated in three ways. First, the fold enrichment of the motifs was compared (Figure 4B and Supplementary Figure S4B). Randomly selected genomic sequences (n = 1000) or non-overlapping matched regions (n = 50) were used as background controls. Four out of five motifs were significantly enriched in the Smad1/5 binding regions, and the MEME2 was the best. TFAP2A (also known as AP-2α) binding motif was a positive control and was found to be enriched in the Smad1/5 binding regions (Supplementary Table S3). No statistically significant differences were observed for motifs of transcription factors known to be expressed and functional in ECs, such as GATA2 (35). In contrast to the study of Chen and colleagues (16), the motifs for SOX2 and POU5F1 (also known as OCT4) were not enriched in the Smad1/5 binding regions, suggesting that different mechanisms or different enhancer complexes are adopted in differentiated ECs compared with mESCs. In addition, the incidence of the MEME motifs in the peaks was calculated. MEME2 occurred in about 45% of all Smad1/5 binding regions in HUVECs and PASMCs (Figure 4C and Supplementary Figure S4C). Moreover, it was enriched in the higher ranked peaks in HUVECs treated with BMP-9 (Supplementary Figure S4D). Finally, the relative distribution of the motif around the peak summits, where Smad1/5 was expected to be located, was analyzed. MEME2 was enriched in the Smad1/5 binding regions, especially around the peak summits, while other MEME motifs were not (Figure 4D and Supplementary Figure S4E and F). We therefore designated MEME2 as GC-SBE because it is similar in sequence to the previously reported GC-rich sequences for BR-Smads (11–13).
Figure 4.
De novo prediction of Smad1/5 binding motif. Total 170 peak regions were analyzed for overrepresented motifs using MEME. (A) MEME2 is displayed as a sequence logo. (B) Enrichment of TFBS in the Smad1/5 binding regions. Fifty sets of non-overlapping matched genomic control sequences were used as background control. Data are given as boxplot. The circles represent outlier values. The black circles indicate the number of matched motifs observed in the Smad1/5 binding regions. (C) MEME2 motif occurs in about 45% of all Smad1/5 binding regions in HUVECs stimulated with BMP-9. (D) Distribution of MEME2 motif around the peak summits. The number of the MEME2 motif around the peak summits was counted and plotted in a 7 bp sliding window against the distance from the summits (within 500 bp from the summits) (blue). The motifs closest to the summits are located within 100 bp from the peak summit (First motif; green). Five separate matched control regions were randomly chosen by CisGenome and used as a control. The number of the MEME2 motif in those regions was counted and the average was plotted (red).
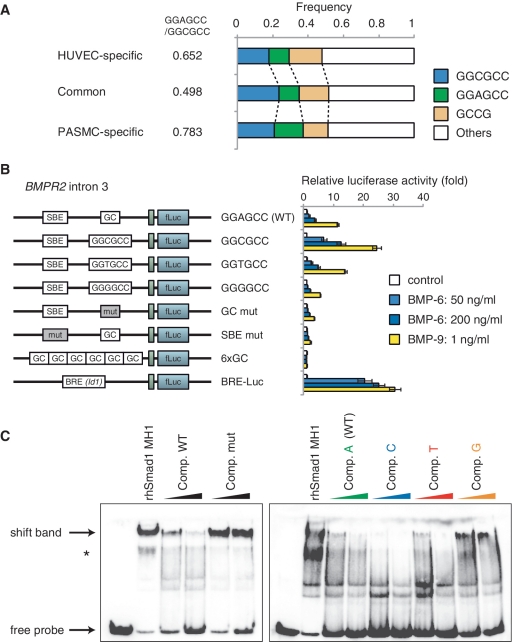
Analysis of the frequency of GC-SBE sequence in Smad1/5 binding regions revealed that GGCGCC sequence was enriched in Smad1/5 binding regions shared with HUVECs and PASMCs, while GGAGCC sequence was enriched in both HUVEC- and PASMC-specific binding regions (Figure 5A). To validate the enhancer activity of the Smad1/5 binding regions and the effects of the newly identified GC-SBE on the cell type specificity, luciferase assays were performed in HUVECs. Both BMPR2 and JAG1 were HUVEC-specific target genes (Supplementary Table S4). Fragments from Smad1/5 binding regions in intron 3 of BMPR2 and the JAG1 promoter, which contain the GGAGCC sequence, were cloned into a luciferase reporter construct (Supplementary Figure S5A). Both BMP-9 and BMP-6 were able to activate these reporters in HUVECs, while BMP-4 induced only weak response in PASMCs (Figure 5B and Supplementary Figure S5B and S5C). Consistent with ChIP data (Figure 1C), the Smad1/5 binding regions induced higher luciferase expression following treatment with BMP-9 compared with BMP-6. Even 1 ng/ml BMP-9 induced stronger luciferase activities in HUVECs than 50 or 200 ng/ml BMP-6 (Figure 5B). We also confirmed that these Smad1/5 binding fragments worked as transcriptional enhancers in the human microvascular endothelial cell line, HMEC-1 (Supplementary Figure S5D).
Figure 5.
Validation of GGAGCC sequence as a novel BMP responsive element. (A) Frequency of GC-SBE sequences in the Smad1/5 binding regions. GGCGCC, GGAGCC and GCCG sequences were enriched in the Smad1/5 binding regions. The ratio of GGAGCC:GGCGCC is indicated. (B) pGL4-BMPR2 reporter constructs were introduced into HUVECs using lentiviral vector system, in order to evaluate their enhancer activity. The cells were stimulated with indicated doses of BMP-9 or BMP-6 and then they were harvested and assayed for luciferase activity at 12 h after stimulation. The data are the mean of triplicate values ± SD. (C) Recombinant human Smad1 MH1 proteins interacted with GGAGCC sequence. (Left panel) rhSmad1 MH1 binding to the probe was competed with a 50-fold molar excess of the unlabeled wild-type competitor (Comp. WT), but not with the mutant competitor (Comp. mut). (Right panel) To evaluate the importance of ‘A’ in GGAGCC sequence, single-point mutant competitors were evaluated. An asterisk indicates background band. Full wild-type probe sequence was ACAGCTCT GGAGCC AGATGGCCTGG.
In order to compare the difference of enhancer activities between GGAGCC and GGCGCC sequence, a point mutation was introduced at the ‘A’ in the GGAGCC sequence. A mutation to GGCGCC induced higher luciferase expression compared with the GGAGCC wild-type. In contrast, a mutation to GGGGCC attenuated BMP responsiveness (Figure 5B and Supplementary Figure S5B). Interestingly, the fragments with GGAGCC sequence did not respond to BMP stimulation in PASMCs, whereas mutation to GGCGCC showed a higher responsiveness (Supplementary Figure S5C). Luciferase assays were also performed in HepG2 cells to examine the cell type specificity of the fragments. Similarly, the GGCGCC mutant responded very well compared with wild-type and the T-mutant, while the G-mutant had no enhancer activity in HepG2 (Supplementary Figure S5E).
We next showed the direct binding of recombinant human Smad1 MH1 (rhSmad1 MH1) to the GGAGCC sequence using EMSAs. The amino acid sequence of rhSmad1 MH1 is identical to the corresponding sequence of mouse Smad1 MH1, which was reported to bind to the GGCGCC sequence (14). rhSmad1 MH1 was able to bind to the GGAGCC probe and this binding was blocked by wild-type oligonucleotide but not by the mutated one (Figure 5C). The effects of single-point mutation in the GGAGCC sequence were also examined. The GGCGCC sequence competed more efficiently than the GGAGCC sequence, suggesting that this sequence had higher affinity for binding to rhSmad1 MH1 (Figure 5C). Thus, these results showed that the GGAGCC sequence is also a direct binding motif for Smad1/5 and that GC-SBE is a generalized form of the previously reported GC-rich sequences.
Both GC-SBE and SBE are required for full BMP responsiveness
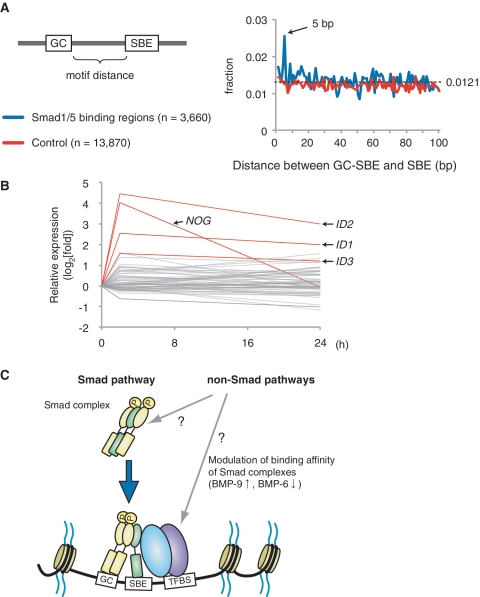
In Drosophila, Dpp (Decapentaplegic; Drosophila BMP orthologs)-responsive elements are shown to contain a GC-rich Mad binding site and a flanking GTCT Medea (Drosophila Smad4) binding site with a 5 bp spacer sequence (36). Indeed, Smad3 binding motifs were significantly enriched in the Smad1/5 binding regions found in our analysis (Figure 4B). The analysis of the spacer length between GC-SBE and SBE revealed that the 5 bp spacer was also prominent in HUVECs (Figure 6A), suggesting that the 5 bp spacer sequence has some beneficial effect for binding of the Smad complex, containing Smad1/5 and Smad4, in mammalian cells too. On the other hand, expressions of genes associated with the GC-SBE/SBE composite motif with 5 bp spacer were not necessarily regulated by BMP-9 stimulation (Figure 6B).
Figure 6.
GC-SBE is required, but not sufficient, for full BMP responsiveness. (A) The distance between GC-SBE and GTCT-AGAC sequence in Smad1/5 binding regions is calculated and plotted (blue). Total 13 870 GC-SBEs in randomly-adapted control regions were used as a control (red). The dashed line indicates the expected average. (B) Graphical summary of expression microarray data of genes with GC-SBE and GTCT-AGAC composite motif with 5 bp spacer. The value of genes with GC-SBE/SBE composite motif with 5 bp spacer is represented as log2-fold change relative to time 0. Several, but not all of these genes were induced more than 2-fold within 2 h, including the well-known Smad1/5 target genes ID1, ID2, ID3 and NOG (Noggin) (red). (C) Schematic representation of Smad1/5 binding. Smad1/5 binding patterns appear to be predetermined by cell-specific differences in baseline chromatin accessibility patterns. Number and distribution of BR-Smad binding sites over the genome are primarily defined by the intensity of the Smad pathway. Each Smad1/5 binding site has different binding affinity for Smad complexes, which is determined by the affinities of GC-SBEs and SBEs. Non-Smad pathways were reported to affect the BR-Smad signaling through degrading Smad complexes or modulating binding affinity of Smad complexes. GC: GC-SBE, TFBS: transcription factor binding site.
Next, the roles of SBE sequences, which were located at different distances from GC-SBE, were examined. Evolutionarily conserved SBE/GC-SBE composite motifs with a 28 bp spacer sequence were found in the BMPR2 intron 3 and the JAG1 promoter (Supplementary Figure S5A). These sequences were able to drive luciferase expression in reporter assays in response to BMP stimulation (Figure 5B and Supplementary Figure S5B). Mutations in either GC-SBE or SBE sequence showed significant attenuation of BMP responsiveness (Figure 5B and Supplementary Figure S5B), indicating that the effective distance between GC-SBE and SBE was not restricted to 5 bp. The GC-SBE was not able to respond to BMP stimulation in luciferase reporter assays, even when present in six copies (Figure 5B). These findings clearly showed that both GC-SBE and SBE were required for full BMP responsiveness. Collectively, our results suggest that the binding affinity of Smad complexes to DNA is defined by the affinities of GC-SBEs and SBEs (Figure 6C).
JAG1 is a direct target gene of Smad1/5 in ECs and transactivates Notch signaling in the neighboring cells
EC-specific target genes contained well-known Notch-signal target genes and signaling components, including HEY1, HEY2, HES1, FOXC1, LFNG, NRARP and JAG1 (Figure 7A and Supplementary Table S4). Synergic effects between Notch and BMP signaling on several Notch target genes, such as HEY1 and CDH2, have been reported previously (13,37). However, little is known about direct expression regulation of Notch ligands by BMP signaling in ECs.
Figure 7.
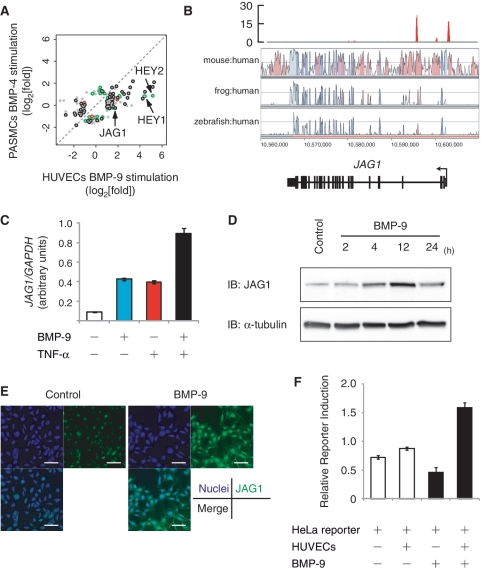
The Notch ligand JAG1 is a direct target gene of Smad1/5 and transactivates Notch signaling in the neighboring cells. (A) Scatter plot representation of differentially regulated genes between HUVECs and PASMCs. Probes of genes with more than 2-fold change in expression relative to time 0 are plotted. If the genes are associated with Smad1/5 binding regions of HUVECs (green), PASMCs (red) or both (black), the plots are colored. Signal intensities of HUVECs treated with BMP-9 for 2 h is plotted on the _X_-axis and those of PASMCs treated with BMP-4 for 2 h is plotted on the _Y_-axis. (B) Visualization of JAG1 locus with the result of BMP-9 ChIP-seq. Red peaks represent ChIP regions (top panel). The conservation plots for mouse/human, frog/human and zebrafish/human are derived from VISTA genome browser (middle panel), which represents the sequence conservation between species. (C) Induction of JAG1 after BMP-9 stimulation in HUVECs. HUVECs were starved overnight, stimulated with 1 ng/ml BMP-9 and/or 10 ng/ml TNF-α for 2 h and subjected to qRT–PCR analysis for JAG1. Values were normalized to the amount of housekeeping GAPDH mRNA. The data are the mean of triplicate values ± SD. (D) HUVECs were starved overnight and stimulated with 1 ng/ml BMP-9 for indicated time periods and subjected to immunoblot analysis to determine the JAG1 protein expression level. α-Tubulin was used as a loading control. (E) Immunocytochemistry of HUVECs treated with or without 1 ng/ml BMP-9 for 24 h. The cells were immunostained with anti-JAG1 antibody (green). Nuclei were labeled with TOTO-3 (blue). Scale bar, 100 µm. (F) Endothelial JAG1 induced by BMP-9 stimulation transactivates Notch signaling in neighboring cells. HeLa cells were transiently transfected with pGL4-12xCSL-luciferase reporter construct and co-cultured with HUVECs. Cells were treated with or without 5 ng/ml BMP-9 for 24 h and subjected to luciferase assay. The data are the mean of triplicate values ±SD.
Two strong Smad1/5 binding regions were identified in the JAG1 locus, in the promoter region at −500 bp from the TSS and in the second intron (Figure 7B), which were verified by ChIP–qPCR (Figure 1C). Both regions worked as transcriptional enhancers in HUVECs (Supplementary Figure S5B). Consistent with the results of the reporter assays, BMP-9 was able to induce expression of JAG1 mRNA (Figure 7C). TNF-α has been shown to induce JAG1 expression in ECs (38). The induction by BMP-9 was equivalent to that of TNF-α and also had some additive effects (Figure 7C). Western blot analysis and immunocytochemistry revealed that the JAG1 protein was also upregulated by BMP-9 stimulation in ECs (Figure 7D and E). This JAG1 mRNA induction was not affected by CHX, and siRNA against SMAD4 (siSmad4) attenuated BMP-9-mediated upregulation of JAG1 (Supplementary Figure S6A and B). These results showed that JAG1 is a direct target gene of BMP-Smad1/5 pathway.
A HeLa reporter cell system was used to verify the function of JAG1 as a Notch ligand. HeLa cells were transfected with the Notch-specific luciferase reporter construct (pGL4-12xCSL-Luc), and thus responsive to Notch activation (26). In the absence of HUVECs, BMP-9 did not induce reporter activity in the transfected HeLa cells (Figure 7F; lanes 1 and 3). In the presence of HUVECs, however, BMP-9 induced strong activation of reporter expression (Figure 7F; lanes 2 and 4), indicating that JAG1 induced by BMP-9 in ECs was able to efficiently transactivate Notch signaling in neighboring cells.
DISCUSSION
In this study, genome-wide maps of Smad1/5 binding regions in human primary cells revealed how BR-Smads recognize and regulate their target genes. Both HUVECs and PASMCs express Smad1, Smad5 and Smad8 (Supplementary Figure S1E). However, redundant functions between Smad1 and Smad5 have been demonstrated in vivo, especially in the vasculature (39). Smad1+/−; Smad5+/− double heterozygous mutant mice are embryonic lethal and display defects, which closely resemble those seen in _Smad1_- or _Smad5_-null mice, whereas Smad1 or Smad5 single heterozygous mice show no overt phenotype. _Smad8_-null mice additionally lacking one copy of Smad1 or Smad5 did not exhibit overt phenotypes, and the tissue disturbances seen in Smad1- or _Smad5-_null embryos are not exacerbated in the absence of Smad8. These findings suggest that Smad1 and Smad5 possess equivalent biological functions especially in the vasculature, while Smad8 is dispensable.
The mapping data of Smad1/5 showed that ∼30% of the binding sites were located in the introns of known genes. Smad1/5 binding peaks of 85.4% overlapped with enhancer regions in HUVECs, where histone modification markers in basal conditions were available. Motif analysis revealed that binding motifs for ETS, AP-1, AP-2 and SP-1 were enriched in Smad1/5 binding regions regardless of the cell types. These motifs were also enriched in the Smad4 binding regions in human keratinocyte HaCaT cells (31). Other motifs occurred only in a small proportion of sequences analyzed. Recently, John and colleagues (40) reported that cell type-specific glucocorticoid receptor binding patterns are comprehensively predetermined by cell-specific differences in baseline chromatin accessibility patterns, with secondary contributions from local sequence features. The similar motif occurrence patterns between HUVECs and PASMCs suggest that the binding regions of BR-Smad are also predetermined in the specific cell types.
Smad1/5 reproducibly bound to some target sites such as ID1 and ID3 loci with comparable enrichment after BMP-9 and BMP-6 stimulation, while the total number of Smad1/5 binding sites was dramatically lower in HUVECs treated with BMP-6 compared to those with BMP-9 (3750 versus 880). Increasing the dose of BMP-6 up to 200 ng/ml was not enough to elicit comparable level of enhancer activities as 1 ng/ml BMP-9 (Figures 2C and 5B). This suggests that each binding site has different binding affinity for Smad complex and that BR-Smad signaling through ALK-2 was not enough to occupy full sets of target sites in ECs. This is consistent with the facts that HHT2 is the result of haploinsufficiency of ALK-1 (6), and that ALK-2 signaling is not able to compensate for ALK-1 mutations in HHT patients even though BMP-9 can signal through ALK-2 (1).
In Drosophila, Ashe et al. (41) have reported that each enhancer element for Mad target genes has a different binding affinity for Smad/Mad. A gene with low-affinity Smad/Mad binding sites is transcribed only in response to high concentrations of Dpp, while a gene with higher affinity sites responds to a low dose of Dpp. Increasing the affinity of the Smad/Mad binding sites in the enhancer of the Ance (also known as Race) resulted in a wider expression pattern in vivo (42). We revealed that a mutation in our consensus GC-SBE sequences attenuates BMP responsiveness of target genes (Figure 5B and Supplementary Figure S5B). In addition, a mutation of the HAMP promoter from GGCGCC to GGTGCC, which was identified in a hemochromatosis patient, impairs the BMP responsiveness in vivo and contributes to the severe phenotype (43). These results suggest that the binding affinity for Smad complex is the sum of the affinities of GC-SBEs, SBEs and other DNA binding proteins like Sox2 and Oct4 in mESCs (16), and that unidentified mutations in the BR-Smad binding regions will be implicated in HHT or PAH.
Collectively, our findings support the notion that BR-Smad binding sites are predetermined in specific cell type and determined by the binding affinity of Smad complex to possible binding sites. It suggests that the strength of the BR-Smad pathway is converted to the number and distribution of BR-Smad binding sites over the genome. It does not necessarily exclude the possibilities that non-Smad pathways play important roles. Non-Smad pathways have been reported to affect the BR-Smad pathway through degrading BR-Smads or modulating binding affinity of Smad complexes [for review, see (44)]. It is possible that they modulate the intensity of BR-Smad pathway and affect the number and distribution of Smad1/5 binding sites in ECs (Figure 6C).
Dysregulation of Notch signaling has been reported to cause AVM [for review, see (45)] that is one of the major pathological features of HHT. JAG1 has been reported to cause differentiation of vascular smooth muscle cell (vSMC) precursor cells and induce vSMC-specific genes in vitro through the JAG1-Notch3 signaling pathway (46,47). EC-specific deletion of Jag1 showed defects in vSMC coverage in mice (38,48). Interestingly, genetic and pharmacological inhibition of ALK-1 signaling showed a severe vascular phenotype including lack of differentiation and recruitment of vSMCs and defects in the maturation phase of angiogenesis (5,49,50). In the clinical settings, thalidomide has been shown to stimulate vessel maturation and have beneficial effects on HHT patients (51). Therefore, our results suggested the important roles of ALK-1-Smad-JAG1 pathway in the pathogenesis of the vascular lesions of the HHT. They also suggest that this pathway will be a novel therapeutic target for treatment of HHT.
ACCESSION NUMBERS
The microarray data from this study have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE27661, and the sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA030442.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KAKENHI grants-in-aid for scientific research on Innovative Area [Integrative Research on Cancer Microenvironment Network (grant number 22112002)]; The Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan [scientific research (S) grant number 20221009 to H.A.]; Genome Network Project from MEXT (to H.A.); Japan Society for the Promotion of Science (JSPS), the Global Center of Excellence Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms); Swedish Cancer Society (grant number 10 0452). Funding for open access charge: KAKENHI [Integrative Research on Cancer Microenvironment Network (grant number 22112002)].
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
The authors are grateful to Drs Aristidis Moustakas and Helen M. Arthur for constructive comments; Kaori Shiina for technical assistance; and members of the Miyazono laboratory for discussion and advice.
REFERENCES
- 1.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J. Biochem. 2010;147:35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DW, Berg JN, Baldwin MA, Gallione CJ, Marondel I, Yoon SJ, Stenzel TT, Speer M, Pericak-Vance MA, Diamond A, et al. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat. Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 3.McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, Jackson CE, Helmbold EA, Markel DS, McKinnon WC, Murrell J, et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat. Genet. 1994;8:345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- 4.Gallione CJ, Richards JA, Letteboer TG, Rushlow D, Prigoda NL, Leedom TP, Ganguly A, Castells A, Ploos van Amstel JK, Westermann CJ, et al. SMAD4 mutations found in unselected HHT patients. J. Med. Genet. 2006;43:793–797. doi: 10.1136/jmg.2006.041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK, Li L, Miyazono K, ten Dijke P, Kim S, et al. Activin receptor-like kinase 1 modulates transforming growth factor-beta 1 signaling in the regulation of angiogenesis. Proc. Natl Acad. Sci. USA. 2000;97:2626–2631. doi: 10.1073/pnas.97.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricard N, Bidart M, Mallet C, Lesca G, Giraud S, Prudent R, Feige J-J, Bailly S. Functional analysis of the BMP9 response of ALK1 mutants from HHT2 patients: a diagnostic tool for novel ACVRL1 mutations. Blood. 2010;116:1604–1612. doi: 10.1182/blood-2010-03-276881. [DOI] [PubMed] [Google Scholar]
- 7.Lebrin F, Goumans M-J, Jonker L, Carvalho RL, Valdimarsdottir G, Thorikay M, Mummery C, Arthur HM, ten Dijke P. Endoglin promotes endothelial cell proliferation and TGF-beta/ALK1 signal transduction. EMBO J. 2004;23:4018–4028. doi: 10.1038/sj.emboj.7600386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heldin C-H, Miyazono K, ten Dijke P. TGF-beta signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Long L, Southwood M, Rudarakanchana N, Upton PD, Jeffery TK, Atkinson C, Chen H, Trembath RC, Morrell NW. Dysfunctional Smad signaling contributes to abnormal smooth muscle cell proliferation in familial pulmonary arterial hypertension. Circ. Res. 2005;96:1053–1063. doi: 10.1161/01.RES.0000166926.54293.68. [DOI] [PubMed] [Google Scholar]
- International PPH Consortium, Lane,K.B., Machado,R.D., Pauciulo,M.W., Thomson,J.R., Phillips,J.A. 3rd, Loyd,J.E., Nichols,W.C. and Trembath,R.C. (2000) Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. The International PPH Consortium. Nat. Genet., 26, 81–84. [DOI] [PubMed]
- 11.Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 12.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 13.Itoh F, Itoh S, Goumans M-J, Valdimarsdottir G, Iso T, Dotto GP, Hamamori Y, Kedes L, Kato M, ten Dijke P. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 2004;23:541–551. doi: 10.1038/sj.emboj.7600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BabuRajendran N, Palasingam P, Narasimhan K, Sun W, Prabhakar S, Jauch R, Kolatkar PR. Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors. Nucleic Acids Res. 2010;38:3477–3488. doi: 10.1093/nar/gkq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 17.Fei T, Xia K, Li Z, Zhou B, Zhu S, Chen H, Zhang J, Chen Z, Xiao H, Han J-D, et al. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 2010;20:36–44. doi: 10.1101/gr.092114.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor-beta signaling. Mol. Cell. Biol. 2009;29:172–186. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–W554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagano Y, Koinuma D, Miyazawa K, Miyazono K. Context-dependent regulation of the expression of c-Ski protein by Arkadia in human cancer cells. J. Biochem. 2010;147:545–554. doi: 10.1093/jb/mvp202. [DOI] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl LJ, Zimber-Strobl U, Bornkamm GW, Honjo T. RBP-L, a transcription factor related to RBP-Jkappa. Mol. Cell. Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.David L, Mallet C, Mazerbourg S, Feige J-J, Bailly S. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood. 2007;109:1953–1961. doi: 10.1182/blood-2006-07-034124. [DOI] [PubMed] [Google Scholar]
- 28.David L, Mallet C, Keramidas M, Lamande N, Gasc J-M, Dupuis-Girod S, Plauchu H, Feige J-J, Bailly S. Bone morphogenetic protein-9 is a circulating vascular quiescence factor. Circ. Res. 2008;102:914–922. doi: 10.1161/CIRCRESAHA.107.165530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrera B, Inman GJ. A rapid and sensitive bioassay for the simultaneous measurement of multiple bone morphogenetic proteins. Identification and quantification of BMP4, BMP6 and BMP9 in bovine and human serum. BMC Cell Biol. 2009;10:20. doi: 10.1186/1471-2121-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdimarsdottir G, Goumans M-J, Rosendahl A, Brugman M, Itoh S, Lebrin F, Sideras P, ten Dijke P. Stimulation of Id1 expression by bone morphogenetic protein is sufficient and necessary for bone morphogenetic protein-induced activation of endothelial cells. Circulation. 2002;106:2263–2270. doi: 10.1161/01.cir.0000033830.36431.46. [DOI] [PubMed] [Google Scholar]
- 31.Koinuma D, Tsutsumi S, Kamimura N, Imamura T, Aburatani H, Miyazono K. Promoter-wide analysis of Smad4 binding sites in human epithelial cells. Cancer Sci. 2009;100:2133–2142. doi: 10.1111/j.1349-7006.2009.01299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev. Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss A, Charbonnier E, Ellertsdottir E, Tsirigos A, Wolf C, Schuh R, Pyrowolakis G, Affolter M. A conserved activation element in BMP signaling during Drosophila development. Nat. Struct. Mol. Biol. 2010;17:69–76. doi: 10.1038/nsmb.1715. [DOI] [PubMed] [Google Scholar]
- 37.Li F, Lan Y, Wang Y, Wang J, Yang G, Meng F, Han H, Meng A, Wang Y, Yang X. Endothelial Smad4 maintains cerebrovascular integrity by activating N-cadherin through cooperation with notch. Dev. Cell. 2011;20:291–302. doi: 10.1016/j.devcel.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Arnold SJ, Maretto S, Islam A, Bikoff EK, Robertson EJ. Dose-dependent Smad1, Smad5 and Smad8 signaling in the early mouse embryo. Dev. Biol. 2006;296:104–118. doi: 10.1016/j.ydbio.2006.04.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.John S, Sabo PJ, Thurman RE, Sung M-H, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashe HL, Mannervik M, Levine M. Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development. 2000;127:3305–3312. doi: 10.1242/dev.127.15.3305. [DOI] [PubMed] [Google Scholar]
- 42.Wharton SJ, Basu SP, Ashe HL. Smad affinity can direct distinct readouts of the embryonic extracellular Dpp gradient in Drosophila. Curr. Biol. 2004;14:1550–1558. doi: 10.1016/j.cub.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 43.Island M-L, Jouanolle A-M, Mosser A, Deugnier Y, David V, Brissot P, Loreal O. A new mutation in the hepcidin promoter impairs its BMP response and contributes to a severe phenotype in HFE related hemochromatosis. Haematologica. 2009;94:720–724. doi: 10.3324/haematol.2008.001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moustakas A, Heldin C-H. Non-Smad TGF-beta signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 45.Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134:2709–2718. doi: 10.1242/dev.004184. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Kennard S, Lilly B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ. Res. 2009;104:466–475. doi: 10.1161/CIRCRESAHA.108.184846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doi H, Iso T, Sato H, Yamazaki M, Matsui H, Tanaka T, Manabe I, Arai M, Nagai R, Kurabayashi M. Jagged1-selective notch signaling induces smooth muscle differentiation via a RBP-Jkappa-dependent pathway. J. Biol. Chem. 2006;281:28555–28564. doi: 10.1074/jbc.M602749200. [DOI] [PubMed] [Google Scholar]
- 48.High FA, Lu MM, Pear WS, Loomes KM, Kaestner KH, Epstein JA. Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc. Natl Acad. Sci. USA. 2008;105:1955–1959. doi: 10.1073/pnas.0709663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niessen K, Zhang G, Ridgway JB, Chen H, Yan M. ALK1 signaling regulates early postnatal lymphatic vessel development. Blood. 2010;115:1654–1661. doi: 10.1182/blood-2009-07-235655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu-Lowe DD, Chen E, Zhang L, Watson KD, Mancuso P, Lappin P, Wickman G, Chen JH, Wang J, Jiang X, et al. Targeting activin receptor-like kinase 1 inhibits angiogenesis and tumorigenesis through a mechanism of action complementary to anti-VEGF therapies. Cancer Res. 2011;71:1362–1373. doi: 10.1158/0008-5472.CAN-10-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lebrin F, Srun S, Raymond K, Martin S, van den Brink S, Freitas C, Breant C, Mathivet T, Larrivee B, Thomas J-L, et al. Thalidomide stimulates vessel maturation and reduces epistaxis in individuals with hereditary hemorrhagic telangiectasia. Nat. Med. 2010;16:420–428. doi: 10.1038/nm.2131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data