Scraping fibrosis: Expressway to the core of fibrosis (original) (raw)
. Author manuscript; available in PMC: 2011 Nov 17.
Published in final edited form as: Nat Med. 2011 May;17(5):552–553. doi: 10.1038/nm0511-552
Abstract
Animal experiments using single organs as models of fibrosis spur therapeutic development toward promising targets, but testing of these therapies in human fibrosis yielded disappointing results and limited efficacy. Finding core pathways relevant in different organs that can become fibrotic will uncover molecules that will prove useful as therapeutic targets in many species, including humans. In ‘Bench to Bedside’, Scott Friedman, Wajahat Mehal and John Iredale discuss this new paradigm in fibrosis research and its potential as a new drug development approach. In ‘Bedside to Bench’, Alison Eddy peruses how the protein encoded by UMOD, a gene linked to variable risk for chronic kidney disease and hypertension in humans, may have a role in fibrosis and kidney disease. Uncovering the normal function of UMOD and its gene variants will shed light on the pathogenesis of chronic kidney disease and aid in the discovery of new targets for kidney fibrosis and hypertension.
Studies of organ fibrosis have flourished over the last decade, identifying a growing number of molecules that regulate the development of fibrosis in experimental models. Fibrotic diseases account for up to 45% of deaths in the developed world, yet there are no approved antifibrotic therapies1. Translating advances in experimental fibrosis to clinical management has been challenging, in part, because there are now many candidate antifibrotic targets, and most are tested in highly controlled experimental settings, usually only in a single organ type and species. The therapeutic implications of these animal studies are therefore only relevant to that specific rodent strain and within the constraints of the experimental design. Moreover, the field is driven by a bias toward publishing the most novel findings and away from confirmatory or relevant negative observations.
With perhaps hundreds of molecules contributing to fibrosis, and many interconnections between them, the field of fibrosis is beginning to resemble an urban sprawl, with a growing number of connections that obscure the expressways to developing successful therapies.
The concept of ‘core’ and ‘regulatory’ pathways in fibrosis may help tackle this conundrum. Whereas a core pathway is essential to convert an initial stimulus to the development of fibrosis, regulatory pathways are those that can influence the core pathway but do not directly convert the initial stimulus into the basic component of fibrosis. Therefore, cells and molecules along the core pathway are essential to fibrosis, and their targeting may be sufficient to limit progression. The regulatory pathways may have substantial effects on fibrosis but will also have greater variability between organs, species and individuals, challenging the value of these targets. For this core-versus-regulatory pathway approach to have utility in fibrosis, criteria should be established for identifying components of these pathways, and, unfortunately, the reductionist approach used in animal models rarely makes this distinction.
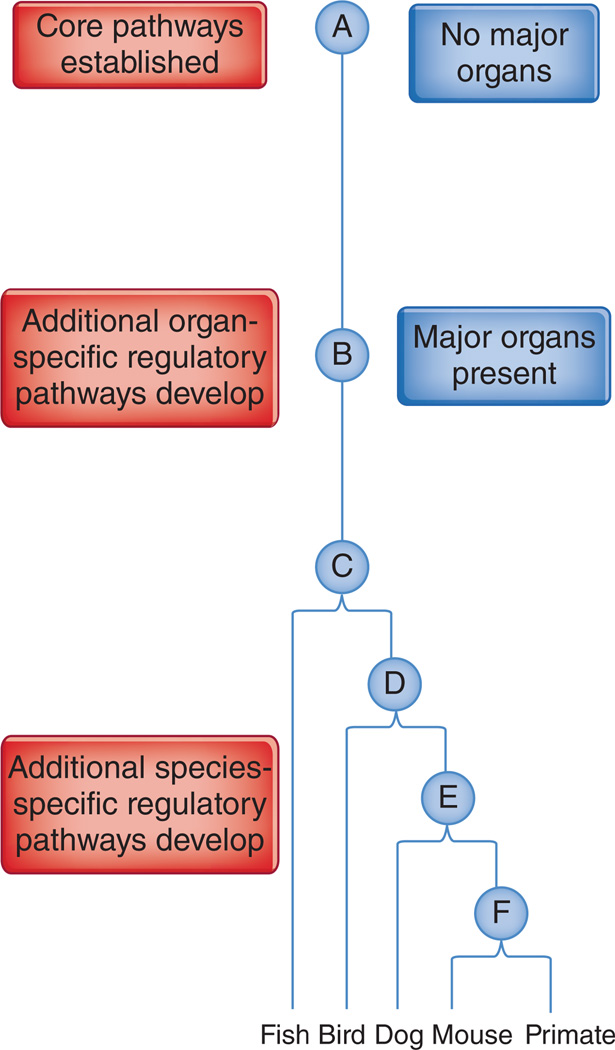
Evidence of evolutionary pathway conservation can help identify core pathways, assuming that core pathways in fibrosis were present at an early evolutionary stage, with regulatory pathways developing later, possibly in an organ- and species-specific manner—a molecule found to be important in a number of different organs or species will probably be part of a core pathway (Fig. 1). This concept is useful given that the differences between organs are evolutionarily older and more substantial than the differences between individual mammalian species. If a pathway is shown to be key in multiple organs in mice, which is easier than doing so in multiple species, it might have predated speciation and be shared by different species.
Figure 1.
Organ specialization predated mammalian speciation and provides a practical approach to uncover core and regulatory pathways as antifibrotic targets. Integrating the concept of core and regulatory pathways into an evolutionary timeline will help to establish a rationale for comparing data among different organs, showing common ancestors at successive points of evolution. At an early evolutionary point, A, an organism did not have major organs—heart, lungs and liver—but then major organs developed, B, followed by the evolution of a common ancestor into all vertebrates, including fish, birds and mammals (C–F). A strict criterion for a core fibrotic pathway would be the contribution of a molecule within a fibrosis pathway in multiple organs, rather than to fibrosis of the same organ in single or different mammals.
Yet regulatory pathways can still be relevant, as they can unearth therapeutic targets that may be inherently more organ specific. For example, natural killer (NK) cell activity is involved in limiting liver fibrosis by killing activated stellate cells2–4, the fibrogenic cells in liver, despite NK cells not being required for the development of liver fibrosis. NK cells may be on a regulatory pathway probably limited to the liver, as they are present in very low numbers in other organs. NK cells could be potential targets to limit fibrosis in individuals with hepatitis C without compromising wound repair at other sites.
A recent study by Barry-Hamilton et al.5 exploring the role of the enzyme lysyl oxidase-2 (LOXL2) in fibrosis represents a comprehensive approach in which they have probably identified a core pathway. It showed a vital role for LOXL2 in mouse experimental models of liver and lung fibrosis, as well as in subcutaneous tissue fibrosis in response to tumor implants. LOXL2 expression also correlated with fibrosis in human tissues, including chronic hepatitis C and idiopathic pulmonary fibrosis. These findings, therefore, meet the above criteria for placing LOXL2 in a core pathway. Although the role of LOXL2 function in a core pathway is based largely on mouse data, on the basis of these data LOXL2 may be vital for liver fibrosis in other species, including humans. This contrasts with other experimental studies that have shown the role of a molecule in fibrosis only in the mouse liver, bringing into question their relevance to human liver fibrosis.
The tendency to perform experiments in one single organ results from the organ-centered organization of clinical medicine. But, as we have argued above, to increase the relevance of molecules and pathways derived from experimental mouse data to humans, a multiorgan approach is vital. Regulatory pathways will be restricted to an organ, and the same comprehensive approach will be required to identify this restriction.
Any data showing antifibrotic activity, regardless of the model and species, could indicate that a molecule is important in a number of species, and the distinction between core and regulatory pathways may be arbitrary. But this is not necessarily the case. Studies exploring the role of a particular molecule in liver fibrosis in rodents using carbon tetrachloride to induce chronic injury can show positive results, which are confirmed in a second model such as that using thioacetamide6. Both these models initiate fibrosis through hepatocyte injury, producing concordant results that may not yet reflect the more complex milieu seen in advanced human fibrosis or cirrhosis. But these findings still provide rigorous data, albeit within the more limited context of a single organ and species, as in a mouse study that suggested the chemokine Ccl5 as a promising antifibrotic target in the liver7.
The use of knockout and transgenic mouse models in which candidate molecules regulating fibrosis have been genetically knocked out or overexpressed poses a challenge to translating initial findings into a different species. Confirming the role of a molecule in another organ type after the initial positive result will be more informative than testing the same organ with a different toxin—concordant results may suggest a core pathway function, whereas discordant results may indicate a role in an organ specific regulatory pathway.
The impressive progress resulting from a focused, organ-specific approach to experimental fibrosis has yielded many molecular targets for therapy, yet, paradoxically, it has also generated uncertainty about the relevance of each molecule to human fibrosis. Viewing fibrosis through the prism of core and regulatory pathways provides one approach to identifying the best ways toward effective antifibrotic drugs. It would be useful to test this hypothesis by considering some of the clinical trials of liver fibrosis therapies conducted to date, which failed to show efficacy, including trials of interleukin-10 (ref. 8), γ-interferon9, α-interferon10 and a peroxisome proliferator–activated receptor-γ ligand, farglitazar11. Would each of these agents have had antifibrotic activity in different organs and species, and thereby have meet the criteria of a core pathway constituent? Will targeting a core pathway constituent yield more efficacy but more off-target effects? For example, transforming growth factor-β1 antagonism has broad antifibrotic activity in many organs and species12, yet its pleiotropic effects limit the safety of systemic administration.
A hybrid strategy combining the identification of core pathway constituents and the development of targeted approaches of cell- or tissue-specific delivery may exploit the coreversus-regulatory paradigm and limit collateral, unwanted effects. As we turn the corner toward antifibrotic therapies, these considerations may provide a more direct route to success.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Contributor Information
Wajahat Z Mehal, Section of Digestive Diseases, Yale University, New Haven, Connecticut, USA.
John Iredale, Medical Research Council Centre for Inflammation Research, Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK.
Scott L Friedman, Division of Liver Diseases, Mount Sinai School of Medicine, New York, New York, USA. wajahat.mehal@yale.edu.
References
- 1.Wynn TA. J. Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morishima C, et al. Hepatology. 2006;43:573–580. doi: 10.1002/hep.21073. [DOI] [PubMed] [Google Scholar]
- 3.Melhem A, et al. J. Hepatol. 2006;45:60–71. doi: 10.1016/j.jhep.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Radaeva S, et al. Gastroenterology. 2006;130:435–452. doi: 10.1053/j.gastro.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 5.Barry-Hamilton V, et al. Nat. Med. 2010;16:1009–1017. doi: 10.1038/nm.2208. [DOI] [PubMed] [Google Scholar]
- 6.Russo FP, et al. Gastroenterology. 2006;130:1807–1821. doi: 10.1053/j.gastro.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Berres ML, et al. J. Clin. Invest. 2010;120:4129–4140. doi: 10.1172/JCI41732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson DR, et al. Hepatology. 2003;38:859–868. doi: 10.1053/jhep.2003.50427. [DOI] [PubMed] [Google Scholar]
- 9.Pockros PJ, et al. Hepatology. 2007;45:569–578. doi: 10.1002/hep.21561. [DOI] [PubMed] [Google Scholar]
- 10.Di Bisceglie AM, et al. N. Engl. J. Med. 2008;359:2429–2441. doi: 10.1056/NEJMoa0707615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHutchison J, et al. Gastroenterology. 2010;138:1365–1373. doi: 10.1053/j.gastro.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Rosenbloom J, Castro SV, Jimenez SA. Ann. Intern. Med. 2010;152:159–166. doi: 10.7326/0003-4819-152-3-201002020-00007. [DOI] [PubMed] [Google Scholar]