A public genome-scale lentiviral expression library of human ORFs (original) (raw)
. Author manuscript; available in PMC: 2012 Feb 1.
Published in final edited form as: Nat Methods. 2011 Jun 26;8(8):659–661. doi: 10.1038/nmeth.1638
Abstract
Functional characterization of the human genome requires tools for systematically modulating gene expression in both loss- and gain-of-function experiments. We describe the production of a sequence-confirmed, clonal collection of over 16,100 human open-reading frames (ORFs) encoded in a versatile Gateway vector system. Utilizing this ORFeome resource, we created a genome-scale expression collection in a lentiviral vector, thereby enabling both targeted experiments and high-throughput screens in diverse cell types.
While recent technological advances provide the means to efficiently scan the human genome to identify genes associated with diseases1-2, the subsequent functional characterization of these genes is a bottleneck in translating these discoveries into mechanistic insights and ultimately into therapeutics. Genome-scale RNA interference reagents have recently been created to enable systematic loss-of-function mammalian genomics3. To perform complementary gain-of-function gene studies, comparable libraries of arrayed cDNAs or open-reading frames (ORFs) are required along with efficient methods to employ these reagents in cell-based assays.
We4-5 and others6-9 have previously reported the construction of genome-scale ORFeome collections. These collections are useful templates for subcloning9 or recombinational transfer8 between vectors and protein production8, and they enable certain applications including interaction mapping10, but there are limits to the direct applications of these collections. They vary dramatically in terms of gene representation, format, and functionality, as well as quality measures such as the extent of clonality, sequence annotation, and experimental validation (see Supplementary Table 1). Tellingly, gain-of-function screening now lags behind the use of RNAi.
Here, we report the creation and characterization of two publicly available genome-scale human ORFeome collections: the human ORFeome version 8.1 Entry Clone Collection (hORFeome V8.1) and the CCSB-Broad Lentiviral Expression Library. Together, these collections are: (i) extensive, comprising 16,172 distinct ORFs mapping to 13,833 genes, (ii) clonal and sequenced, as each ORF plasmid is derived from a single bacterial colony and nearly all clones are fully sequenced, (iii) versatile, due to use of Gateway recombinational cloning11-12 (iv) enabling of cell-based functional screens, as the Expression Library encodes these clones in a lentiviral expression vector that produces consistent titers and gene expression levels and permits delivery to most cell types, and (v) available via ORFeome Collaboration (Supplementary Note 1).
We assembled these collections in four phases: First we expanded our previous collections to 19,281 ORFs in polyclonal format largely using existing protocols4-5; second, we derived clonal plasmid isolates from single bacterial colonies; third, we sequenced these clonal isolates and used the sequence data to choose clones for inclusion in hORFeome V8.1; and fourth we transferred clones to a lentiviral expression vector to create the CCSB-Broad Lentiviral Expression Library.
We expanded our library by transferring recently available ORFs from Mammalian Gene Collection (MGC)9 cDNAs into the Gateway system using directed PCR4-5 to create Entry vector clones while removing stop codons (Fig. 1a, top). To maximize throughput during this initial phase, clones were represented as a non-clonal pool of bacteria derived from recombinational cloning of PCR products. We next resolved this polyclonal library into clonal isolates using a robust and efficient workflow (Fig. 1a, middle) in which we isolated two colonies per ORF bacterial stock (see online Methods, Supplementary Note 2) from which we prepared ORF templates for sequencing.
Figure 1. Overview of hORFeome V8.1.
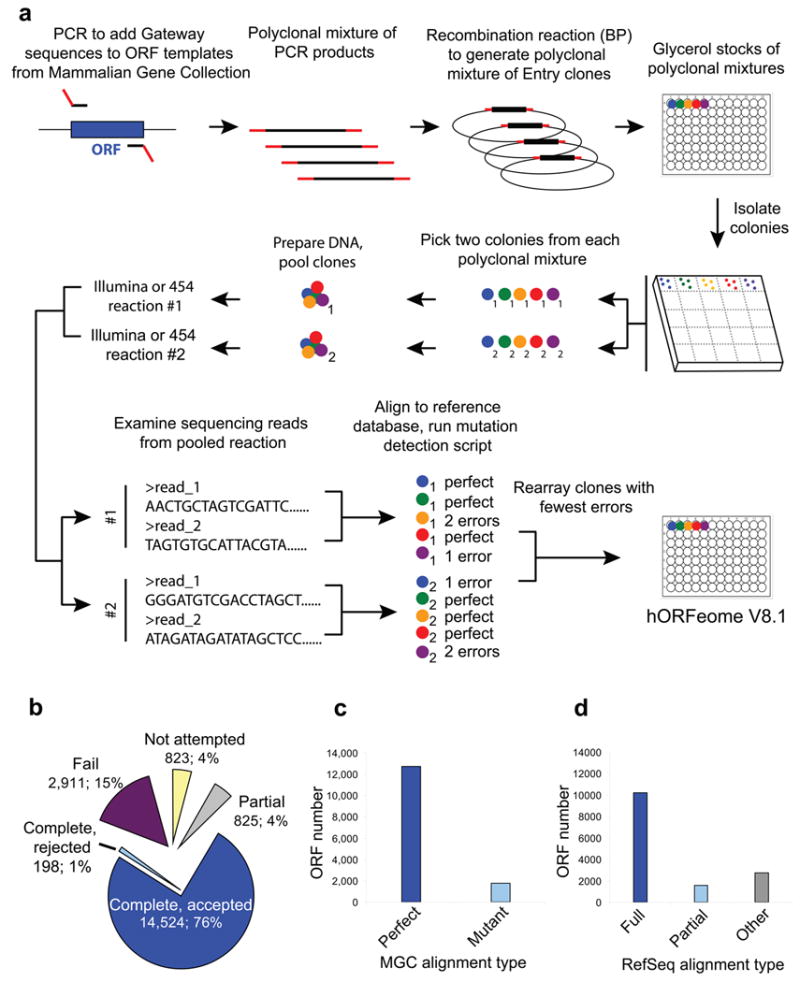
(a) Schematic of hORFeome V8.1 creation. Templates from the Mammalian Gene Collection (MGC) were transferred into the Gateway system via PCR and recombinational cloning, resolved as clonal isolates, fully sequenced and rearrayed. (b) Sequencing outcomes for 19,281 ORF samples in polyclonal format, from which single colonies were isolated. 14,524 ORFs were fully sequenced and accepted into the final collection (Complete, accepted). 198 ORFs were fully sequenced, but rejected for lacking a start codon (Complete, rejected). 825 ORFs were partially sequenced, including undetermined nucleotides (Partial). 823 ORFs were made clonal but were intentionally not sequenced, since these ORFs were isoforms of other ORFs in the sequencing pool and could cause unambiguous read mapping (Not attempted). See Supplementary Figure 3 for more details. (c) Alignment of the 14,524 completely sequenced clones with MGC templates. 12,736 clones have identical sequence as templates or have one synonymous error only (Perfect), and another 1,788 clones have additional mutations (Mutant). (d) Alignment of the 14,524 completely sequenced clones with NCBI RefSeq transcripts. 10,216 ORFs represent full length coding sequences with > 99% homology (Full), 1,545 ORFs were partial length coding sequences with > 85% homology (Partial) and 2,763 clones fell into other categories (Other). See Supplementary Figure 4 for more details.
We developed an optimized process to leverage next-generation sequencing and efficient alignment algorithms13 to efficiently sequence large numbers of ORF clones at high coverage (Fig. 1a, bottom). Clonal isolates for each ORF were pooled. Using Illumina sequencing technology, we compared efficiencies of sequencing full vectors versus purified ORF inserts only (Supplementary Fig. 1a). While purified ORF inserts yielded higher median sequence coverage, the added clone manipulation led to substantially greater coverage variability (Supplementary Fig. 1b-g, Supplementary Table 2) so we proceeded to sequence pools of full entry-clone plasmids. For some clone pools, we employed an alternative approach in which we PCR-amplified ORF sequences from individual bacterial colonies and sequenced the amplicons in a multiplexed, pooled format previously reported14 using 454 technology. Both methods were effective at sequencing ORF clones (Supplementary Figure 2), but our protocol based on Illumina technology yielded higher yields at lower cost per attempted clone (data not shown), and was therefore used to sequence the majority (84%) of the collection.
To assemble ORF sequences, reads from each clone pool were aligned to MGC reference sequences. Adequate reads were obtained to produce full ORF alignments for > 27,000 clonal isolates from 14,722 polyclonal ORFs, at the fold-coverage required for accurate base-calling (Supplementary Fig. 3). ORF sequences were annotated for mismatches, insertions, and deletions (Supplementary Tables 3,4). To evaluate the sequence accuracy of multiplexed Illumina and 454 sequencing combined with our automated alignment algorithms, we re-sequenced >121,000 nucleotides from 287 ORFs by the Sanger method, and found a confirmation rate of >99.99% of nucleotides. For each original ORF stock, the clonal isolate that most closely matched the MGC reference sequence was selected for inclusion in the hORFeome V8.1 collection. 198 clones with missing start codons were omitted.
Of 14,524 retained sequenced ORFs (Figure 1b), 82% (12,736) were either sequence-identical to the MGC reference or had one synonymous error, and comprise the majority of the hORFeome V8.1 collection (Fig. 1c, Supplementary Fig. 3). Another component of the V8.1 collection, denoted as the hORFeome V8.1 Mutant Subcollection, consists of 1,788 ORFs that had more than one synonymous or any non-synonymous mutations or other errors, and were retained since these plasmids may prove useful in some applications. We supplemented the fully sequenced set of ORFs with 825 clones comprising the hORFeome V8.1 Partially Sequenced Subcollection, including 597 clones from our recently described subcollection of kinases and kinase-related ORFs (clonal isolates, end-read Sanger sequencing in 2 directions)15 (see Supplementary Note 3) and 228 clones that were sequenced using next-generation technology over only part of the intended MGC ORF sequences. Finally, we denote the 823 clonal versions of isoforms that were removed prior to pooled sequencing as the hORFeome V8.1 Unsequenced Subcollection. Overall, hORFeome V8.1 includes 16,172 clonal ORFs, mapping to 13,833 human genes, of which 14,524 clones (90%) for 12,940 genes (94%) are fully sequenced (Supplementary Fig. 3, Supplementary Tables 3,4).
We next determined which currently annotated human transcripts are represented in hORFeome V8.1. The 14,524 fully sequenced library clones were mapped to National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) coding transcripts, and we found that 10,216 ORFs map with > 99% homology and constitute full length coding sequences (Fig. 1d, Supplementary Fig. 4). 1,545 additional ORFs represent partial coding sequences. The remaining 2,763 sequenced ORFs map to non-coding transcripts or to RefSeq transcripts with lower homology or are not currently found in RefSeq. Since the original MGC cDNA source templates for these clones were derived from expressed cellular transcripts, some of these non-full length clones may represent un- or mis-annotated but physiologically relevant transcripts. Indeed, incomplete knowledge of the transcriptome is a major challenge to obtaining a comprehensive ORF resource (Supplementary Note 4).
hORFeome V8.1 enables many applications as it permits rapid ORF shuttling into any Gateway-compatible expression vector. To enable large-scale screening of this collection in mammalian cells, we developed, optimized and validated a series of Gateway-compatible mammalian expression vectors (pLX series, Supplementary Fig. 5) encoding numerous desirable elements (see online Methods). We elected to shuttle the entire hORFeome V8.1 collection into the pLX304-Blast-V5 vector to create the CCSB-Broad Lentiviral Expression Library (Fig. 2a).
Figure 2. Overview and performance of the CCSB-Broad Lentiviral Expression Library.
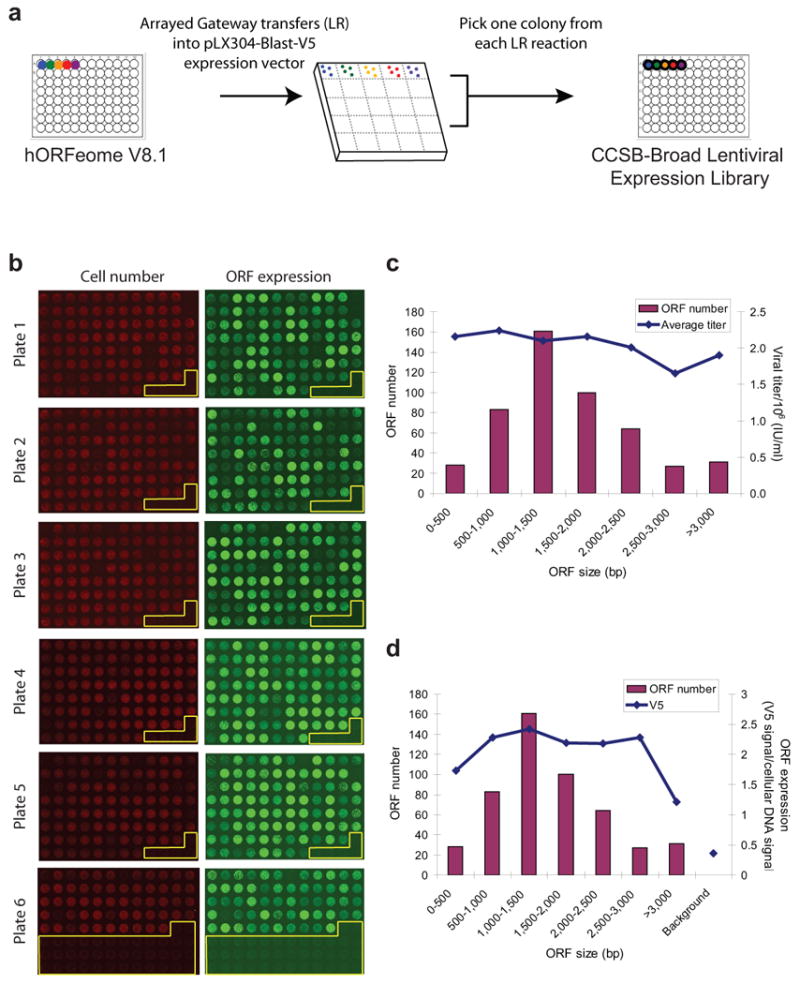
(a) Schematic of the creation of the CCSB-Broad Lentiviral Expression Library. pLX304-Blast-V5 is a custom lentiviral vector validated for high-throughput screening encoding Blasticidin (Blast) resistance and a C-terminal V5-epitope tag. (b) Evaluation of high-throughput ORF transduction and expression in A549 lung cancer cell lines. The micrographs show images of cells stained for cellular DNA (to assess cell number) and with antibodies recognizing the V5 epitope (to assess ORF expression) after lentiviral infection and three days of growth in blasticidin. Wells in which no virus was added are highlighted with yellow outline. (c) Distribution of ORF sizes and average viral titer as a function of ORF size. (d) ORF expression as a function of ORF size. ORFs larger than 3 kb showed a decreased yet detectable above-background level of expression. Background was assessed from cells expressing a control vector without V5 expression.
We conducted a pilot experiment on 509 ORF clones to assess: (i) protocols to transfer the entry library, (ii) high-throughput production of DNA and virus, and (iii) ORF expression in A549 cells (Fig. 2b-d, Supplementary Figs. 6-9). Plasmid DNA production and viral packaging were achieved in 96-well format with consistent DNA yields and titers averaging 2.1 × 106 infectious units (IU)/ml (Fig. 2c, Supplementary Fig. 7a). Titers were preserved across all ORF sizes (Fig. 2c, Supplementary Fig. 8a). We assessed ORF expression via quantification of V5-epitope tag expression and observed that approximately 90% of ORF lentiviruses induced expression signals greater than 2 standard deviations above the control mean (Fig. 2b, d, Supplementary Figs. 7b, 8b, 9).
Using the optimized protocols, we then produced the CCSB-Broad Lentiviral Expression Library in the pLX304-Blast-V5 vector, successfully isolating a single bacterial colony from 98.5% of reactions (15,935 total clones). To estimate the accuracy of the final collection of expression vectors, we performed end-read sequencing of 325 colonies and confirmed 98.2% accurate transfers (see online Methods). The utility of this resource for systematic functional genomic screens in mammalian cells is illustrated by recent results from a screen of a pilot subset of this collection (597 genes), which identified novel mediators of resistance to RAF inhibition in melanoma15. Additional pilot experiments confirm that this resource enables other readouts including immunofluorescence (Supplementary Figure 10).
In summary, we report here the construction of the most fully sequenced, flexible and annotated version of the human ORFeome to date. The entire collection, comprising both source (entry) clones and lentivirus vector expression clones, is available without restriction through the ORFeome Collaboration (Supplementary Note 1). We anticipate that these collections will greatly facilitate the systematic functional assessment of human genes that mediate cellular phenotypes.
Supplementary Material
1
Supplementary Figure 1 Pilot experiments to optimize pooling strategy for next-generation sequencing of ORF clones.
Supplementary Figure 2 Coverage histograms of sequencing hORFeome V8.1.
Supplementary Figure 3 Flowchart of hORFeome V8.1 creation.
Supplementary Figure 4 Alignment results of 14,524 completely sequenced clones with current NCBI RefSeq transcripts.
Supplementary Figure 5 Plasmid maps of pLX lentiviral expression vectors created as part of this study.
Supplementary Figure 6 Confirmation of viral preparations.
Supplementary Figure 7 Determination of virus titer and ORF expression.
Supplementary Figure 8 Virus titer and ORF expression are maintained across a wide range of ORF lengths.
Supplementary Figure 9 Western blot showing expressed ORFs.
Supplementary Figure 10 Viral preparations enable immunofluoresence high throughout screens.
Supplementary Table 1a Clonal and sequenced ORF Gateway entry clone collections
Supplementary Table 1b Comparison of Nomura and CCSB-Broad ORF collections
Supplementary Table 2 Illumina sequencing pilot data
Supplementary Table 3 Overview of next generation sequencing results
Supplementary Table 4 Annotated list of hORFeome V8.1 and CCSB-Broad Lentiviral Expresson Library.
Supplementary Note 1 Availability of clones and distribution procedures
Supplementary Note 2 Pilot experiments to determine number of colonies to isolate per polyclonal ORF.
Supplementary Note 3 Supplementing hORFeome V8.1 with kinase ORFs.
Supplementary Note 4 Challenges of completing the human ORFeome
Supplementary Note 5 Detailed high-throughput protocol of single colony isolation.
Supplementary Note 6 Details of pooled sequencing protocol optimization experiments.
Supplementary Note 7 Computing virus titers.
Supplementary Note 8 Li-COR in-cell Western and immunoblotting.
Acknowledgments
We acknowledge B. Piqani, I. Budianto, D. Szeto, T. Hirozane-Kishikawa, V. Swearingen and A. MacWilliams who participated in the generation of various intermediate ORFeome libraries, T. Nieland who offered technical advice, S. Hoang who provided automation support, J. Bochicchio, S. Young, A. Berlin, C. Russ and the Broad Institute Genetic Sequencing Platform who assisted with sequencing and alignment of reads, M. Garber who assisted with ORF sequence annotation, and J. Zhao, T. Roberts and T. Golub who participated in the generation of the kinase sub-collection. This work was supported by Broad Institute Scientific Planning and Allocation of Resources Committee (SPARC) funding, The Ellison Foundation (D.E.H., M.V.), DFCI Institute Sponsored Research funds to CCSB and CCGD and NIH R33 CA128625 (W.C.H., D.E.R., D.E.H). M.V. is a “Chercheur Qualifié Honoraire” from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
Author Contributions: J.S.B., Xia.Y. and D.E.R. wrote the manuscript and designed and supervised the process of creating clonal, sequenced ORFs and expression vectors from starting polyclonal ORF pools via Illumina sequencing. D.E.H, K.S.-A. and M.V. designed and supervised the process of cloning ORFs from MGC cDNA templates to generate Gateway entry clones as well as creating clonal, sequenced ORFs via 454 sequencing. Xin.Y., D.B., L.G., and R.R.M. created starting polyclonal ORF pools from MGC cDNA templates. T.H., Y.S., C.F., and C.L. performed bioinformatic analyses. R.L. produced the clonal ORF library and did LR reactions, transformations, colony picking and ORF DNA preparations. J.S.B., S.R.T., H.H., R.R.M., K.S.-A., and C.M.J. created the kinase sub-library and performed pilot experiments. O.A. and J.S.B. created and tested pLX vectors. J.S.B., T.B., T.H., C.F., C.L. and T.M.G analyzed sequencing data. T.B., T.M.G., Xia.Y. and D.E.R. conducted BLAST analysis of ORF sequences. C.N., Xia.Y. and S.S. created virus from ORFs. C.N., S.R.T., C.M.J. and Xia.Y. performed ORF expression assays. M.V., W.C.H., D.E.H. and D.E.R. supervised the project.
Competing Interest Statement: No competing interests to declare
References
- 1.Meyerson M, Gabriel S, Getz G. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 2.1000 Genomes Project Consortium et al. Nature. 2010;467:1061–1073. [Google Scholar]
- 3.Moody SE, Boehm JS, Barbie DA, Hahn WC. Cur Opin Mol Ther. 2010;12:284–293. [PubMed] [Google Scholar]
- 4.Rual JF, et al. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamesch P, et al. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolfs A, et al. PLoS One. 2008;3:e1528. doi: 10.1371/journal.pone.0001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bechtel S, et al. BMC Genomics. 2007;8:399. doi: 10.1186/1471-2164-8-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goshima N, et al. Nat Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- 9.MGC Project Team et al. Genome Res. 2009;19:2324–2333. doi: 10.1101/gr.095976.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rual JF, et al. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 11.Hartley JL, Temple GF, Brasch MA. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walhout AJ, et al. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salehi-Ashtiani K, et al. Nat Methods. 2008;5:597–600. doi: 10.1038/nmeth.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannessen CM, et al. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillier L, Green P. PCR Methods Appl. 1991;1:124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]
- 17.Moffat J, et al. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 18.Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. J Gen Virol. 1991;72:1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 19.Zufferey R, Donello JE, Trono D, Hope TJ. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
Supplementary Figure 1 Pilot experiments to optimize pooling strategy for next-generation sequencing of ORF clones.
Supplementary Figure 2 Coverage histograms of sequencing hORFeome V8.1.
Supplementary Figure 3 Flowchart of hORFeome V8.1 creation.
Supplementary Figure 4 Alignment results of 14,524 completely sequenced clones with current NCBI RefSeq transcripts.
Supplementary Figure 5 Plasmid maps of pLX lentiviral expression vectors created as part of this study.
Supplementary Figure 6 Confirmation of viral preparations.
Supplementary Figure 7 Determination of virus titer and ORF expression.
Supplementary Figure 8 Virus titer and ORF expression are maintained across a wide range of ORF lengths.
Supplementary Figure 9 Western blot showing expressed ORFs.
Supplementary Figure 10 Viral preparations enable immunofluoresence high throughout screens.
Supplementary Table 1a Clonal and sequenced ORF Gateway entry clone collections
Supplementary Table 1b Comparison of Nomura and CCSB-Broad ORF collections
Supplementary Table 2 Illumina sequencing pilot data
Supplementary Table 3 Overview of next generation sequencing results
Supplementary Table 4 Annotated list of hORFeome V8.1 and CCSB-Broad Lentiviral Expresson Library.
Supplementary Note 1 Availability of clones and distribution procedures
Supplementary Note 2 Pilot experiments to determine number of colonies to isolate per polyclonal ORF.
Supplementary Note 3 Supplementing hORFeome V8.1 with kinase ORFs.
Supplementary Note 4 Challenges of completing the human ORFeome
Supplementary Note 5 Detailed high-throughput protocol of single colony isolation.
Supplementary Note 6 Details of pooled sequencing protocol optimization experiments.
Supplementary Note 7 Computing virus titers.
Supplementary Note 8 Li-COR in-cell Western and immunoblotting.