Frequent Mutation of Isocitrate Dehydrogenase (IDH)1 and IDH2 in Cholangiocarcinoma Identified Through Broad-Based Tumor Genotyping (original) (raw)
The results of mutational profiling of 287 patients with gastrointestinal cancer are presented, identifying for the first time IDH1 mutations in a significant subset of patients with intrahepatic cholangiocarcinoma.
Keywords: Biliary tract cancer, Cholangiocarcinoma, IDH1, IDH2, 2-Hydroxyglutarate, Mutation
Abstract
Cancers of origin in the gallbladder and bile ducts are rarely curable with current modalities of cancer treatment. Our clinical application of broad-based mutational profiling for patients diagnosed with a gastrointestinal malignancy has led to the novel discovery of mutations in the gene encoding isocitrate dehydrogenase 1 (IDH1) in tumors from a subset of patients with cholangiocarcinoma. A total of 287 tumors from gastrointestinal cancer patients (biliary tract, colorectal, gastroesophageal, liver, pancreatic, and small intestine carcinoma) were tested during routine clinical evaluation for 130 site-specific mutations within 15 cancer genes. Mutations were identified within a number of genes, including KRAS (35%), TP53 (22%), PIK3CA (10%), BRAF (7%), APC (6%), NRAS (3%), AKT1 (1%), CTNNB1 (1%), and PTEN (1%). Although mutations in the metabolic enzyme IDH1 were rare in the other common gastrointestinal malignancies in this series (2%), they were found in three tumors (25%) of an initial series of 12 biliary tract carcinomas. To better define IDH1 and IDH2 mutational status, an additional 75 gallbladder and bile duct cancers were examined. Combining these cohorts of biliary cancers, mutations in IDH1 and IDH2 were found only in cholangiocarcinomas of intrahepatic origin (nine of 40, 23%) and in none of the 22 extrahepatic cholangiocarcinomas and none of the 25 gallbladder carcinomas. In an analysis of frozen tissue specimens, IDH1 mutation was associated with highly elevated tissue levels of the enzymatic product 2-hydroxyglutarate. Thus, IDH1 mutation is a molecular feature of cholangiocarcinomas of intrahepatic origin. These findings define a specific metabolic abnormality in this largely incurable type of gastrointestinal cancer and present a potentially new target for therapy.
Introduction
High-throughput tumor genotyping is becoming an integral part of the initial clinical management of cancer patients [1, 2]. The ability to direct targeted cancer agents based on tumor-specific molecular abnormalities has had a clear and positive impact on the management of patients with non-small cell lung, breast, colon, skin, and gastrointestinal tumors [3–7]. Evaluating a broad range of key cancer gene mutations across diverse cancers has the potential for identifying new therapeutically relevant mutations. Whereas genetic signatures in colorectal and pancreatic cancers have been well defined [8, 9], this approach can expand our knowledge of less-studied gastrointestinal malignancies such as biliary tract carcinomas.
Biliary tract carcinomas encompass gallbladder carcinoma and bile duct cancer (cholangiocarcinoma). Nearly 12,000 new cases are identified in the U.S. annually and only 10% of patients present with early-stage disease that can be cured through surgical resection [10, 11]. Even with combination cytotoxic chemotherapy, the prognosis for patients remains poor and the median survival duration is ∼1 year [12]. New and effective molecularly targeted therapies are urgently needed [13]. Although known mutations in the adenomatous polyposis coli (APC), BRAF, deleted in pancreatic carcinoma locus 4 (DPC4), epidermal growth factor receptor (EGFR), Kirsten-ras (KRAS), phosphatidylinositol 3-kinase, catalytic, alpha (PIK3CA), and p53 tumor suppressor (TP53) genes in cholangiocarcinoma and/or gallbladder carcinoma may help prioritize patient assignment to clinical trials of new drugs [14–19], the full spectrum of relevant mutations across the biliary tract carcinoma subtypes has not been clearly defined.
Somatic mutations in the cytoplasmic and peroxisomal isocitrate dehydrogenase gene IDH1 and its mitochondrial counterpart IDH2 have been identified through large genomic screens of human leukemias, glioblastomas, and sarcomas. At the same time, experimental studies have implicated these mutations as driver events in oncogenesis. These mutations cause a single amino acid change at a conserved arginine residue within the isocitrate binding site of IDH1 (R132) or IDH2 (R172, R140), resulting in decreased enzymatic activity for oxidative decarboxylation of isocitrate to α-ketoglutarate [20–22]. Although the oncogenic mechanism is not certain, these mutations confer a novel enzymatic function leading to the NADPH-dependent reduction of α-ketoglutarate to the proposed oncometabolite R(–)-2-hydroxyglutarate [22–24]. Recurrent IDH1 and IDH2 mutations have been identified in a very limited number of tumor types, including ∼70% of diffuse or secondary gliomas—including diffuse astrocytomas (grade II), anaplastic astrocytomas (grade III), secondary glioblastomas (grade IV), oligodendrogliomas (grade II), anaplastic oligodendrogliomas (grade III), oligoastrocytomas (grade II), and anaplastic oligoastrocytomas (grade III)—approximately 20% of acute myelogenous leukemia (AML) cases, and, most recently, the majority of central and periosteal cartilaginous tumors [20, 23, 25–36]. Collectively, these findings define a distinct subset of tumor types for which IDH1 and IDH2 mutations may provide new insights into pathogenesis and therapeutic targeting.
This study presents the results of mutational profiling of 287 patients across gastrointestinal cancer, identifying for the first time IDH1 mutations in a significant subset of patients with intrahepatic cholangiocarcinoma. Our findings further highlight the important role of molecular diagnostic testing for discovery of novel mutations of diagnostic and therapeutic relevance.
Materials and Methods
Patients and Samples
Tumors from patients at the Massachusetts General Hospital (MGH) Cancer Center who had been diagnosed with a gastrointestinal malignancy underwent mutational profiling. Subjects were identified and consented for testing of their tumor samples under a protocol with Partners Healthcare Institutional Review Board approval. The initial screening cohort consisted of 180 cases of colorectal adenocarcinoma, 43 cases of pancreatic adenocarcinoma, 32 cases of gastroesophageal adenocarcinoma, 16 cases of hepatocellular carcinoma, 12 cases of biliary tract carcinoma, and four cases of small intestine adenocarcinoma. Mutational profiling of biliary tract carcinomas was further investigated retrospectively using an expanded cohort consisting of pathological specimens from 52 additional cases of cholangiocarcinoma and 23 additional cases of gallbladder carcinoma. Cholangiocarcinoma cases were designated as intrahepatic, perihilar, common bile duct, or distal bile duct based on independent review of the clinical records by two clinicians (A.X.Z., K.K.T.). Frozen cholangiocarcinoma tissue specimens used for analysis of the IDH1 and IDH2 enzymatic product were obtained from the MGH Gastrointestinal Tumor Bank by crossmatching against the list of patients studied in the expanded cohort.
Genotyping Analysis
Cancer gene mutational analysis was performed in a Clinical Laboratory Improvement Amendments–certified laboratory using formalin-fixed, paraffin-embedded tissue. Nucleic acids were extracted and common cancer gene mutations were evaluated using a custom Applied Biosystems (ABI, Carlsbad, CA) Prism SNaPshot Multiplex system, as previously reported [1]. The addition of AKT1 p.E17K and IDH1 p.R132C=G=H=L=S mutational assays provided screening for 130 somatic mutations across 15 cancer genes (online supplemental Table 1). Briefly, genetic regions of interest were polymerase chain reaction coamplified. Mutational hotspots were then evaluated through a single-base extension step in which pools of allele-specific primers annealed adjacent to the target nucleotide. Extension was performed in the presence of dideoxynucleotide bases, each labeled with a distinct reporter fluorophore to identify the extended base (96°C for 30 seconds; 25 cycles of 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 30 seconds). Labeled products were separated using an ABI Prism 3730 DNA Analyzer and the data were interpreted with GeneMapper Analysis Software (Life Technologies/Applied Biosystems, Carlsbad, CA). For the IDH1 R132X, IDH2 R140X, and IDH2 R172X mutations, the primers used are listed in online supplemental Table 2.
2-Hydroxyglutarate Metabolite Detection
Frozen tumor tissue from five patients with cholangiocarcinoma was weighed and homogenized in ice cold 80% (v/v) methanol using a 1:10 (w/v) dilution of 100 mg tissue per 1 mL buffer. Covaris® Adaptive Focused Acoustics™ (Covaris, Inc., Woburn, MA) was used to generate tissue homogenates that were subsequently centrifuged at 4°C for 15 minutes. Supernatants were diluted 1:1 with 100 μL deionized water containing internal standard. Samples were then analyzed using liquid chromatography mass spectrometry as previously described [22].
Statistical Analysis
Gene mutation frequencies among different cancer types were compared using the Fisher's exact test. _p_-values ≤.05 were considered statistically significant. In the analysis of the prospective cohort, we adjusted _p_-values for multiple statistical tests to control the false-discovery rate at 5% [37]. No such adjustment was performed in the analysis of the validation group.
Results
Patient Characteristics
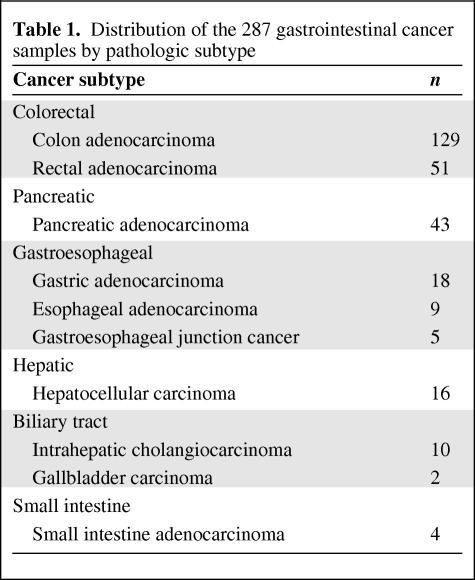
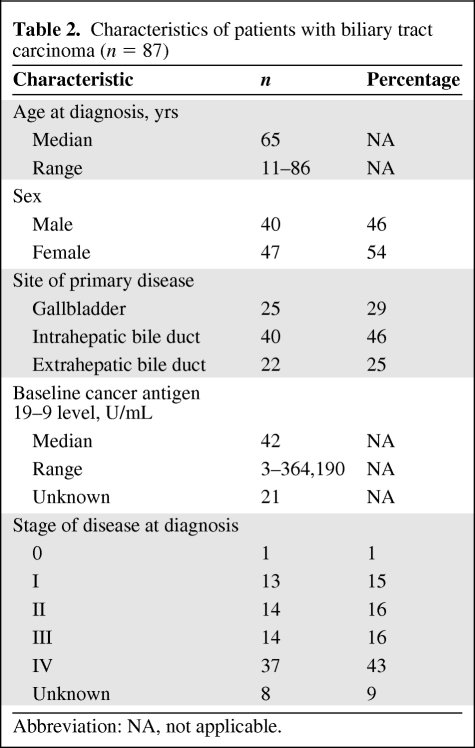
Tumors from 287 patients diagnosed across a spectrum of different gastrointestinal malignancies underwent cancer gene mutational profiling during the course of clinical management at our institution. The cohort of tumors included colorectal, pancreatic, gastroesophageal, hepatocellular, biliary tract, and small intestine carcinomas (Table 1). Following discovery of IDH1 mutations in the initial cohort, an additional 52 cases of cholangiocarcinoma and 23 cases of gallbladder carcinoma were genotyped. The characteristics of the patients with biliary tract carcinoma evaluated in this project are listed in Table 2.
Table 1.
Distribution of the 287 gastrointestinal cancer samples by pathologic subtype
Table 2.
Characteristics of patients with biliary tract carcinoma (n = 87)
Mutational Profiling of Gastrointestinal Cancers
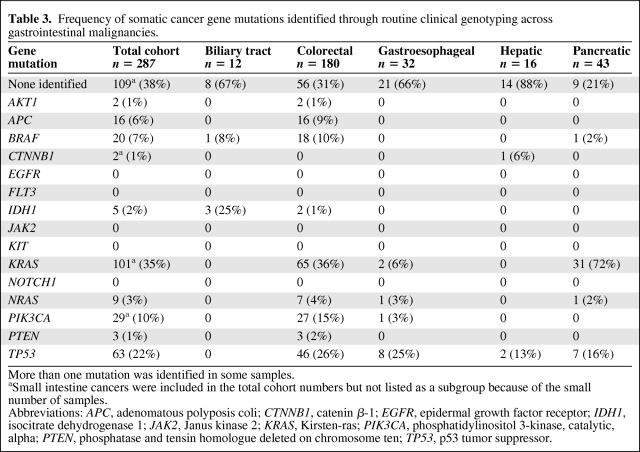
A high-throughput tumor genotyping platform (SNaPshot) has been developed at our institution to simultaneously query for 130 site-specific mutations distributed across 15 established cancer genes. The frequency of mutations in profiled cancer genes in the initial screen of 287 gastrointestinal cancers is summarized in Table 3. Across the overall group, the most widely mutated oncogene was KRAS, identified in 35% of all cases. Although KRAS mutations prevailed in pancreatic and colorectal cancers, they were infrequent in gastroesophageal cancers. Mutations in the PIK3CA gene that encodes the p110 kDa catalytic subunit of PI3K were found in 10% of all gastrointestinal cancers, with the highest prevalence in colorectal cancers. Oncogene mutations that were identified at low frequencies across multiple gastrointestinal cancer types included BRAF and NRAS, whereas AKT1 and catenin β-1 (CTNNB1) mutations were exceedingly rare. The SNaPshot assay provided coverage of only the most common mutation sites within each of the tumor suppressor genes tested [1]. Within this limited scope of coverage, TP53 was the most prevalently and widely mutated tumor suppressor gene whereas mutations in APC were specific to colorectal cancers (Table 3). Finally, oncogenic mutations in the tested mutational hotspot regions of EGFR, FLT3, JAK2, KIT, and NOTCH1 were not observed.
Table 3.
Frequency of somatic cancer gene mutations identified through routine clinical genotyping across gastrointestinal malignancies.
Consistent with reports that IDH1 and IDH2 gene mutations are rare in tumor types other than gliomas, AML, and cartilaginous tumors [20, 36, 38–40], we identified IDH1 mutations in only ∼2% of the total gastrointestinal cohort (Table 3). However, although the relative frequency of IDH1 mutations was extremely low in colorectal cancer (1%), it was unexpectedly found in three of 12 biliary tract carcinomas.
Detailed Analysis of IDH1 and IDH2 Mutations in Biliary Tract Carcinoma
To confirm this observation, a larger cohort of biliary tract carcinomas was assembled. In total, we analyzed tumors from 25 patients with gallbladder carcinoma and 62 tumors from patients with cholangiocarcinoma, including 40 intrahepatic cholangiocarcinoma patient samples and 22 extrahepatic cholangiocarcinoma patient samples (perihilar, common bile duct, or distal bile duct origin). There were no significant differences in the age, gender, baseline cancer antigen 19–9 level, or stage of disease among patients with intrahepatic cholangiocarcinoma, extrahepatic cholangiocarcinoma, and gallbladder carcinoma (data not shown).
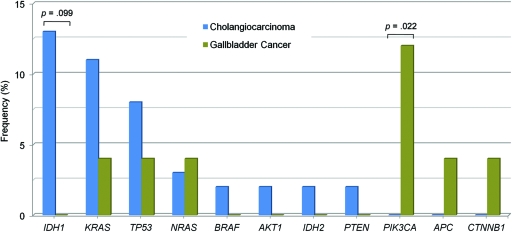
A more extensive mutational evaluation was performed on these specimens, including evaluations of mutations in IDH2 at codons R172 and R140, which were not assayed in our original panel. Diverse cancer gene mutations were identified in gallbladder carcinomas and cholangiocarcinomas in the total group of tumor samples, as shown in Figure 1 and online supplemental Table 3. The overall mutation frequency was similar in the two kinds of biliary tract tumors. The presence of PIK3CA mutations in gallbladder carcinomas was statistically significant when compared with the mutational profile of cholangiocarcinomas (p = .022), as previously reported [41].
Figure 1.
Mutational profile in biliary tract carcinomas. The frequency of cancer genes mutated in cholangiocarcinoma (blue bars; n = 62) versus gallbladder carcinoma (green bars; n = 25) is shown. Nucleic acids were extracted from formalin-fixed, paraffin-embedded tumor tissue and were tested for mutations using a single-base extension approach.
The presence of IDH1 mutations in biliary tract carcinomas initially identified within the clinical screening cohort of 12 patients was subsequently confirmed in the expanded cohort totaling 87 patient samples. Most notably, IDH1 was the most frequently mutated gene in cholangiocarcinomas (eight of 62 samples, 13%) (Fig. 1), and mutations occurred exclusively in intrahepatic tumors. No such mutations were identified in 25 specimens of gallbladder carcinoma, although this difference did not reach statistical significance (p = .099). The primary IDH1 mutation in cholangiocarcinomas resulted in the codon change p.R132C (n = 5), followed by p.R132L (n = 2) and p.R132G (n = 1) (online supplemental Table 3). Additionally, an IDH2 p.R172W mutation was found in a single cholangiocarcinoma sample. Interestingly, neither the common glioma mutation p.R132H of IDH1 nor the common AML mutation at codon R140 of IDH2 was found in cholangiocarcinomas.
IDH1 and IDH2 Mutations Predominate in Intrahepatic Cholangiocarcinomas
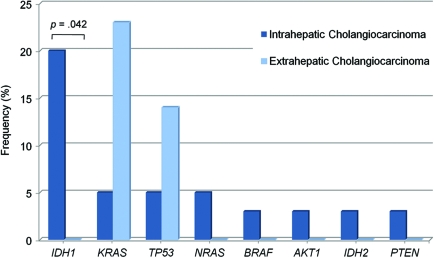
Cholangiocarcinoma is a heterogeneous disease that is classified by the discrete anatomical section of the bile duct in which the tumor arises. Intrahepatic tumors occur in ducts within the liver whereas extrahepatic tumors include those that arise in the perihilar ducts, the common bile duct, and the distal bile duct. There was a statistical trend (p = .086) for KRAS mutations predominating in extrahepatic versus intrahepatic cholangiocarcinoma samples (Fig. 2 and online supplemental Table 4). Most strikingly, mutations in IDH1 and IDH2 combined were both exclusive and predominant molecular features of intrahepatic cholangiocarcinoma samples, found in 23% (nine of 40) of cases (p = .042 for IDH1).
Figure 2.
The distribution and frequency of profiled mutations across cholangiocarcinoma subtypes. Patient cholangiocarcinoma samples were designated as either intrahepatic (dark-blue bars; n = 40) or extrahepatic (light-blue bars; n = 22) based on surgical record and/or pathological review. Extrahepatic cholangiocarcinomas included tumors arising in the perihilar duct (n = 11), common bile duct (n = 4), or distal bile duct (n = 7) region.
Abbreviations: IDH, isocitrate dehydrogenase; KRAS, Kirsten-ras; PTEN, phosphatase and tensin homologue deleted on chromosome ten; TP53, p53 tumor suppressor.
The clinicopathological features of the nine patients whose tumors harbored mutations in IDH1 or IDH2 were reviewed (online supplemental Table 5). Of these, five patients were male and four were female. Six patients had stage IV disease, two patients had stage I disease, and the disease stage of one patient was unknown. None of the patients had any known underlying risk factor associated with mutational status, including parasitic infections, primary sclerosing cholangitis, biliary duct cysts, hepatolithiasis, or thorotrast exposure. In addition, there was no association with inflammatory bowel disease, hepatitis C virus, hepatitis B virus, cirrhosis, or alcohol or tobacco use. Pathological review performed on stained tissue sections revealed that three samples were well differentiated and six were moderately differentiated. All tumors showed well-formed glands embedded within moderate to abundant stromal tissue. One tumor focally showed clear cytoplasm, and all tumors lacked both intra- and extracellular mucin. The non-neoplastic liver showed no evidence of cirrhosis in any case. Therefore, within this small group, no unique histologic or clinical features were associated with cholangiocarcinomas harboring IDH1 or IDH2 mutations.
2-Hydroxyglutarate Accumulation Is Associated with IDH1 and IDH2 Mutations
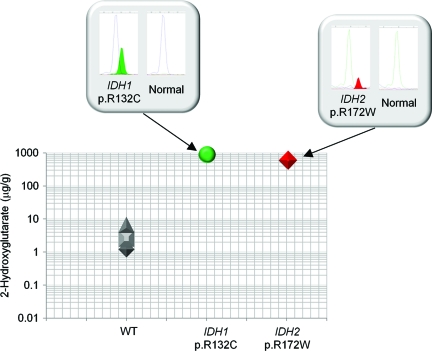
A known gain-of-function activity of mutant IDH1 and IDH2 protein is the production and subsequent tissue accumulation of the metabolite 2-hydroxyglutarate [22–24]. This biological effect of IDH1 as well as IDH2 mutations in cholangiocarcinoma patients was evaluated using frozen tissue obtained through surgical resection for five of the 62 cohort samples (Fig. 3). In three wild-type IDH1 and IDH2 samples, analysis found an average of 3.41 μg of 2-hydroxyglutarate per gram of tumor tissue (1.199 μg/g, 2.497 μg/g, and 6.534 μg/g). In a cholangiocarcinoma sample with an IDH1 p.R132C mutation, 2-hydroxyglutarate levels were elevated 248-fold above normal (845.9 μg/g). High accumulation of 2-hydroxyglutarate (588.5 μg/g) was similarly found in a cholangiocarcinoma sample harboring the IDH2 p.R172W mutation. Therefore, gain-of-function activity of 2-hydroxyglutarate production and accumulation is a likely biological consequence of mutant IDH1 as well as IDH2 in cholangiocarcinoma patients.
Figure 3.
Mutations in the genes encoding isocitrate dehydrogenase (IDH1 and IDH2) in cholangiocarcinoma are associated with 2-hydroxyglutarate metabolite accumulation. 2-Hydroxyglutarate levels were measured in frozen cholangiocarcinoma specimens using liquid chromatography mass spectrometry analysis. Three tumor samples were wild-type (WT) IDH1 and IDH2, one was IDH1 p.R132C mutation positive, and one was IDH2 p.R172W mutation positive. SNaPshot genotyping electropherograms are shown in the boxes with the mutation peak filled.
Discussion
This study provides the results of a broad-based clinical tumor genotyping approach that was applied across patients diagnosed with a gastrointestinal malignancy. While interrogating 130 mutations within 15 cancer driver genes using our SNaPshot platform, at least a single gene mutation was identified in 178 of 287 tumor samples (62%). We observed that the prevalence of the tested mutations varied significantly across disease subtypes. A primary advantage of applying broad-spectrum genotyping equally across cancer groups is the potential for identifying therapeutically relevant mutations not previously known in a particular cancer type. In our study, novel IDH1 and IDH2 mutations in intrahepatic cholangiocarcinomas were found at high frequency (nine of 40 tumors, 23%), but they were not found in extrahepatic cholangiocarcinomas (zero of 22 tumors). The vast majority of patients with cholangiocarcinoma present with advanced stage disease, associated with a poor prognosis. Therefore, these observations provide an important first step in assessing whether IDH1 and IDH2 abnormalities can provide a new therapeutic target that can improve clinical management of these patients.
Despite the best current therapy for patients with unresectable or metastatic cholangiocarcinoma, the median survival time remains <1 year [11, 12]. Recent phase II studies evaluating targeted agents that inhibit EGFR or mitogen-activated protein kinase/extracellular signal–related kinase kinase have demonstrated some evidence of antitumor activity in unselected biliary tract carcinoma patients [13]. Our data suggest that cholangiocarcinomas are also molecularly diverse according to sites within the biliary tract. Across the spectrum of cancer genes represented on our genotyping platform, IDH1 and IDH2 were the most frequently mutated genes in tumors arising in the biliary tract within the liver. Our study therefore extends the list of tumors in which mutations in IDH1 and IDH2 may be the driving event in tumorigenesis, including AML, central and periosteal cartilaginous tumors, and a subset of gliomas, and this may lead to new targeted therapies. Furthermore, the presence of an IDH1 or IDH2 mutation may assist in distinguishing intrahepatic from extrahepatic cholangiocarcinoma at the molecular level.
The mechanistic role of mutant IDH1 and IDH2 in tumorigenesis is unclear, but it has most recently been attributed to the gain-of-function activity of R(–)-2-hydroxyglutarate production through the NADPH-dependent reduction of α-ketoglutarate [22–24, 42]. The excessive accumulation of 2-hydroxyglutarate associated with IDH1 and IDH2 mutations inhibits the binding of α-ketoglutarate to a number of dioxygenases that require this substrate for physiological activity [43]. Prolyl-hydroxylases that function to hydroxylate and promote the degradation of the hypoxia inducible factor (HIF)-1α subunit of the HIF-1 transcription factor are α-ketoglutarate dependent, and _IDH1_-mutant gliomas have been closely associated with induced HIF-1α protein levels and target gene expression [21]. Aberrantly hypermethylated CpG islands in gene promoter regions have also been tightly associated with IDH1 and _IDH_2 mutations in a subset of AML and glioma cases, leading to transcriptional silencing across many genes and impaired differentiation [32, 44]. These effects presumably occur through inhibition of α-ketoglutarate–dependent enzymes that control DNA and histone methylation, and may therefore promote cancer through epigenetic changes [32, 43]. Inhibition of IDH1 and IDH2 may become an effective strategy for cancer treatment in patients selected through tumor mutational profiling.
Conclusion
This study identified for the first time a high frequency of mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 in cholangiocarcinomas specifically of intrahepatic origin. In two cholangiocarcinoma samples tested, these mutations were associated with accumulation of the novel 2-hydroxyglutarate metabolite. The frequency of IDH1 and IDH2 gene mutations was greater than the combined frequency of activating mutations in AKT1, KRAS, NRAS, and BRAF in this cancer subtype, making it the most prevalent drug targetable gene alteration in intrahepatic cholangiocarcinoma.
See the accompanying commentary on pages 5–8 of this issue.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Vanessa Scialabba and Jae Han for their technical assistance. We also thank Mohammad Miri for his assistance in tissue procurement from the MGH Tissue Repository, which was used in the metabolite analysis. This work was supported by internal MGH Cancer Center funds.
Footnotes
(C/A)
Consulting/advisory relationship
(RF)
Research funding
(E)
Employment
(H)
Honoraria received
(OI)
Ownership interests
(IP)
Intellectual property rights/inventor or patent holder
(SAB)
Scientific advisory board
Author Contributions
Conception/Design: A. John Iafrate, Kenneth K. Tanabe, Andrew X. Zhu, Darrell R. Borger, Vikram Deshpande, Dora Dias-Santagata, Eunice L. Kwak, David P. Ryan, Jeffrey W. Clark, Valeria R. Fantin, Marek Ancukiewicz, Aram F. Hezel, David P. Schenkein, Leif W. Ellisen
Provision of study material or patients: Kenneth K. Tanabe, Andrew X. Zhu, Eunice L. Kwak, David P. Ryan, Jeffrey W. Clark
Collection and/or assembly of data: Andrew X. Zhu, Darrell R. Borger, Vikram Deshpande, Valeria R. Fantin, Kimberly S. Straley, Hannah M. Liebman, Hector U. Lopez, Kenneth Fan
Data analysis and interpretation: Andrew X. Zhu, Darrell R. Borger, Vikram Deshpande, Valeria R. Fantin, Marek Ancukiewicz, David P. Schenkein, Kimberly S. Straley, Hannah M. Liebman, Hector U. Lopez, Kenneth Fan
Manuscript writing: A. John Iafrate, Kenneth K. Tanabe, Andrew X. Zhu, Darrell R. Borger, Vikram Deshpande, Dora Dias-Santagata, Eunice L. Kwak, David P. Ryan, Jeffrey W. Clark, Valeria R. Fantin, Marek Ancukiewicz, Aram F. Hezel, David P. Schenkein, Kimberly S. Straley, Leif W. Ellisen, Hannah M. Liebman, Hector U. Lopez, Kenneth Fan
Final approval of manuscript: A. John Iafrate, Kenneth K. Tanabe, Andrew X. Zhu, Darrell R. Borger, Vikram Deshpande, Dora Dias-Santagata, Eunice L. Kwak, David P. Ryan, Jeffrey W. Clark, Valeria R. Fantin, Marek Ancukiewicz, Aram F. Hezel, David P. Schenkein, Kimberly S. Straley, Leif W. Ellisen, Hannah M. Liebman, Hector U. Lopez, Kenneth Fan
References
- 1.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 6.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 7.Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchholz M, Gress TM. Molecular changes in pancreatic cancer. Expert Rev Anticancer Ther. 2009;9:1487–1497. doi: 10.1586/era.09.107. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 11.Shaib YH, Davila JA, McGlynn K, et al. Rising incidence of intrahepatic cholangiocarcinoma in the United States: A true increase? J Hepatol. 2004;40:472–477. doi: 10.1016/j.jhep.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 12.Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 13.Zhu AX, Hezel AF. Development of molecularly targeted therapies in biliary tract cancers: Reassessing the challenges and opportunities. Hepatology. 2011;53:695–704. doi: 10.1002/hep.24145. [DOI] [PubMed] [Google Scholar]
- 14.Saetta AA, Papanastasiou P, Michalopoulos NV, et al. Mutational analysis of BRAF in gallbladder carcinomas in association with K-ras and p53 mutations and microsatellite instability. Virchows Arch. 2004;445:179–182. doi: 10.1007/s00428-004-1046-9. [DOI] [PubMed] [Google Scholar]
- 15.Tannapfel A, Sommerer F, Benicke M, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanada K, Tsuchida A, Iwao T, et al. Gene mutations of K-ras in gallbladder mucosae and gallbladder carcinoma with an anomalous junction of the pancreaticobiliary duct. Am J Gastroenterol. 1999;94:1638–1642. doi: 10.1111/j.1572-0241.1999.01155.x. [DOI] [PubMed] [Google Scholar]
- 17.Riener MO, Bawohl M, Clavien PA, et al. Rare PIK3CA hotspot mutations in carcinomas of the biliary tract. Genes Chromosomes Cancer. 2008;47:363–367. doi: 10.1002/gcc.20540. [DOI] [PubMed] [Google Scholar]
- 18.Hahn SA, Bartsch D, Schroers A, et al. Mutations of the DPC4/Smad4 gene in biliary tract carcinoma. Cancer Res. 1998;58:1124–1126. [PubMed] [Google Scholar]
- 19.Suto T, Habano W, Sugai T, et al. Aberrations of the K-ras, p53, and APC genes in extrahepatic bile duct cancer. J Surg Oncol. 2000;73:158–163. doi: 10.1002/(sici)1096-9098(200003)73:3<158::aid-jso9>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 20.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin G, Reitman ZJ, Spasojevic I, et al. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS One. 2011;6:e16812. doi: 10.1371/journal.pone.0016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green CL, Evans CM, Hills RK, et al. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood. 2010;116:2779–2782. doi: 10.1182/blood-2010-02-270926. [DOI] [PubMed] [Google Scholar]
- 26.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner K, Damm F, Göhring G, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 28.Abbas S, Lugthart S, Kavelaars FG, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: Prevalence and prognostic value. Blood. 2010;116:2122–2126. doi: 10.1182/blood-2009-11-250878. [DOI] [PubMed] [Google Scholar]
- 29.Schnittger S, Haferlach C, Ulke M, et al. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood. 2010;116:5486–5496. doi: 10.1182/blood-2010-02-267955. [DOI] [PubMed] [Google Scholar]
- 30.Boissel N, Nibourel O, Renneville A, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: A study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28:3717–3723. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- 31.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 32.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amary MF, Bacsi K, Maggiani F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 38.Bleeker FE, Lamba S, Leenstra S, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 39.Kang MR, Kim MS, Oh JE, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 40.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 41.Deshpande V, Nduaguba A, Zimmerman SM, et al. Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma. BMC Cancer. 2011;11:60. doi: 10.1186/1471-2407-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data