Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data (original) (raw)
Abstract
Summary: More and more cancer studies use next-generation sequencing (NGS) data to detect various types of genomic variation. However, even when researchers have such data at hand, single-nucleotide polymorphism arrays have been considered necessary to assess copy number alterations and especially loss of heterozygosity (LOH). Here, we present the tool Control-FREEC that enables automatic calculation of copy number and allelic content profiles from NGS data, and consequently predicts regions of genomic alteration such as gains, losses and LOH. Taking as input aligned reads, Control-FREEC constructs copy number and B-allele frequency profiles. The profiles are then normalized, segmented and analyzed in order to assign genotype status (copy number and allelic content) to each genomic region. When a matched normal sample is provided, Control-FREEC discriminates somatic from germline events. Control-FREEC is able to analyze overdiploid tumor samples and samples contaminated by normal cells. Low mappability regions can be excluded from the analysis using provided mappability tracks.
Availability: C++ source code is available at: http://bioinfo.curie.fr/projects/freec/
Contact: freec@curie.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Cancer genomes often display copy number alterations (CNAs) and/or losses of heterozygosity (LOH) (Hanahan and Weinberg, 2011). Genetic abnormalities in specific regions may be related to the aggressiveness of a cancer and be associated with clinical outcomes (Caren et al., 2010; Suzuki et al., 2000).
To detect CNA and LOH regions, single-nucleotide polymorphism (SNP) arrays have been recently much in use (Popova et al., 2009). Furthermore, next-generation sequencing (NGS) has been moving to replace SNP-arrays in prediction of CNAs (Boeva et al., 2010). A recent study presented ExomeCNV, a tool to predict CNAs and LOH using exome sequencing data (Sathirapongsasuti et al., 2011). However, detection of LOH regions and, more generally, prediction of genotype status (copy number and allelic content) of an altered region using whole-genome sequencing data has remained unsolved. The main challenges to doing so are non-uniform read coverage of genomic positions [for example, due to different mappability and GC-content (Boeva et al., 2010)] and alignment bias (reference allele coverage is usually higher than the coverage of the alternative allele). Thus, the resulting signal is noisier and more difficult to process than in the case of SNP arrays.
Here, we present Control-FREEC (Control-FREE Copy number and allelic content caller)—a tool that annotates genotypes and discovers CNAs and LOH. Control-FREEC inherits many features from FREEC (Boeva et al., 2010) (assessment of copy number variation and evaluation of contamination by normal cells) as well as the general methodology of the GAP algorithm for SNP arrays (Popova et al., 2009). Control-FREEC takes as an input aligned reads, then constructs and normalizes the copy number profile, constructs the B-allele frequency (BAF) profile, segments both profiles, ascribes the genotype status to each segment using both copy number and allelic frequency information, then annotates genomic alterations. If a control (matched normal) sample is available, Control-FREEC discerns somatic variants from germline ones.
2 METHODS
Workflow: the workflow of Control-FREEC consists of three steps: (i) calculation and segmentation of copy number profiles; (ii) calculation and segmentation of smoothed BAF profiles; (iii) prediction of final genotype status, i.e. copy number and allelic content for each segment (for example, A, AB, AAB, etc.).
- (i) Calculation of copy number profiles is mainly done as described in our previous publication (Boeva et al., 2010). The most important features of the procedure are: (a) possibility to use GC-content and mappability profiles to normalize read count if a control sample is unavailable; (b) proper characterization of overdiploid genomes; (c) correction for possible contamination by normal cells when constructing the copy number profile of a tumor genome. The new tool Control-FREEC can also be used on non-mammalian genomes and includes many new user control settings, such as (a) defining the program's behavior in low mappability regions (http://bioinfo.curie.fr/projects/freec/tutorial.html); (b) choosing the minimal number of consecutive windows required to call a CNA.
- (ii) We characterize the allelic content via the BAF introduced previously for SNP arrays (Popova et al., 2009). We limit the list of genomic positions that we consider to evaluate allelic content to known SNPs only (Sherry et al., 2001). By the B allele, we mean the alternative variant in SNP database (dbSNP). SNPs that are homozygous in the genome being considered give no information about allelic content (in SNP arrays they are denoted as non-informative); therefore putatively homozygous positions are discarded. A position is discarded if the probability of having variation due to sequencing errors under the condition of actual homozygosity is greater than a specified threshold (Supplementary Materials).
We calculate the total coverage and B-allele coverage for each known putatively heterozygous SNP position. For each window i, we calculate the median of the BAF values: Med_j_ = median(abs(x_ij_−0.5)), where {x_ij_} are BAF values of the remaining SNP positions. We segment {Med_j_} using the same lasso-based algorithm as used for copy numbers (Harchaoui and Lévy-Leduc, 2008). - (iii) We predict genotype status for each genomic segment independently, by choosing the allelic content that corresponds to the maximal log-likelihood, given the copy number detected previously.
First, we combine breakpoints issued from both copy number and median BAF segmentations to get genomic segments with presumably one status. Second, copy number status of each segment is detected as described previously (Boeva et al., 2010). If the CNA is present in most of the cells, there is no ambiguity in determining exact copy number of the region (see Supplementary Materials for more details on the strategy in the case of presence of subclones or normal contamination). Third, given the copy number of the region, we fit Gaussian mixture models (GMMs) with fixed means to the observed BAF values and select the model that provides the highest log-likelihood. For example, for a region with a copy number of two, we fit a two component model (mixture of ‘AA’ and ‘BB’ alleles) and a three component model (‘AA’, ‘AB’ and ‘BB’, with a condition on the minimal weight of ‘AB’). The component means in the GMM depend on the level of contamination by normal DNA (Supplementary Materials).
Input and output: the input consists of a SAM pileup (http://samtools.sourceforge.net/pileup.shtml) and a dbSNP file. The control dataset is optional if a reference genome is provided. The output contains a list of CNAs and LOH regions as well as read count, copy number, BAF and genotype information for each window. If a control (matched normal) dataset is available, each event is annotated as somatic or germline.
3 RESULTS
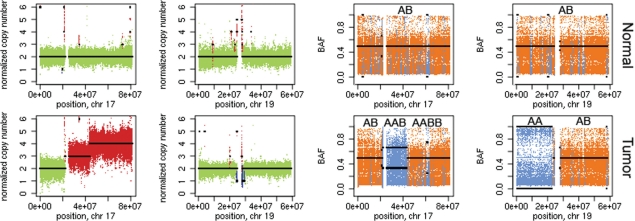
We applied Control-FREEC to detect CNAs and LOH regions in a tumor/normal dataset for a neuroblastoma patient (~ 30x-coverage, unpublished data). Control-FREEC detected somatic CNA and LOH regions covering 75% of the tumor genome (Fig. 1) and was able to identify the genotype status despite contamination of the tumor sample by normal cells (estimated percent of tumor cells was 60%).
Fig. 1.
Control-FREEC calculates copy number and BAF profiles and detects regions of copy number gain/loss and LOH regions. Tumor chromosomes 17 and 19 (bottom panels) versus ‘normal’ chromosomes (top panels; unpublished data). Predicted BAF and copy number profiles are shown in black. Gains, losses (left panels) and LOH (right panels) are shown in red, blue and light blue, respectively.
Our results agreed with the SNP-array analysis output. We obtained 95.4% consistency between the results of Control-FREEC and GAP (Popova et al., 2009), which we applied to SNP array data generated for the same tumor sample (Supplementary Materials).
4 CONCLUSION
Control-FREEC is a tool for automatic detection of CNAs and LOH regions using NGS data. It accurately calls genotype status even when no control experiment is available and/or the genome is polyploid. It corrects for GC-content and mappability biases. In the case of tumor samples, Control-FREEC is able to evaluate the level of contamination by normal cells. The software is written in C++ and freely available.
Funding: ‘Projet Incitatif et Collaboratif Bioinformatique et Biostatistiques’ of the Institut Curie; Ligue Nationale Contre le Cancer.
Conflict of Interest: none declared.
Supplementary Material
Supplementary Data
REFERENCES
- Boeva V., et al. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics. 2011;27:268–269. doi: 10.1093/bioinformatics/btq635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caren H., et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc. Natl Acad. Sci. USA. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harchaoui Z., Lévy-Leduc C. Catching change-points with lasso. Adv. Neural Inform. Process. Syst. 2008;22:617–624. [Google Scholar]
- Popova T., et al. Genome Alteration Print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays. Genome Biol. 2009;10:R128. doi: 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathirapongsasuti J.F., et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics. 2011;27:2648–2654. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T., et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., et al. An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res. 2000;60:5382–5385. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data