HOXC4 homeoprotein efficiently expands human hematopoietic stem cells and triggers similar molecular alterations as HOXB4 (original) (raw)
Abstract
Background
Expansion of hematopoietic stem cells represents an important objective for improving cell and gene therapy protocols. Retroviral transduction of the HoxB4 homeogene in mouse and human hematopoietic stem cells and hematopoietic progenitors is known to promote the cells’ expansion. A safer approach consists in transferring homeobox proteins into hematopoietic stem cells taking advantage of the natural ability of homeoproteins to cross cell membranes. Thus, HOXB4 protein transfer is operative for expanding human hematopoietic cells, but such expansion needs to be improved.
Design and Methods
To that aim, we evaluated the effects of HOXC4, a protein encoded by a HOXB4 paralog gene, by co-culturing HOXC4-producing stromal cells with human CD34+ hematopoietic cells. Numbers of progenitors and stem cells were assessed by in vitro cloning assays and injection into immuno-deficient mice, respectively. We also looked for activation or inhibition of target downstream gene expression.
Results
We show that the HOXC4 homeoprotein expands human hematopoietic immature cells by 3 to 6 times ex vivo and significantly improves the level of in vivo engraftment. Comparative transcriptome analysis of CD34+ cells subjected or not to HOXB4 or HOXC4 demonstrated that both homeoproteins regulate the same set of genes, some of which encode key hematopoietic factors and signaling molecules. Certain molecules identified herein are factors reported to be involved in stem cell fate or expansion in other models, such as MEF2C, EZH2, DBF4, DHX9, YPEL5 and Pumilio.
Conclusions
The present study may help to identify new HOX downstream key factors potentially involved in hematopoietic stem cell expansion or in leukemogenesis.
Keywords: hematopoietic stem cell, hematopoiesis, stem cell transplantation, cell therapy
Introduction
All blood cells derive from multipotent hematopoietic stem cells (HSC) that have the capacity of both long-term self-renewal and differentiation, according to body needs. Because of their scarcity, expansion of HSC represents a major challenge for improving engraftment efficiency for cell or gene therapy applications. Ex vivo expansion of human hematopoietic cells currently relies on the use of high concentrations of cytokines and growth factors. However, the value of this practice is limited since it often leads to irreversible differentiation of HSC in the culture. An alternative approach consists in using transcription factors involved in HSC self-renewal or maintenance. Among them, the HOXB4 homeoprotein was identified as a major expansion factor of mouse and human HSC after retroviral transduction of the HoxB4 coding sequence.1–3 Although that gene was first described as non-leukemogenic, a recent study revealed that transduction of HoxB4 into the HSC of large animals could lead to the late emergence of acute myeloid leukemias.4 Thus, retrovirus-mediated genetic alterations of HSC along with constitutive expression of human HOXB4 can be hazardous for therapeutic applications. To overcome this problem, we established an alternative expansion method taking advantage of the property of homeoproteins to translocate spontaneously and reversibly through membranes and reach the cytoplasm and nucleus.5,6 Long-term culture of human CD34+ immature cells in the presence of the homeoprotein induces expansion of HSC and hematopoietic progenitors of the myeloid and lymphoid lineages. Expanded cells have an enhanced capacity to repopulate in vivo and to maintain their pluripotentiality.7–9
Nevertheless, whatever the technology used, the HOXB4-mediated expansion of HSC and progenitors is always somewhat lower in humans than in mice. We, therefore, decided to examine whether using HOXC4 would improve expansion efficacy. Actually, retroviral transduction of HOXC4, a paralog of the HOXB4 gene, had been shown to cause the expansion of human hematopoietic progenitors.10 In the present study, we demonstrate that HOXC4 protein transfer into human CD34+ hematopoietic cells by way of co-culture with MS-5 stromal cells engineered to actively secrete this homeoprotein, induces 3- to 6-fold expansion of HSC and hematopoietic progenitors.
The human genes regulated by homeoproteins during hematopoiesis are mostly unknown. We, therefore, chose to search for potential effectors of HOXB4 and HOXC4 using comparative transcriptome analysis of CD34+ human cells following exposure to these factors. We reasoned that, since HOXB4 and HOXC4 display important molecular analogies and have similar temporal and spatial expression patterns during embryogenesis, both molecules should influence the expression of the same set of genes. We show herein that the transcriptomes from CD34+ cells exposed to HOXB4 or HOXC4 are virtually identical. Gene expression profiling revealed that various sets of genes encoding key hematopoietic factors and signaling pathway molecules (KLF10, HNRPDL, IKZF, and hypoxia, myc, IGF-1, 14-3-3 and angiopoietin-1 signaling) were either activated or repressed after cell exposure to these homeoproteins. Moreover, certain molecules identified herein (MEF2C, EZH2, DBF4, DHX9, YPEL5, Pumilio) are involved with stem cell fate or expansion in other models, thus corresponding to important targets for further studies.
Design and Methods
Construction of the HOXC4 vector
The HOXC4 cDNA was a gift from Dr. Daga.10 The mouse immunoglobulin κ-chain leader sequence for protein secretion was inserted upstream of the HoxC4 sequence. That construct was then cloned into the TRIP vector plasmid, as described elsewhere.11
Lentiviral vector production and transduction
HOXC4-, HOXB4-, or enhanced green fluorescent protein (EGFP)-containing lentiviral vectors were generated as described previously.7 Vector particle concentrations were normalized according to the p24 human immunodeficiency virus (HIV-1) capsid protein content of the supernatants. Infectious particles were also counted using serial dilutions of supernatants onto wild-type human embryonic kidney (HEK)-293T cells. Mouse stromal MS-5 cells grown to 70% confluence were transduced for 24 h with 2.5 μg/mL of p24 from recombinant vectors. Cells were then washed and cultured for at least five passages before use.
Cell lines
The cell lines MS-5 wild-type, MS-5/GFP (MS-5 transduced with a lentiviral vector containing the EGFP cDNA, referred to as control), MS-5/HOXC4 and MS-5/HOXB4 (MS-5 transduced with vectors containing the human HoxC4 or HoxB4 cDNA, respectively) were grown in alpha-minimum essential medium (α-MEM) containing 10% fetal calf serum (FCS) (Invitrogen, Cergy Pontoise, France).
Isolation, immuno-labeling and cultures of CD34+ cells
Human immature hematopoietic cell isolation, labeling, culture and cloning assays were performed as already described,7–9 and fully presented in the Online Supplementary Design and Methods section.
Cell expansion analysis
The ‘relative fold expansion’ was calculated as the fold-increase in HSC expansion in the presence of MS-5/HOXC4 or MS-5/HOXB4 cells, divided by that in the presence of MS-5/GFP cells. The ‘absolute fold expansion’ was calculated as the total number of HSC recovered per culture at day X, divided by that at day 0.
Mouse assays
Human cord blood CD34+ cells (5×103 cells) were co-cultured for 4 weeks with MS-5/GFP, MS-5/HOXB4, or MS-5/HOXC4. Cells were collected and the whole CD34+ cells issued from the initial 5×103 cell input were sorted then injected into 3-Gy irradiated NOD/LtSz-scid/scid (NOD-SCID) mice. More details on the numbers of injected cells are presented in the Online Supplementary Design and Methods section. Cord blood CD34+ cells from day 0 (5×103 cells, along with 105 CD34neg irradiated accessory cells) were used as controls. Seven to 8 weeks later, nucleated cells from the bone marrow of transplanted animals were analyzed by flow cytometry for the presence of human cells. Mice were considered positive when at least 1% of human cells were detected among mouse bone marrow cells. Animal experiments were approved by the Ethical Committee of the French Agriculture Department.
Cytofluorometry
The presence of human cells in NOD-SCID mouse bone marrow was determined using phycoerythrin (PE)-conjugated antibody to human CD45 (clone J33). These cells were immunophenotyped with allophycocyanin (APC)-conjugated anti-CD34 (clone 581), fluorescein isothiocyanate (FITC)-conjugated anti-CD14 (clone RMO14), anti-CD15-FITC (clone 80H5), and phycoerythrin-cyanine 5 (PC5)-conjugated anti-CD19 (clone J3-119) antibodies (Beckman Coulter, Villepinte, France).
5,6-carboxyfluorescein-diacetate-succinimidyl-ester labeling
For 5,6-carboxyfluorescein-diacetate-succinimidyl-ester (CFSE) labeling, CD34+ cells were incubated with 2.5 μM CFSE (Molecular Probes, Eugene, OR, USA) for 10 min at 37°C then quenched with cold FCS. Labeled cells were then co-cultured with MS-5/HOXB4, MS-5/HOXC4, or MS-5/GFP and analyzed on days 3 and 6 using a FACSCalibur (Becton-Dickinson, Le Pont de Claix, France).
Western blots
Methods for western blot analysis of cell products are presented in the Online Supplementary Design and Methods section.
Microarray and hybridization
Human CD34+ cord blood (4×106) cells were co-cultured for 24 h with irradiated EGFP-, HOXB4- or HOXC4-transduced MS-5 cells. Cells were lysed and total RNA extracted using the Trizol RNA method (Invitrogen). Purified RNA samples were processed for use on DNA arrays prepared by Dr. Gidrol’s team (CEA, Genopole Evry, France) from the Mediante database. The integrity of the RNA samples was verified using agarose gel electrophoresis and the Bioanalyzer 2100 (Agilent, Massy, France). RNA samples were reverse-transcribed using random primers, and aminoallyl-dUTP was incorporated for indirect labeling with Cy3 and Cy5. Three competitive hybridizations to the arrays were performed in duplicate using a dye-swap strategy.
GeneChip data analysis
Arrays were scanned and features were extracted using Genepix 6.0 software (MDC/Axon Instruments, Sunnyvale, CA, USA). Data analysis, including intensity-dependent Lowess normalization of the raw data, was performed using Bioconductor software (R package version 1.9). Ingenuity Pathways Analysis (IPA, Redwood City, CA, USA) was used to study the HOXB4-and HOXC4-mediated gene regulation profile in human CD34+ cells.
Microarray data are available in the Gene Expression Omnibus (GEO), accession number GSE24379.
Gene expression analysis
RNA samples were reverse-transcribed with SuperScript II (Invitrogen) reverse transcriptase by using random hexamers at 42°C. The cDNA obtained were used for semi-quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) of 28 genes, using the primers described in Online Supplementary Table S1. Cycling conditions were: 95°C for 10 s, 60°C for 7 s, and 72°C for 10 s, for a total of 40 cycles.
Results
Recombinant MS-5/HOXC4 cells actively secrete the HOXC4 protein
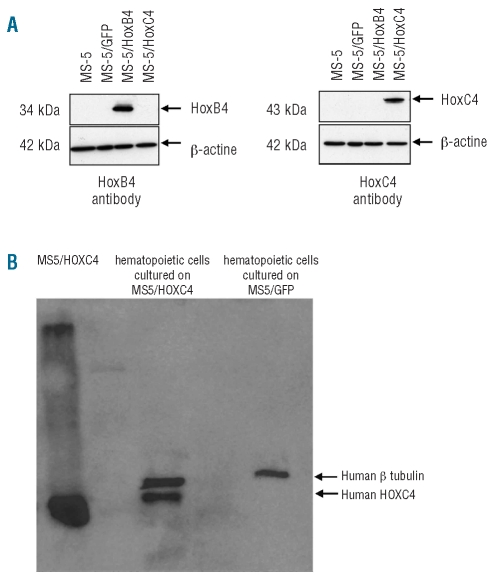
To transfer the human HOXC4 protein into HSC, we used a strategy consisting of co-culturing human CD34+ hematopoietic cells with MS-5 cells engineered to secrete HOXC4, as we reported previously with HOXB4.7 To this end, we first established the MS-5/HOXC4 stromal cell line that could supply a continuous source of this homeoprotein. After lentiviral gene transduction, most MS-5 cells stably expressed the HOXC4 or the HOXB4 protein (as shown by western blotting, Figure 1A) or GFP protein control (fluorescence analysis, not shown). MS-5/HOXC4 cells were identical to non-transduced MS-5 cells and to MS-5/HOXB4 and MS-5/GFP cells in appearance, growth rate, and phenotypic markers. The presence of the HOXC4 homeoprotein in the cells co-cultured with secreting stromal cells was detected by western blotting (Figure 1B). To avoid stromal cell contamination, hematopoietic cells were co-cultured with Transwell permeable supports.
Figure 1.
Western blot analysis of (A) HOXC4 protein by MS-5 engineered cells. HOXC4 was present at the expected size only in the MS-5 cells transduced by the HOXC4-coding vector; (B) HOXC4 protein transduction into human hematopoietic target cells co-cultured with HOXC4-secreting MS-5 cells: whole hematopoietic cell lysate revealed the presence of HOXC4 at the expected size.
HOXC4 induces expansion of human hematopoietic cells
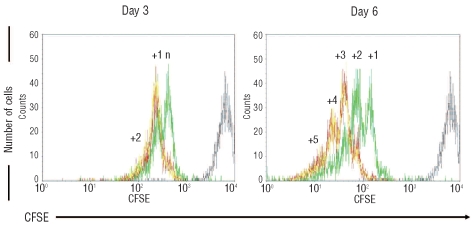
In order to establish the ability of HOXC4 to expand human HSC and hematopoietic progenitors in vitro, we first analyzed the division profiles of CD34+ cells exposed to HOXC4, by comparison with HOXB4 or none. Cell division kinetics during cultures was assessed by measuring fluorescence after CFSE cell staining (Figure 2). At day 3, CD34+ cells co-cultured with MS-5/HOXC4 cells yielded at least one more cell generation than CD34+ cells co-cultured with MS-5/GFP cells, and at day 6 a mean of two or three supplementary divisions was observed, alike with MS-5/HOXB4. Thus, HOXC4 induced the expansion of CD34+ cells, indicating that this homeoprotein rapidly activates the transcription of genes involved in the regulation of cell division.
Figure 2.
CD34+ cell division kinetic profiles during co-cultures. Representative flow cytometry profiles of CFSE-labeled CD34+ cells co-cultured with MS-5/HOXB4 (red line), MS-5/HOXC4 (yellow line), or MS-5/GFP (green line). The position of the non-dividing cells corresponds to fluorescence intensity of the cells at Day-0 (gray line). N = number of cell divisions between day-0 and the first day-3 peak; the number above each peak represents additional numbers of divisions, according to the fluorescence intensity.
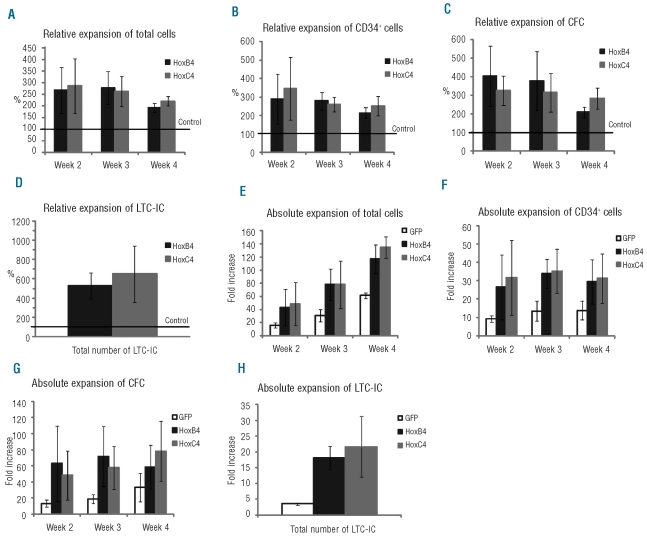
To evaluate cell proliferation in vitro, hematopoietic cells in long-term cultures were assessed every week over 4–5 weeks for the number of: (i) total cells, (ii) total CD34+ cells, (iii) total clonogenic progenitors (colony-forming cells), (iv) total immature progenitors (long-term culture-initiating cells), and (v) HSC in vivo defined as immature SCID-repopulating cells (SRC). These values were used to calculate the expansion rates of these different populations. For total cells, CD34+ cells, colony-forming cells and long-term culture-initiating cells, relative (Figure 3A-D) and absolute (Figure 3E-H) expansion levels observed in the presence of MS-5/HOXC4 cells were comparable to those observed with MS-5/HOXB4. For total cells, CD34+ cells, and colony-forming cells, the best relative expansion rates were observed at weeks 2 and 3 of co-culture. Absolute expansion of total cells was maximal at week 4 of co-culture, probably due to the progressive accumulation of maturing cells in the cultures. Of note, colonies from colony-forming cells generated in the presence of HOXC4 were similar in size and in cell composition to those from control cultures. Finally, the expansion rates of the progenitors were equivalent to those obtained in the presence of MS-5/HOXB4 using a similar co-culture method (Online Supplementary Figure S1).
Figure 3.
Relative and absolute expansion of total cells, CD34+ cells, colony-forming cells (CFC), and long-term culture-initiating cells (LTC-IC). CD34+ cells were co-cultured with MS-5/HOXB4, MS-5/HOXC4, or MS-5/GFP. Histograms (A-D) represent the fold expansion relative to MS-5/GFP (defined as 100%). Error bars represent ± standard deviation. The Y axis of (E) to (H) indicates fold expansion over the initial numbers of cells studied.
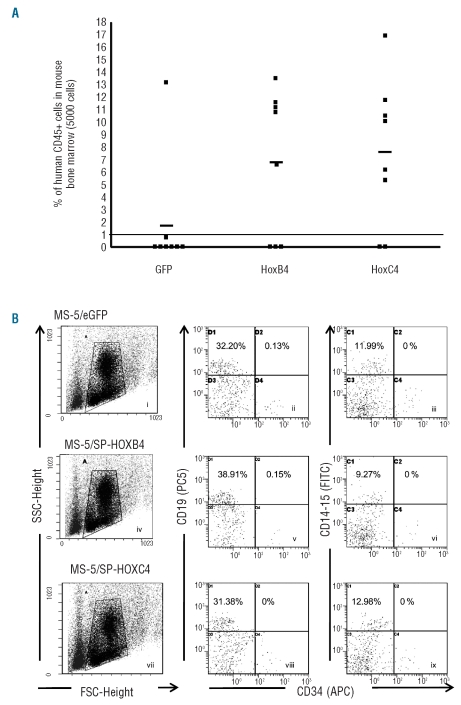
To determine the effect of HOXC4 on SRC expansion, CD34+ cells (5000 cells plated at day 0) were expanded for 4 weeks in the presence of HOXC4 and the progeny was injected to NOD-SCID mice. Bone marrow from injected mice was analyzed for human stem cell engraftment 7–8 weeks after injection. SCR co-cultured with MS-5/HOXC4 were significantly expanded when compared to cells co-cultured with MS-5/GFP (6/8 versus 1/8 positive animals, respectively) (P<0.05). By contrast, equivalent engraftment ability was observed when compared to cells cultured in the presence of MS-5/HOXB4 (5/8 positive animals), Moreover, the repopulating capacity in terms of proportion of human cells per mouse was clearly greater in cultures with MS-5/HOXC4 than in cultures with MS-5/GFP (7.8% versus 1.8%, respectively) (Figure 4A). These findings demonstrate that HOXC4 induces clear-cut ex vivo expansion of SRC in culture, while maintaining their capacity of long-term reconstitution in animals. Phenotypic analysis revealed that the induction of SRC expansion in vitro by HOXC4 did not change their differentiation program in vivo: the proportions of human myeloid cells (CD14+CD15+), B lymphoid cells (CD19+), and CD34+ cells were not altered when compared to control cells (Figure 4B). Finally, we looked for a potential synergistic effect of HOXB4 and HOXC4 on HSC and progenitor cells. We found that hematopoietic cells exposed to both HOXB4 and HOXC4 (by co-culture with MS-5/HOXB4/C4, a cell line transduced with both HOXB4 and HOXC4 cDNA) just displayed moderate improvement in the rate of cell expansion, compared with HSC exposed to HOXB4 or to HOXC4 alone (Online Supplementary Figure S1). This suggested an additive effect of HOXB4 and HOXC4 activities rather than true synergy.
Figure 4.
Ex vivo expansion of SRC co-cultured with MS-5/HOXB4, MS-5/HOXC4, or MS-5/GFP control cells. (A) Proportion of human cells in the bone marrow of NOD-SCID mice transplanted with the total progeny from 5000 initial human CD34+ cells, co-cultured for 4 weeks with MS-5/HOXB4 (n = 8), MS-5/HOXC4 (n = 8), or MS-5/GFP (n = 8). Each symbol represents results from a single transplanted mouse. The three horizontal lines indicate the average percentages of chimerism among injected mice. (B) Representative flow cytometry profiles showing multilineage repopulation of engrafted human CD34+ cells after 4 weeks of co-cultures with MS-5/GFP (i–iii), MS-5/HOXB4 (iv–vi), or MS-5/HOXC4 (vii–ix) cells. Dot plots i, iv, and vii define gates for lymphoid and myeloid cells. The X axis indicates forward angle scatter (FSC), Y axis indicates side angle scatter (SSC). Dot plots ii, v, and viii are gated on CD19 lymphoid cells. Dot plots iii, vi, and ix are gated on CD14/CD15 myeloid cells. Percentages indicate positive cells for the defined antigens.
Determination of early molecular targets of HOXB4 and HOXC4 in immature hematopoietic cells
Transcriptome experiments
At the molecular level, the mechanisms of action of Hox genes during hematopoiesis remain elusive. Although reports have described the involvement of Hox genes in some molecular pathways in mice,12,13 the HOX protein targets in human hematopoietic cells are mostly unidentified. Since HOXB4 and HOXC4 are encoded by paralogous genes that display similar effects on HSC and progenitors, we presumed that both factors regulate the same target genes during human HSC expansion. To identify early factors and signaling pathways involved in cell expansion consecutive to the action of HOXB4 or HOXC4, we performed a differential transcriptome analysis of RNA samples from cord blood-derived CD34+ cells cultured for 24 h in the presence of MS-5/HOXB4 or MS-5/HOXC4 cells. MS-5/GFP cells were used as a control in separate co-cultures to exclude non-specific changes in gene expression which may be associated with the presence of lentivirus-transduced MS-5 cells. RNA were hybridized with micro-array slides containing probes that represented 21,353 different human genes. Duplicate, both-sense experiments were performed to compare transcriptomes either directly (cells exposed to HOXB4 versus HOXC4) or indirectly (cells exposed to HOXB4 versus GFP versus HOXC4), as illustrated in Online Supplementary Figure S2.
Differential analysis of hematopoietic cell transcriptomes
Global variations of gene expression obtained by hybridization of probe-containing slides with the cDNA from the various CD34+ cultured cells are deposited in GEO database (accession number: GSE24379). HOXB4 or HOXC4 was considered to have an influence on gene expression when the _P_-value between HOX and control micro-array results was less than 0.05 and the log2-fold change was higher than 0.5. Changes in gene expression observed in the arrays were confirmed by RT-qPCR performed on a series of 28 selected genes encompassing all the scale of changes (Online Supplementary Figure S3). Overall, the transcriptomes from hematopoietic cells exposed to HOXB4 or HOXC4 were virtually similar. Indeed, direct comparison of transcriptomes of CD34+ cells exposed either to HOXB4 or HOXC4 did not show any difference in gene expression profiles. This was confirmed by indirect transcriptome comparison which just revealed some insignificant differences.
When applying a fold-change of 0.5, we found that 422 genes were up-regulated and 167 genes down-regulated after exposure to HOXB4/C4 with regard to GFP. A list of these genes is available in Online Supplementary Table S2 (and Table S2 xlsx).
Further analysis of the transcripts whose expression was altered after contact with HOXB4/C4 proteins (versus GFP control) comprised: first, identifying cardinal functions and canonical molecular pathways that involve the identified genes and second, comparing these gene expression patterns with those reported in the literature in other cell models.
Identification of cardinal functions, pathways and stem cell regulatory factors whose expression was deregulated by HOXB4/C4
To understand the biological effects of HOXB4/C4 on human HSC, all the 422 up-regulated and 167 down-regulated genes were submitted to ingenuity pathway analysis.
IPA analysis showed that the two major cell functions called “Cell cycle” and “Cancer” were mainly involved in the HOXB4/C4-treated cells as well as, to a lesser extent, important functions such as “Cellular growth and proliferation”, “RNA post-transcriptional modifications”, “Protein synthesis” and “DNA replication, recombination and repair” (Online Supplementary Figure S4A). Moreover, numerous genes involved in cell cycle progression (such as Cyclins A2, D2, E2; CDC CDK2; Myc; Max; HDAC2) were positively regulated following cell exposure to HOXB4/C4 whereas some genes involved in cell cycle reduction (p57/Kip2; Cyclin D binding protein 1) were negatively regulated (Online Supplementary Table S3).
We also used ingenuity pathway analysis to identify the molecular pathways implicated in HOXB4/C4-mediated stem cell expansion. Thus, we identified key molecules belonging to specific pathways such as “Role of BRCA1 in DNA damage response”, “Hypoxia signaling”, “Cell cycle G1/S and G2/M checkpoints”, or “Myc mediated apoptosis signaling” (Online Supplementary Figure S4B and Table S4).
The percentages of genes whose expression was modified in each of these pathways are presented in Online Supplementary Figure S4C: the expression of between 18% (for BRCA1) and 8% (for angiopoietin) of the total genes from each pathway was up-or down-regulated.
Elsewhere, it must be emphasized that the expression of 14 genes belonging to the heat-shock-encoding proteins family or related genes (listed in Online Supplementary Table S5), among which Hspa1a, was up-regulated by HOXB4/C4. This observation underlines the potential importance of Hsp molecules in the regulation of hematopoiesis by HOX proteins.
Finally, the expression of molecules previously known to be involved in stem cell fate or expansion in other models (MEF2C, EZH2, DBF4, DHX9, YPEL5, Pumilio) was also regulated by HOXB4/C4 in our model.
Comparison of gene expression changes with those reported in other models
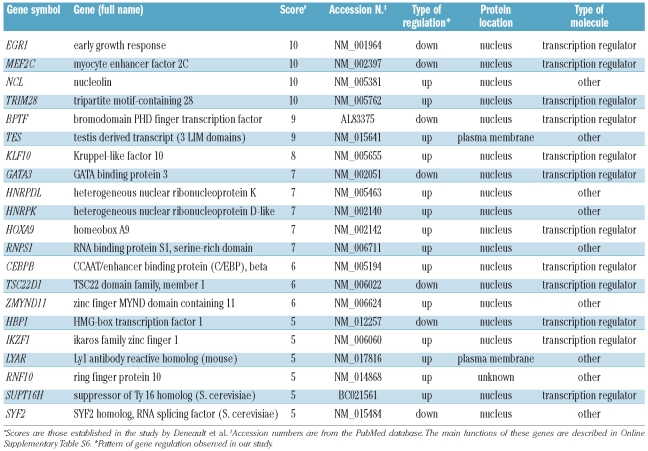
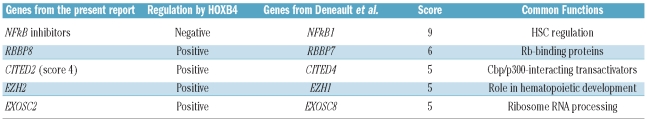
The study of gene regulation by HOXB4 or HOXC4 in a variety of cellular models may reveal important common pathways and factors that are regulated by these homeoproteins. To find key genes involved in stem cell renewal, Deneault et al.14 performed a wide review of the literature and established a database of nuclear factors in which the selected genes were scored on the basis of their level of expression, particularly in immature cell fractions. According to this criterion, genes encoding these factors were ranked from 1 to 10 relative to their potential importance for stem cell maintenance and growth. We compared the HOXB4/C4 up- and down-regulated genes listed in Online Supplementary Table S2 with the genes selected by Deneault et al. whose score was 5 or more. In this way we identified 21 genes in common with those from Deneault’s study (Table 1A). The main functions of these genes are described in Online Supplementary Table S6. Among these, KLF10 and HNRPDL are of particular interest since they belong to a series of 18 genes known to confer a repopulation advantage to HSC. In our study, HOXB4/C4 also induced changes in the expression of some genes closely related to genes reported by Deneault et al. (Table 1B). Among them, six (namely two genes encoding NFκB inhibitors, RBBP, Cited, EZH2, and EXOSC) belong to families that encode key factors for stem cell regulation.
Table 1A.
Genes whose expression was modulated by exposure of CD34+ cells to HOXB4 or HOXC4, and whose expression was also modified in the study by Deneault et al.14
Table 1B.
Representative examples of genes whose expression was modulated by HOXB4 in our study and which are homologous to those reported in the study by Deneault et al.
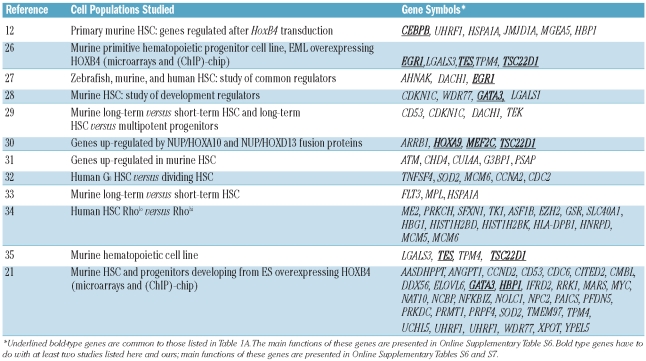
We also compared our data with published data from divergent hematopoietic and embryonic stem cell models, particularly studies including gene expression analysis of cells that over-expressed HOXB4. To that aim, we examined 12 studies reporting the modulation of gene expression in HSC. (Table 2). These genes are of particular relevance since they were described to be part of the HSC signature design in several models.
Table 2.
Genes whose expression was altered in our study as a result of CD34+ cell exposure to HOXB4/C4, compared with data from the literature acquired using different cell models.
Discussion
There is a large body of evidence demonstrating the important role of HOX factors in hematopoiesis. The four Hox paralog group 4 members, Hoxa4, b4, c4, and d4, encode proteins with highly conserved homeodomains. Hoxb4 gene disruption only induces very subtle hematopoietic defects,15 possibly because of a compensatory activity of the other three Hox genes. Recently, Iacovino et al.16 showed that each Hox paralog group 4 member similarly promotes proliferation and inhibits differentiation of hematopoietic progenitors derived from the c-kit+/CD41+ cell fraction from murine embryoid bodies, leading to expansion of undifferentiated hematopoietic cells in vitro. This indicates a potentially identical self-renewal activity for all members of the Hox paralog group 4 genes in hematopoiesis derived from murine embryonic stem cells. In addition to a possible redundancy within the Hox paralog group 4, there are several examples of inter-paralog redundancies in hematopoiesis. This is demonstrated by the fact that mice that underwent simultaneous disruption of several homeogenes such as Hoxb3 + Hoxb417 and Hoxa9 + Hoxb3 + Hoxb418 have important deficits in HSC proliferative capacity. However, these findings did not ascertain that the four Hox paralog group 4 members regulate the proliferation of human HSC via the same mechanisms.
HOXB4 and HOXC4 proteins have similar effects on the behavior of human hematopoietic progenitors and stem cells
We have previously shown that the continuous presence of the HOXB4 protein in long-term co-cultures results in the expansion of the most immature HSC identifiable in human, i.e. long-term culture-initiating cells and SRC, and of more mature progenitors.7 In order to improve in vitro HSC expansion, and to circumvent potential drawbacks such as those observed by Zhang et al.,4 we transduced the HOXC4 protein into human HSC using the same method as for HOXB4.7 We provide evidence that, like HOXB4, the human HOXC4 protein behaves as a growth factor able to enhance the number of hematopoietic progenitors and stem cells. We had shown that in presence of the HOXB4 protein, the number of SRC was amplified by approximately 2.1-fold after 4 weeks of cultures when compared to the number of SRC from CD34+ cells cultured in the absence of HOXB4. Here, we demonstrate that SRC submitted to HOXC4 or to HOXB4 are expanded and maintained similarly. HOXC4-expanded primitive HSC could not be distinguished from non-expanded HSC as they retained full multilineage repopulating ability, and the mature cells derived from HOXC4-expanded HSC had normal phenotypes. Hence, HOXC4 homeoprotein, like HOXB4, may be useful in clinical applications involving cell transfer.
Regarding our experimental model, one may wonder whether HOXB4- or HOXC4-dependent expansion is due to the homeoprotein activity in HSC or to other factors attributed to HOX-expressing MS-5 cells. We previously showed that MS-5 cells engineered to secrete HOXB4 had no phenotypic changes.7,9 In the present study, we extended this observation to _HOXC4_-transduced MS-5 cells as well. This strongly argues for a direct role of the secreted homeoproteins on human HSC. Additional studies using purified recombinant HOX proteins would be needed to confirm the proteins’ direct effect on HSC. Unfortunately, these proteins are difficult to purify, and improvements in the production and stabilization of homeoproteins are required to allow the use of purified proteins instead of co-cultures.
HOXB4 and HOXC4 regulate the same early genes in an identical manner
We chose to study gene expression after short-term exposure of CD34+ cells to the homeoproteins because we believed that the main events regulating HSC expansion would be rapidly initiated by the action of HOX transcription factors on target genes. It must be emphasized that only a small fraction of the CD34+ cells are actually true repopulating cells. The gene expression changes evaluated here could, therefore, just in part reflect the changes present in these repopulating cells. The similarity of the gene expression regulation patterns observed in HOXB4- and HOXC4-treated cells suggests that they operate through the same mechanisms of action on the same target genes.
HOXB4 and HOXC4 regulate genes important in various cell growth pathways
Analysis of important pathways revealed that HOXB4 and HOXC4 induce notable variations in the expression of key molecules involved in cell growth, differentiation, and transformation. Importantly, variations were observed in the processes and pathways regulating the cell cycle (checkpoint regulations, BTG-containing protein regulation, RAN signaling), translation initiation (EIF1 signaling), tumor suppression (BRCA2 and p53 signaling), and HSC metabolism (hypoxia, Myc, IGF-1 and angiopoietin-1 signaling). The involvement of these signaling pathways is of particular importance, as discussed below. First, hypoxia has been implicated in the control of the metabolism and growth of numerous stem cells, and HSC in particular.19 Second, it has been suggested that HOXB4 induces stem cell expansion through c-myc expression, resulting in enhancement of HSC self-renewal.20 In agreement, the study by Oshima et al.21 and ours showed that c-myc (and its partner max in our study) was up-regulated in HSC exposed to HOXB4 (or to HOXC4). Third, IGF-1 signaling is required for the maintenance or growth of various stem cells, particularly mesenchymal stem cells,22 spermatogonia,23 and hematopoietic stem and progenitors cells, and IGF-1 is involved in normal erythropoiesis, granulopoiesis, and lymphopoiesis. Fourth, angiopoietin-1 was identified as an indispensable factor for the quiescent and anti-apoptotic states of HSC, mediated by binding to its receptor Tie2 on stem cells.24
Collectively, these findings are coherent with the hypothesis that HOXB4 and HOXC4 stimulate the initiation of stem cell expansion by activating a number of early genes involved in the cell cycle, which, in turn, allow the progression of the cell cycle. HOXB4 and HOXC4 also modulate the expression of other genes or pathways that have important functions in the hematopoietic process: in particular, they up-regulate the genes coding for the growth hormone, Ikaros, and several members of the ATP-binding cassette family, and down-regulate mpl and GATA3 genes. Moreover, HOXB4 and HOXC4 modulate the expression of N-FATC2IP, which regulates N-FAT expression, and KU-70, DOCK2, and Ezrin, whose products interact with the hematopoietic-specific factor VAV-1.
Some genes regulated by HOXB4 and HOXC4 proteins in our study are also found in other models of stem cell regulation
A comparison of our results with those of Deneault et al.14 revealed 21 genes (Table 1A) with similar expression patterns (score ≥5). Further examination of these genes revealed that some are well known for their role in stem cell metabolism. Notably, KLF10 and HNRPDL belong to the gene set defined by these authors as able to induce substantial enhancement of HSC activity. KLF10 is particularly relevant since it encodes a zinc-finger transcription factor that induces early expression of transforming growth factor-β. Thus, it may take part in microenvironment activity on stem and leukemia cells in bone marrow. Importantly, KLF10 is a target of VHL,25 a factor involved in the ubiquitination and degradation of the hypoxia-inducible factor (HIF), which plays a central role in oxygen-mediated regulation of gene expression, particularly during hematopoiesis.
Milsom et al.15 suggested that HOXB4 could exert its effects by inhibiting tumor necrosis factor (TNF)-α signaling in HSC. Actually, the use of a gene disruption mouse model for Fanconi anemia allowed that team to conclude that over-expression of HOXB4 reversed the hypersensitivity of Fanconi anemia HSC to TNF-α and rescued the in vivo engraftment capacity of these cells, probably mediated by the down-regulation of TNF-α receptor expression in _HoxB4_-transduced cells. In contrast to that study, our model did not allow us to conclude that TNF-α pathway genes are regulated by HOXB4 or HOXC4, possibly because our human HSC expansion model is notably different from the Fanconi anemia mouse stem cell rescue model by Milsom et al., and therefore involves different pathways. Importantly, we noted that the modified expression of 33 genes in our study is shared by genes in the study by Oshima et al. described as potential direct targets of HOXB4.21
Gene expression variations reported in various publications often lead to divergent or contradictory results because the models used in the studies are different. Our model is the only one that takes into consideration the effects of HOXB4 and HOXC4 proteins on gene expression in human HSC. The results presented here show that gene expression variations in our transcriptomes concerned important genes and pathways shared with other studies, but also genes and pathways apparently specific to our model. Importantly, we found that the expression of genes involved in stem cell fate was modulated by the exposure of HSC to HOXB4 or HOXC4. Among these genes, some (Mef2C, Ezh2, Dbf4, Dhx9, Ypel5, Pumilio) are also involved in the fate of stem cells from various tissues in several species of mammals, fishes, and insects. We are currently performing functional studies of some of these genes to determine their potential involvement in expansion of HSC or regulation of hematopoiesis.
In conclusion, we have demonstrated that the HOXC4 homeoprotein displays clear-cut positive effects on growth and maintenance of human HSC and hematopoietic progenitors, similar to those previously reported with HOXB4 in other models. This is corroborated by almost identical transcriptomes of HOXB4- and HOXC4-treated cells. Target genes of homeoproteins may represent good candidates for the development of new stem cell therapy strategies using the products of HOX target genes instead of homeoproteins themselves. These not yet clearly identified factors could be safer and more potent for the expansion, self-renewal, and maintenance of human HSC. Some of them may also be involved in leukemic stem cell expansion or maintenance.
Footnotes
Funding: this work was supported by the French Ligue Nationale Contre le Cancer (LNCC) [EL2009.LNCC/SF2], and by the Association pour la Recherche sur le Cancer (ARC) [A08/1/1048].
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, et al. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9(14):1753–65. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 2.Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109(1):39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 3.Schiedlmeier B, Klump H, Will E, Arman-Kalcek G, Li Z, Wang Z, et al. High-level ectopic HOXB4 expression confers a profound in vivo competitive growth advantage on human cord blood CD34+ cells, but impairs lymphomyeloid differentiation. Blood. 2003;101(5):1759–68. doi: 10.1182/blood-2002-03-0767. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XB, Beard BC, Trobridge GD, Wood BL, Sale GE, Sud R, et al. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118(4):1502–10. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez F, Joliot A, Bloch-Gallego E, Zahraoui A, Triller A, Prochiantz A. Antennapedia homeobox as a signal for the cellular internalization and nuclear addressing of a small exogenous peptide. J Cell Science. 1992;102(4):717–22. doi: 10.1242/jcs.102.4.717. [DOI] [PubMed] [Google Scholar]
- 6.Maizel A, Bensaude O, Prochiantz A, Joliot A. A short region of its homeodomain is necessary for engrailed nuclear export and secretion. Development. 1999;126(14):3183–90. doi: 10.1242/dev.126.14.3183. [DOI] [PubMed] [Google Scholar]
- 7.Amsellem S, Pflumio F, Bardinet D, Izac B, Charneau P, Romeo PH, et al. Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nat Med. 2003;9(11):1423–7. doi: 10.1038/nm953. [DOI] [PubMed] [Google Scholar]
- 8.Haddad R, Caignard A, Visentin G, Vigon I, Fichelson S, Amsellem S. The HOXB4 homeoprotein improves ex vivo generation of functional human NK-cell progenitors. Leukemia. 2007;21(8):1836–9. doi: 10.1038/sj.leu.2404725. [DOI] [PubMed] [Google Scholar]
- 9.Haddad R, Pflumio F, Vigon I, Visentin G, Auvray C, Fichelson S, et al. The HOXB4 homeoprotein differentially promotes ex vivo expansion of early human lymphoid progenitors. Stem Cells. 2008;26(2):312–22. doi: 10.1634/stemcells.2007-0721. [DOI] [PubMed] [Google Scholar]
- 10.Daga A, Podesta M, Capra MC, Piaggio G, Frassoni F, Corte G. The retroviral transduction of HOXC4 into human CD34(+) cells induces an in vitro expansion of clonogenic and early progenitors. Exp Hematol. 2000;28(5):569–74. doi: 10.1016/s0301-472x(00)00135-1. [DOI] [PubMed] [Google Scholar]
- 11.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101(2):173–85. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 12.Schiedlmeier B, Santos AC, Ribeiro A, Moncaut N, Lesinski D, Auer H, et al. HOXB4’s road map to stem cell expansion. Proc Natl Acad Sci USA. 2007;104(43):16952–7. doi: 10.1073/pnas.0703082104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milsom MD, Schiedlmeier B, Bailey J, Kim MO, Li D, Jansen M, et al. Ectopic HOXB4 overcomes the inhibitory effect of tumor necrosis factor-{alpha} on Fanconi anemia hematopoietic stem and progenitor cells. Blood. 2009;113(21):5111–20. doi: 10.1182/blood-2008-09-180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deneault E, Cellot S, Faubert A, Laverdure JP, Frechette M, Chagraoui J, et al. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137(2):369–79. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brun AC, Bjornsson JM, Magnusson M, Larsson N, Leveen P, Ehinger M, et al. Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood. 2004;103(11):4126–33. doi: 10.1182/blood-2003-10-3557. [DOI] [PubMed] [Google Scholar]
- 16.Iacovino M, Hernandez C, Xu Z, Bajwa G, Prather M, Kyba M. A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 2009;18(5):783–92. doi: 10.1089/scd.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjornsson JM, Larsson N, Brun AC, Magnusson M, Andersson E, Lundstrom P, et al. Reduced proliferative capacity of hematopoietic stem cells deficient in Hoxb3 and Hoxb4. Mol Cell Biol. 2003;23(11):3872–83. doi: 10.1128/MCB.23.11.3872-3883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnusson M, Brun AC, Lawrence HJ, Karlsson S. Hoxa9/hoxb3/hoxb4 compound null mice display severe hematopoietic defects. Exp Hematol. 2007;35(9):1421–8. doi: 10.1016/j.exphem.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Guitart AV, Hammoud M, Dello Sbarba P, Ivanovic Z, Praloran V. Slow-cycling/quiescence balance of hematopoietic stem cells is related to physiological gradient of oxygen. Exp Hematol. 2010;38(10):847–51. doi: 10.1016/j.exphem.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Satoh Y, Matsumura I, Tanaka H, Ezoe S, Sugahara H, Mizuki M, et al. Roles for c-Myc in self-renewal of hematopoietic stem cells. J Biol Chem. 2004;279(24):24986–93. doi: 10.1074/jbc.M400407200. [DOI] [PubMed] [Google Scholar]
- 21.Oshima M, Endoh M, Endo TA, Toyoda T, Nakajima-Takagi Y, Sugiyama F, et al. Genome-wide analysis of target genes regulated by HoxB4 in hematopoietic stem and progenitor cells developing from embryonic stem cells. Blood. 2011;117(15):e142–50. doi: 10.1182/blood-2010-12-323212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez C, Oskowitz A, Pochampally RR. Epigenetic reprogramming of IGF1 and leptin genes by serum deprivation in multipotential mesenchymal stromal cells. Stem Cells. 2009;27(2):375–82. doi: 10.1634/stemcells.2008-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YH, Chin CC, Ho HN, Chou CK, Shen CN, Kuo HC, et al. Pluripotency of mouse spermatogonial stem cells maintained by IGF-1- dependent pathway. FASEB J. 2009;23(7):2076–87. doi: 10.1096/fj.08-121939. [DOI] [PubMed] [Google Scholar]
- 24.Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118(2):149–61. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov SV, Ivanova AV, Salnikow K, Timofeeva O, Subramaniam M, Lerman MI. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370(4):536–40. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HM, Zhang H, Schulz V, Tuck DP, Forget BG. Downstream targets of HOXB4 in a cell line model of primitive hematopoietic progenitor cells. Blood. 2010;116(5):720–30. doi: 10.1182/blood-2009-11-253872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi I, Ono H, Moritomo T, Kano K, Nakanishi T, Suda T. Comparative gene expression analysis of zebrafish and mammals identifies common regulators in hematopoietic stem cells. Blood. 2010;115(2):e1–9. doi: 10.1182/blood-2009-07-232322. [DOI] [PubMed] [Google Scholar]
- 28.Mascarenhas MI, Parker A, Dzierzak E, Ottersbach K. Identification of novel regulators of hematopoietic stem cell development through refinement of stem cell localization and expression profiling. Blood. 2009;114(21):4645–53. doi: 10.1182/blood-2009-06-230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forsberg EC, Prohaska SS, Katzman S, Heffner GC, Stuart JM, Weissman IL. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genet. 2005;1(3):e28. doi: 10.1371/journal.pgen.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmqvist L, Pineault N, Wasslavik C, Humphries RK. Candidate genes for expansion and transformation of hematopoietic stem cells by NUP98-HOX fusion genes. PLoS One. 2007;2(1):e768. doi: 10.1371/journal.pone.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent DG, Copley MR, Benz C, Wohrer S, Dykstra BJ, Ma E, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113(25):6342–50. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- 32.Graham SM, Vass JK, Holyoake TL, Graham GJ. Transcriptional analysis of quiescent and proliferating CD34+ human hemopoietic cells from normal and chronic myeloid leukemia sources. Stem Cells. 2007;25(12):3111–20. doi: 10.1634/stemcells.2007-0250. [DOI] [PubMed] [Google Scholar]
- 33.Zhong JF, Zhao Y, Sutton S, Su A, Zhan Y, Zhu L, et al. Gene expression profile of murine long-term reconstituting vs. short-term reconstituting hematopoietic stem cells. Proc Natl Acad Sci USA. 2005;102(7):2448–53. doi: 10.1073/pnas.0409459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, Grindle SM, et al. Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol. 2005;3(8):e254. doi: 10.1371/journal.pbio.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argiropoulos B, Yung E, Xiang P, Lo CY, Kuchenbauer F, Palmqvist L, et al. Linkage of the potent leukemogenic activity of Meis1 to cell-cycle entry and transcriptional regulation of cyclin D3. Blood. 2010;115(20):4071–82. doi: 10.1182/blood-2009-06-225573. [DOI] [PubMed] [Google Scholar]